Abstract
The two endonucleases, Rad27 (yeast Fen1) and Dna2, jointly participate in the processing of Okazaki fragments in yeasts. Mus81–Mms4 is a structure-specific endonuclease that can resolve stalled replication forks as well as toxic recombination intermediates. In this study, we show that Mus81–Mms4 can suppress dna2 mutational defects by virtue of its functional and physical interaction with Rad27. Mus81–Mms4 stimulated Rad27 activity significantly, accounting for its ability to restore the growth defects caused by the dna2 mutation. Interestingly, Rad27 stimulated the rate of Mus81–Mms4 catalyzed cleavage of various substrates, including regressed replication fork substrates. The ability of Rad27 to stimulate Mus81–Mms4 did not depend on the catalytic activity of Rad27, but required the C-terminal 64 amino acid fragment of Rad27. This indicates that the stimulation was mediated by a specific protein–protein interaction between the two proteins. Our in vitro data indicate that Mus81–Mms4 and Rad27 act together during DNA replication and resolve various structures that can impede normal DNA replication. This conclusion was further strengthened by the fact that rad27 mus81 or rad27 mms4 double mutants were synergistically lethal. We discuss the significance of the interactions between Rad27, Dna2 and Mus81–Mms4 in context of DNA replication.
INTRODUCTION
During the replication of chromosomal DNA, replication forks (RFs) are likely to encounter many potential obstacles such as damaged lesions or tightly bound proteins, resulting in the formation of stalled or collapsed RFs. In order to maintain genome stability, cells have developed mechanisms to reestablish impaired RFs. One mechanism contributing to RF restart is homologous recombination, which requires multiple enzymatic activities (1,2). When progression of RFs is blocked, they regress and form a Holliday junction (HJ)-like structure that arises from the annealing of regressed leading and lagging strands (1,3).
Collapsed or regressed RFs must be processed to restart DNA replication, and can be restored with the aid of RecQ-family helicases, which include Sgs1 (budding yeast), Rqh1 (fission yeast), BLM, WRN and RTS (humans) helicases (4). They promote reversion of regressed RFs (4–6). The heterodimeric Mus81–Mms4 complex has been implicated in processing of stalled and blocked RFs as well as in the processing of recombination intermediates (2,7,8). Mus81–Mms4 is a structure-specific endonuclease originally identified in a sgs1 synthetic-lethal screen as a component that acts in parallel or redundant pathways with Sgs1 (9–16). In vitro experiments showed that the Mus81–Mms4 complex cleaved nicked Holliday junctions, D loop, RFs, and 3′ flaps that could form in vivo during the repair of damaged RFs (11,12,14–16). The common structural feature of Mus81–Mms4 substrates is the presence of three- or four-way junctions containing a 5′ end at the junction, which serves to direct the cleavage reaction by Mus81–Mms4 (9,11,12,17,18). Furthermore, mus81 mutants exhibited hypersensitivity to methyl methanesulfonate (MMS), cisplatin, hydroxyurea (HU) and UV, all of which can lead to either the stalling or the collapse of RFs (19–23). In general, inactivation of genes involved in fork processing displayed several types of genome instability, such as increased rates of both mitotic and meiotic recombination, gross chromosomal rearrangements and chromosome loss (4,5,24–27). Thus, failure to repair impaired RFs puts cells at high risks of genome instability, contributing in part to the development of human diseases such as cancers (4). In humans, Bloom syndrome, Werner syndrome and Rothmund–Thomson syndrome are caused by mutations in the BLM, WRN and RTS genes, respectively.
Flap endonuclease 1 (Fen1) is another structure-specific endonuclease that resolves single-stranded flap DNA intermediates. With this activity, it participates in virtually all DNA transactions such as DNA replication, repair and recombination (28–30). In addition, many proteins interact functionally and genetically with Fen1 (29). During DNA replication, Fen1 is required to quantitatively remove the RNA primer in Okazaki fragments (28–30). RNase H, which can remove most of the initiator RNA endonucleolytically, leaves one ribonucleotide residue upstream of the RNA–DNA junction. The removal of the last ribonucleotide requires Fen1 exonuclease activity (28,30). Alternatively, Fen1 can cleave a 5′ flap containing RNA–DNA generated by DNA polymerase (pol) δ strand displacement activity, to directly generate ligatable nicks. In this case, the entire RNA primer can be removed in the absence of RNase H activity (28,30). When a flap is long enough to be coated with replication protein A (RPA), Dna2 endonuclease activity becomes essential because the RPA-coated flaps are resistant to Fen1 activity, but stimulate Dna2-catalyzed cleavage. Such processing events generate short flaps devoid of RPA, which can be processed rapidly by Fen1 (31). Thus, RPA provides a mechanism by which the processing of flaps can switch from Dna2 to Fen1 (31). The concerted action of Dna2 and Fen1 efficiently generates nicks that can be sealed by DNA ligase 1 (31).
It is worthwhile to mention that MUS81 or MMS4 interacted genetically with several genes involved in DNA replication. For example, deletion of RNH202 (a subunit of RNase H2) in a mus81 background increased its sensitivity to camptothecin and to HU, while these drugs had no significant effect on rnh202Δ alone (32). This result suggests that RNase H2 and Mus81–Mms4 act in parallel to overcome camptothecin- and HU-induced DNA damage. Furthermore, rad27Δcells lacking yeast Fen1 (yFen1) suffer severe synthetic growth defects or lethality in combination with mutations in mus81 or mms4 (33). Both RNase H2 and Fen1 are known to play critical roles in processing of Okazaki fragments in eukaryotes. However, the manner in which Mus81–Mms4 participates in Okazaki fragment processing is presently unclear. In this report, we have investigated functional and physical interactions between Dna2, Fen1 and Mus81–Mms4. Our findings indicate that Mus81–Mms4 and Rad27 act together via direct interactions during processing of Okazaki fragments and resolution of damaged RFs.
MATERIALS AND METHODS
Enzymes, antibodies, DNA and nucleotides
Oligonucleotides used in this study were commercially synthesized by Genotech (Daejeon, Korea) and their sequences are listed in Table 1. All oligonucleotides were gel-purified prior to use. Nucleoside triphosphates were obtained from Sigma-Aldrich, and [γ-32P] adenosine triphosphates (ATP) (>5000 Ci/mmol) was purchased from IZOTOP (Budapest, Hungary). Restriction endonucleases and polymerase chain reaction (PCR) polymerases were purchased from either New England Biolabs (Beverly, MA) or Enzynomics (Daejeon, Korea). The pRS plasmids were purchased from New England Biolabs. The pET28 vectors used for protein expression in Escherichia coli were from Novagen (Darmstadt, Germany). Isopropyl β-d-1-thiogalactopyranoside (IPTG) and X-gal were from ElpisBiotech (Daejeon, Korea). Imidazole was from Acros Organics (Geel, Belgium). The uracil analog, 5-fluoroorotic acid (5-FOA) was obtained from Duchefa Biochemie (Haarlem, Netherland). Antibodies used for immunoprecipitation or western blotting were obtained from; anti-His (Qiagen, Valencia, CA, USA), PAP (an antibody:peroxidase conjugate, Sigma-Aldrich), and anti-FLAG (Sigma-Aldrich).
Table 1.
Oligonucleotides used to construct DNA substrates in this study
| No. | Nucleotide sequences (length in nt) |
|---|---|
| 1 | 5′-GAAAACATTATTAATGGCGTCGAGCGTCCGTAGGCACAAGGCGAACTGCTAACGG-3′ (55) |
| 2 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTATTTTTTTTTTTTTTTTTTTT-3′ (45) |
| 3 | 5′-CGGACGCTCGACGCCATTAATAATGTTTTC-3′ (30) |
| 4 | 5′-CGAACAATTCAGCGGCTTTAACCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (52) |
| 5 | 5′-GAAAACATTATTAATGGCGTCGAGCTAGGCACAAGGCGAACTGCTAACGG-3′ (50) |
| 6 | 5′-CGCATCCTATCAGTTCGTATGCAGTGTCCGGTTAAAGCCGCTGAATTGTTCG-3′ (52) |
| 7 | 5′-ACTGCATACGAACTGATAGGATGCG-3′ (25) |
| 8 | 5′-GAAAACATTATTAATGGCGTCGAGC-3′ (27) |
| 9 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTA-3′ (25) |
| 10 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTAG-3′ (26) |
| 11 | 5′-GCTTTAACCGGACGCTCGACGCCATTAATAATGTC-3′ (38) |
Preparation of substrates
The DNA substrates used in this study included 3′ flap (3′F), RF, RF leading strand regressed (RLe), nicked Holliday junction (nHJ) and 5′ double flap (5′DF). DNA substrates for nucleases were prepared using the synthetic oligonucleotides listed in Table 1. The oligonucleotides used to construct DNA substrates are indicated in each figure with encircled numbers at their schematic structure. Briefly, one of the three or four strands (10 pmol) was first 5′-end labeled with [γ-32P] ATP (10 pmol) by polynucleotide kinase according to the manufacturer’s protocol. The labeled oligonucleotides were then annealed to other oligonucleotides (20 pmol) in buffer (5 mM HEPES-NaOH/pH7.5, 300 mM NaCl) using a PCR machine (95°C for 5 min, 65°C for 30 min and −0.5°C/min to 25°C). All annealed substrates were purified by 10% polyacrylamide gel electrophoresis (PAGE) prior to use.
Multi-copy suppression of dna2K1080E lethality
The plasmid pRS325-MUS81/MMS4 was transformed into yJA1B+JK65 (MATα ade2-101 ura3-52 lys2-801 trpΔ63 his3-Δ200 leu2-Δ1 GAL+ dna2Δ::HIS3) harboring both pRS316-DNA2 and pRS314-dna2K1080E. In pRS325-MUS81/MMS4, expression of Mus81 and Mms4 proteins were driven by their native promoters. The transformants were grown on SD-His-Trp-Leu plates. Single colonies from transformants were grown in liquid media, and the cells were spotted in 10-fold serial dilutions onto SD-His-Trp-Leu plates with or without 5-fluoroorotic acid (5-FOA). The plates were incubated for 3 days at 30°C. Loss of pRS316-DNA2 in the presence of 5-FOA in the media allowed us to measure whether the expression of Mus81–Mms4 (in pRS325) suppressed the dna2K1080E mutant allele (in pRS314).
Construction of expression vectors
All recombinant proteins were expressed as native or fusion proteins in E. coli by the use of the pET21c and pET28a (Novagen). Constructions of all expression vectors were performed similarly as follows: PCR fragments were obtained with appropriate pairs of primers, and amplified fragments were purified using QIA-quick gel extraction kit (Qiagen). All fragments amplified contained a unique restriction site at each end to allow directional cloning into a relevant vector. The authenticity of all constructed plasmids was confirmed by sequencing inserted DNA fragments. In order to produce hexahistidine-tagged Mms4 at its N-terminus, for example, a pair of primers, 5′-GAA TTC ATG AGC CG ATC GTT GAT-3′ and 5′-ACG CGT CGA CTC ATT CAA TAG TAT CAT T-3′ (EcoRI and SalI sites used for cloning are underlined, respectively) were used to amplify the MMS4 gene with yeast genomic DNA as template. The amplified fragments were digested with EcoRI and SalI, and cloned into pET28a cleaved with EcoRI and SalI, generating pET-His6-Mms4. The pET-Mus81 was prepared by the use of 5′-GGA ATT CCA TAT GGA ACT CTC ATC AAA C-3′ and 5′-GCG GCC GCC TA AGT TTA CCA AAA GC (NdeI and NotI site, respectively). The amplified fragments were then cloned into NdeI and NotI sites of pET21c, to generate pET-Mus81. To express the Mus81–Mms4 complex, the MUS81 gene was amplified from the pET-Mus81 construct above with primers 5′-ACA GCA TGC AGA TCT CGA TCC CGC GAA-3′ and 5′-TCA GCA TGC AAA AAA CCC CT-3′ (SphI). The amplified fragments were cloned into a SphI site of pET28- His6-Mms4, producing pET28–His6–Mms4–Mus81.
To express Rad27 (yeast Fen1) as a fusion protein (Rad27-FLAG) with a FLAG tag at its C-terminus, the RAD27 gene was PCR-amplified using yeast genomic DNA as template with the primers; 5′-GGA ATT CAA ACA AAA AGA ACA GGG A-3′ and 5′-CCG CTC GAG TCA CTT ATC GTC ATC GTC CTT GTA GTC TCT TCT TCC CTT TGT GAC-3′ (EcoRI and XhoI). The amplified fragment was cloned into EcoRI and XhoI sites of pRS426, generating pRS426-Rad27-FLAG. The cloned fragment contained the native Rad27 promoter. To express Rad27 with a C-terminal tagged hexahistidine (Rad27-His6), the RAD27 gene was amplified with the primers; 5′-GGA ATT CAT GGG TAT TAA AGG TTT G-3′ and 5′-CCG CTC GAG TCT TCT TCC CTT TGT GAC-3′ (EcoRI and XhoI). The amplified PCR fragments were digested with EcoRI and XhoI, and cloned into pET21a cleaved with EcoRI and XhoI, resulting in pET21a-Rad27-His6. All truncated forms of Rad27 were cloned into plasmid pGEX4T-1, and produced as a fusion protein with GST fused to its N-terminus. They included Rad271–366, Rad27261–382, Rad27261–317, Rad27318–382, Rad27318–350, Rad27351–382 and Rad27335–366 (the subscript indicates positions of amino acids).
Purification of Mus81–Mms4 complex and Rad27
Escherichia coli BL21-RIL cells (Stratagene) containing pET28–His6–Mus81–Mms4 were grown at 37°C until A600 = 0.5, and expression was induced with 0.5 mM IPTG. The cells (4 l) were grown for an additional 2 h at 25°C and collected by centrifugation. Cells (7.5g, wet weight) were resuspended in 100-ml buffer A150 (25 mM HEPES-NaOH/pH 7.5, 10% glycerol, 0.01% NP-40, 1 mM EDTA, 1 mM dithiothreitol, 150 mM NaCl and 0.1 mM phenylmethylsulfonate). The subscript in buffer A150 indicates concentration of NaCl in millimoles. Cells were sonicated five times for 1 min with a 5-min cooling interval, and the lysates were centrifuged at 100 000g for 30 min. The resulting extract (4 mg/ml, 100 ml) was loaded onto a phosphocellulose column (5 ml, Φ 1.5 × 5 cm) equilibrated with buffer A150. The column was washed with 25-ml buffer A150, and proteins eluted with a linear NaCl gradient (50 ml) from 0.15 to 1 M in buffer A without DTT and EDTA. Mus81–Mms4 eluted at ∼650 mM NaCl, and peak protein fractions were pooled. The pooled proteins (1.4 mg/ml, 15 ml) were incubated with Ni2+–NTA beads (1 ml, Qiagen) in the presence of 10 mM imidazole for 2 h; the mixture was then poured into a column (Φ 0.7 × 5 cm), and then washed with 10-ml buffer A500 plus 10 mM imidazole, followed by washing with 10-ml buffer A500 plus 50 mM imidazole. The Mus81–Mms4 complex was eluted with buffer A500 plus 200 mM imidazole. Eluted peak fractions (0.38 mg/ml, 1.5 ml) were pooled, and a portion (0.25 ml) was subjected to glycerol gradient centrifugation (5 ml, 15–35% glycerol in 25 mM HEPES-NaOH/pH 7.5, 500 mM NaCl, 0.01% NP40) at 45 000 r.p.m. for 24 h in a Beckman SW55Ti rotor.
For purification of the Rad27-His6 protein, E. coli BL21-RIL cells (2 l) harboring pET21-Rad27-His6 were grown at 37°C until the A600 = 0.5, followed by the addition of 0.5 mM IPTG to induce protein expression. The cells were incubated for an additional 4 h at 25°C, and then harvested. The resulting cell pellet (4.5 g, wet weight) was resuspended in 65-ml buffer A100 without DTT and EDTA. Crude extracts were prepared as described above for Mus81–Mms4. The extract (3.2 mg/ml, 65 ml) was loaded onto a 1.5-ml Ni2+–NTA (Φ 1 × 5 cm) column, followed by successive washings with 10-ml buffer A100 and 10-ml buffer A100 plus 50 mM imidazole without DTT and EDTA. Bound proteins were eluted with buffer A100 plus 200 mM imidazole. The peak protein fractions (0.42 mg/ml, 15 ml) were pooled and loaded onto a Heparin column (1 ml, Φ 0.7 × 5 cm) equilibrated with buffer A100. The column was then washed with 10-ml buffer A200, and bound proteins eluted with buffer A plus 500 mM NaCl. Half (0.25 ml) of the pooled fraction (1.23 mg/ml, 0.5 ml) was subjected to glycerol gradient sedimentation as described for the isolation of Mus81–Mms4. Glycerol gradient fractions obtained from the Mus81–Mms4 and Rad27 preparations were analyzed in sodium dodecyl sulfate (SDS)-PAGE, followed by Coomassie blue staining. The protein yield was quantified using bovine serum albumin (BSA) as standard (Bio-Rad).
Derivatives of GST-Rad27 fusion proteins were purified using GST-beads (Amersham Biosciences) as recommended by the manufacturer.
Nuclease assays
Standard Mus81–Mms4 nuclease assays were performed in reaction mixtures (10 µl) containing 25 mM Tris–HCl/pH 8.0, 100 mM NaCl, 10 mM MgCl2, 0.2 mM DTT, 0.1 mg/ml BSA, 0.1% NP-40, 5% glycerol, 10 fmol of DNA substrates and enzyme as indicated. Reaction mixtures were incubated at 30°C for 30 min, and then deproteinized with 0.2% SDS (final concentration) and 10 µg proteinase K, followed by incubation for 10 min at 37°C. After adding 2 µl of 6X stop buffer (60 mM EDTA/pH 8.0, 40% sucrose, 0.6% SDS, 0.25% bromophenol blue and 0.25% xylene cyanol), mixtures were electrophoresed at 150 V through a 12% PAGE in 0.5× TBE (45 mM Tris, 45 mM boric acid and 1 mM EDTA). Gels were dried on a DEAE-cellulose paper (Whatman) and autoradiographed. Labeled DNA products were quantified by a Phosphorimager (Molecular Dynamics, Inc.). Rad27 nuclease assays were carried out as previously described [Kim et. al. (39)], except that the amount of DNA substrate used was 10 fmol. When required, Mus81–Mms4 proteins were diluted in buffer (25 mM HEPES-NaOH at pH 7.5, 500 mM NaCl, 0.25 mg/ml BSA, 0.01% NP40 and 20% glycerol), while Rad27 proteins were diluted in the same buffer but with 150 mM NaCl.
Determination of kinetics parameters
Kinetic analyses were repeated in triplicate using increasing amounts (5, 10, 25, 50, 100, 200 and 500 fmol) of a double-flap substrate prepared with oligonucleotides 5, 10 and 11 (template, upstream primer and labeled downstream primer, respectively; see Table 1 for the sequences and sizes). Reactions were carried out with 1 fmol of Rad27 and 30 fmol of Mus81–Mms4 containing 90 mM NaCl per reaction. These reaction conditions led to reliable levels of products at early time points. Reaction mixtures (20 µl) were assembled on ice, followed by preincubation on ice for 10 min. Reactions were initiated by incubation at 37°C. Aliquots were withdrawn at 8 min and then added to 4 µl of 6X stop buffer. The amount of products formed was analyzed as described above. Kinetic parameters were obtained based on the Michaelis–Menten equation. V = d[P]/dt, where [P] is the amount of products in nanomoles. The concentration of [P] was calculated using the equation, [P] = Icleaved/(Iuncleaved + Icleaved) × [S], where [S] is concentration of substrate used, and Icleaved and Iuncleaved are band intensities of products formed and substrates left, respectively. The initial velocity was plotted against [S], and the values Km and Vmax were calculated by nonlinear regression using Sigma Plot (Systat Software Inc.) to avoid distortion of the experimental errors, which can occur during reciprocal transformation of the data.
Immunoprecipitation
In order to detect in vivo interactions between Mus81–Mms4 and Rad27, a BJ2168-Mus81-TAP strain was constructed by inserting the Mus81-TAP into its native genomic locus in BJ2168 cells (MATa ura3-52 trp1-Δ63 leu2Δ prb1-1122 pep4-3 prc1-407 GAL2). These cells were grown to A600 = 2.0–3.0 in an appropriate selective media. BJ2168-Mus81-TAP/Rad27-FLAG strains were constructed by replacing chromosomal RAD27 with Rad27-FLAG. Cell lysates from BJ2168-Mus81-TAP/Rad27-FLAG were prepared by disrupting cells using a bead beater (Biospec product) in IP buffer (25 mM HEPES-NaOH/pH 7.5, 150 mM NaCl, 10% glycerol, 0.05% NP-40, and 1 mM PMSF). Clarified lysates were incubated with 200 µl (50% slurry) of FLAG-agarose (Sigma) at 4°C for 2 h with rocking. After a brief centrifugation, the beads were washed five times with IP buffer prior to western blot analyses with PAP or FLAG antibodies.
In vitro interactions between Mus81–Mms4 and Rad27 were carried out as follows; recombinant proteins (5 pmol of each) were mixed in IP buffer (0.1 ml), and the mixture was incubated on ice for 1 h. Polyclonal Rad27 antibodies and protein-A agarose beads (5 µl each) were added and the mixture was incubated at 4°C for 2 h with rocking. After incubation, beads were collected by centrifugation, and washed eight times with IP buffer (0.5 ml for each wash), followed by western blot analyses with anti-His antibodies.
GST-pull-down assay
For the purification of GST-Rad27 and its derivatives, E. coli BL21-RIL cells harboring plasmids (pGEX4T-1-Rad27X–Y, where X and Y indicate the amino acids present in the Rad27 truncated fragment) were grown at 37°C to an A600 of 0.5: Protein expression was induced with 1 mM IPTG for 4 h at 25°C. Cells were harvested and resuspended in lysis buffer (25 mM HEPES-NaOH/pH 7.5, 10% glycerol, 0.01% NP-40, 150 mM NaCl and 0.1 mM PMSF). Lysates were then mixed with 0.1 ml glutathione-agarose beads (Amersham Biosciences) and the mixtures incubated at 4°C for 1 h. The GST-agarose beads were collected by centrifugation, and washed once with lysis buffer (1 ml) plus 1 M NaCl and once with lysis buffer (1 ml). The amount of GST-Rad27 bound to beads was quantified using BSA as standard (Bio-Rad) in a Coomassie-stained 10% SDS-polyacrylamide gel. Beads containing 100 pmol of GST-Rad27 were mixed with 1 pmol of Mus81–Mms4 in binding buffer (25 mM HEPES-NaOH/pH 7.5, 10% glycerol, 0.01% NP-40, 150 mM NaCl), and then incubated at 4°C for 1 h. Beads were then collected by centrifugation, washed eight times with IP buffer (0.5 ml each time), and then subjected to 10% SDS–PAGE for immunoblotting with anti-His monoclonal antibodies. When necessary, GST-Rad27 was eluted from the GST beads with 20 mM glutathione to the beads.
RESULTS
Multi-copy expression of MUS81–MMS4 suppresses lethality of dna2 helicase mutant
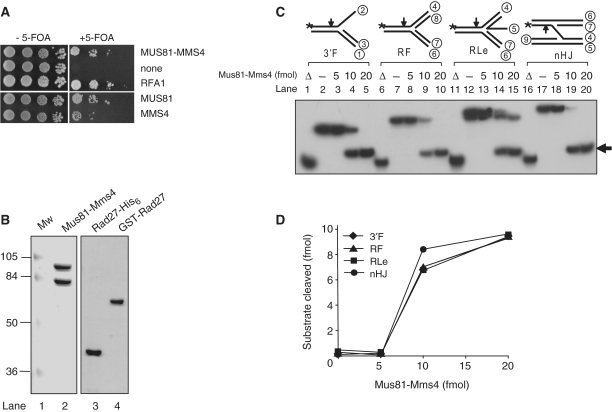
Previously, we showed that overexpression of Mph1 rescued the lethality of dna2K1080E (34), a mutant allele that abolished the helicase activity of Dna2 (35). We noted that the helicase domains of Mph1 and FancM shared some homology at their N-terminal region (36). In addition, the C-terminal region of FancM contained amino acid sequences that are conserved in ERCC4 and Mus81 family of endonucleases (17). This raised the possibility that Mph1 and Mus81 may function together to play a role similar to that of FancM. This possibility was supported by our finding that the protein–protein interaction was detected between Mph1 and Mus81 with yeast two hybrid assays (data not shown). We then assumed that Mus81 was very likely to interact functionally with Dna2 like Mph1. To test this possibility, we examined the ability of Mus81–Mms4 to suppress the lethality of dna2K1080E mutant in a manner similar to that observed with Mph1. For this purpose, we constructed multi-copy plasmids expressing Mus81 and Mms4, individually and in combination, under the control of their native promoters. As shown in Figure 1, transformants expressing one of the two proteins or both significantly suppressed the lethality of dna2K1080E. Expression of either Mus81 (MUS81) or Mms4 (MMS4) alone also resulted in significant suppression of dna2K1080E. The extent of suppression by the Mus81–Mms4 complex (MUS81–MMS4) was nearly as efficient as that observed with overexpression of the large subunit of RPA (RFA1, positive control; ref. 31), whereas an empty vector control (none) failed to do so.
Figure 1.
(A) suppression of dna2K1080E by multi-copy expression of Mus81–Mms4. The dna2Δ strain harboring pRS316-DNA2 and pRS314-dna2K1080E was transformed with pRS325-MUS81/MMS4 (MUS81–MMS4), pRS325 (Vector, vector only control), or pRS325-RFA1 (RFA1, positive control). Transformants were grown on SD-Leu-Trp-His plates. Drop-dilution assays were carried out in 10-fold serial dilutions (104, 103, 102 and 101 cells) and spotted on SD-Leu-Trp-His plates in the absence (–5-FOA) or presence (+5-FOA) of 5-fluoroorotic acid. Cells were grown at 30°C for 3 days. (B) Purification of recombinant Mus81–Mms4 and Rad27 proteins. SDS–PAGE analysis of purified Mus81–Mms4, Rad27-His6 and GST-Rad27 proteins. Mus81–Mms4 (1.6 µg, lane 2), Rad27-His6 (1.4 µg, lane 3) and GST-Rad27 (1.4 µg, lane 4) were subjected to a 10% SDS–PAGE analysis, followed by Coomassie blue staining. Mw, molecular weight marker. (C) Cleavage of four different DNA substrates by purified Mus81–Mms4. Reactions were carried out in standard reaction mixtures containing 10 fmol of each substrate as described in ‘Materials and Methods’ section. The substrates tested included 3′F, 3′ flap; RF, replication fork; RLe, regressed leading-strand replication fork; nHJ, nicked Holliday junction. The structure of each substrate is schematically illustrated at the top of the autoradiograph, and oligonucleotides used to construct the DNA substrates are indicated by encircled numbers (Table 1). Increasing amounts (5, 10 and 20 fmol) of Mus81–Mms4 were added to reaction mixtures that were incubated for 30 min at 30°C. Reactions were terminated with 0.2% SDS, 10 µg proteinase K, followed by incubation for 15 min at 37°C. The products were subjected to a 12% PAGE in 0.5× TBE at 150 V. The arrows on each substrate indicate the sites of cleavage. Asterisk indicates the position of 32P-labels at 5′ DNA ends. Open triangle indicates boiled substrate control. Minus sign indicates no enzyme control. (D) The amount (fmol) of cleavage product formed in (C) was plotted against the amounts of Mus81–Mms4 used.
Purification of recombinant Mus81–Mms4 complex and Rad27
To gain insight into the mechanism by which Mus81–Mms4 suppressed the dna2 helicase mutation, we purified the Mus81–Mms4 complex as described in ‘Materials and Methods’ section and investigated whether it could affect the endonuclease activities of either Dna2 or Fen1. The Mus81–Mms4 complex and two forms of Rad27 (Rad27-His6, Rad27 with a hexahistidine tag at its C-terminus; GST-Rad27, Rad27 with a GST tag at its N-terminus) were purified to near homogeneity (>95%, Figure 1B). They were devoid of nonspecific nuclease activities that interfered with our nuclease assays (see below). GST-Rad27 had virtually no endonuclease activity, whereas Rad27-His6 possessed wild-type-like activity (data not shown). This is consistent with the observation that the N-terminal regions of Fen1 were critical to the formation of its catalytic site (37).
The purified recombinant Mus81–Mms4 complex was active and cleaved a variety of substrates, including those containing a 3′F, RF, RLe and nHJ structures (Figure 1C). We noted that Mus81–Mms4 displayed a marked sigmoidal response to the level of enzyme added. Cleavage products were hardly observed with <5 fmol of Mus81–Mms4, whereas saturation levels of cleavage products were formed with 10 fmol or more of the enzyme produced (Figure 1D). The cleavage products migrated similarly in the polyacrylamide gel used in this study. The cleavage of the labeled strands in these substrates produced largely 20-nt (with the 3′F substrate) or 22-nt long ssDNA products (with all the other substrates), indicating that the cleavage had occurred 5 nt away from nicks present in each substrate (data not shown). We also tested other substrates including a 5′F. Y-structure, intact Holliday junction and RF (RLa), and found that the Mus81–Mms4 complex poorly or hardly cleaved these substrates; the RLa substrate was weakly (100-fold less than the RLe substrate) cleaved (data not shown), while others were hardly cleaved. These findings are in keeping with the substrate specificity determined in the previous reports (14,38).
Mus81–Mms4 stimulates endonuclease activity of Rad27
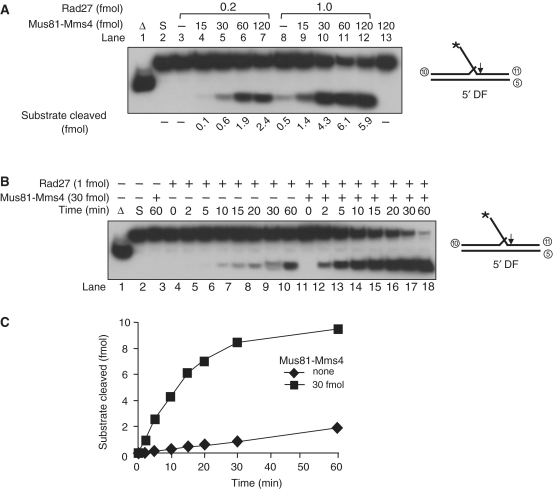
Since other multi-copy suppressors of dna2 mutants like Mgs1, Mph1 or Vts1 markedly stimulated the endonuclease activity of either Dna2 or Rad27 or both (34, 39,40), we decided to examine whether Mus81–Mms4 was able to stimulate the nuclease activities of Rad27 and Dna2. For this purpose, we first tested the ability of Mus81–Mms4 to stimulate Fen1 activity using 5′DF substrate with a 5′ 13- and 3′ 1-nucleotide flap, a physiological substrate for Fen1 (41). When reaction mixtures containing fixed levels (0.2 and 1 fmol) of Rad27 were supplemented with increasing amounts (15, 30, 60 or 120 fmol) of Mus81–Mms4, there was a marked stimulation of Rad27-catalyzed cleavage of the 5′DF substrate (Figure 2A). With 0.2 fmol of Rad27 alone, virtually no cleavage products were formed (Figure 2A, lane 3). However, the addition of Mus81–Mms4 markedly stimulated Rad27 activity (Figure 2A, compare lanes 3 and 4–7). We observed an ∼12-fold stimulation by Mus81–Mms4 at higher levels of Rad27 (1 fmol) (Figure 2A, lanes 8–12). In the absence of Rad27, no cleavage products were detected even with the highest level (120 fmol) of Mus81–Mms4 (Figure 2A, lane 13), in keeping with the stringent substrate specificity of Mus81–Mms4. We also analyzed the products formed in a high-resolution gel, and noted no changes in the cleavage site of Fen1 (data not shown). We found that Mus81–Mms4 also stimulated the 5′ to 3′ exonuclease activity of Rad27 when a nicked duplex substrate was used (data not shown). In conclusion, these findings indicate that Mus81–Mms4 is able to stimulate the activity of Rad27 without altering its specificity.
Figure 2.
The Mus81–Mms4 complex stimulates the endonuclease activity of Rad27. (A) Standard reactions for Rad27 were carried out with two different levels (0.2 or 1 fmol) of Rad27 in the presence of increasing amounts (15, 30, 60 and 120 fmol) of Mus81–Mms4. Reaction mixtures contained 25 mM Tris–HCl/pH 8.0, 2 mM MgCl2, 2 mM DTT, 0.25 mg/ml BSA and 10 fmol of the 5′DF substrate. The schematic structure of the substrate is as shown, and oligonucleotides used to construct the substrate are indicated by encircled numbers (Table 1). The reaction mixtures, after addition of indicating enzyme levels, were incubated for 15 min at 37°C. Reactions were terminated with 0.2% SDS, 10 µg proteinase K, followed by an additional incubation for 15 min at 37°C. The reaction products were analyzed similarly as described in Figure 1C. The arrows on the substrates indicate sites of cleavage. Asterisk indicates the position of 32P-labels at 5′ DNA ends. Open triangle indicates boiled substrate control. S, substrate only control. (B) A time-course experiment of Rad27 endonuclease activity in the absence (–) or presence (+, 30 fmol) of Mus81–Mms4. The reaction mixtures containing the 10 fmol of 5′DF substrates were added with 1 fmol of Rad27, and the mixtures were incubated for varying periods as indicated. The cleavage products were analyzed in a native 10% polyacrylamide gel. (C) The amount (fmol) of cleavage products formed in (B) was plotted against incubation time.
Subsequently, we performed a time-course experiment with a fixed amount of Rad27 in the absence and presence of Mus81–Mms4. The rate of products formed by Rad27 (1 fmol) in the presence of Mus81–Mms4 (30 fmol) was 10- to 20-fold greater than that measured in its absence (Figure 2B and C). Mus81–Mms4 alone was totally inactive (Figure 2B, lane 3). Considering that elevated levels of Fen1 resulted in suppression of dna2 mutations (42), our findings that the Mus81–Mms4 complex stimulated the activity of Rad27 could explain why Mus81–Mms4 suppressed a dna2 mutation when expressed in a multi-copy plasmid (see below for further discussion). We also discovered that Mus81–Mms4 stimulated the endonuclease activity of Dna2, but weakly (2- to 3-fold at most) (data not shown). Thus, we could not rule out the possibility that the weak stimulation of Dna2 by Mus81–Mms4 could partially contribute to the suppression of the dna2 mutation.
Effect of Mus81–Mms4 on the kinetic parameters of the Rad27 cleavage
We next determined the reaction kinetic parameters, Km and Vmax, as described in ‘Materials and Methods’ section. The addition of Mus81–Mms4 resulted in a significant increase (∼2.5-fold) in Vmax (and thus kcat), and a reduction (∼4-fold) of Km (Table 2). Thus, the catalytic efficiency (kcat/Km) of Rad27 increased markedly (∼11-fold) in the presence of Mus81–Mms4 (Table 2). The increase in Vmax suggests that the protein–protein interaction between Mus81–Mms4 and Rad27 may induce conformational change in Rad27, which in turn leads to an increased rate of catalysis. The decrease in Km may reflect the fact that Mus81–Mms4 stabilizes the binding of Rad27 to the substrate and contributes to its increased affinity for DNA substrates. The two kinetic parameters determined suggest that the mechanism by which Mus81–Mms4 stimulates Rad27 activity could be similar to the mechanism by which PCNA stimulates the activity of Rad27. It was shown that PCNA increased Vmax 2-fold, but decreased Km ∼12 fold of Rad27 (43). It has been postulated that the direct physical interaction of PCNA with Fen1 leads to the tethering of Fen1 at DNA cleavage site and contributes to the increased affinity of Fen1 for DNA substrates.
Table 2.
Kinetic parameters for Rad27 endonuclease activity in the presence of Mus81–Mms4
| Vmax (nM/s × 10-3) | Km (nM) | kcat (s−1 × 10−2) | kcat/Km (s−1 M−1 × 107) | |
|---|---|---|---|---|
| Rad27 alone | 3.64 ± 0.75 | 25.33 ± 5.51 | 7.28 ± 1.49 | 0.30 ± 0.08 |
| Rad27+Mus81/Mms4 | 9.58 ± 2.96 | 5.96 ± 3.10 | 19.17 ± 5.91 | 3.47 ± 0.82 |
Rad27 stimulates Mus81–Mms4 cleavage activity
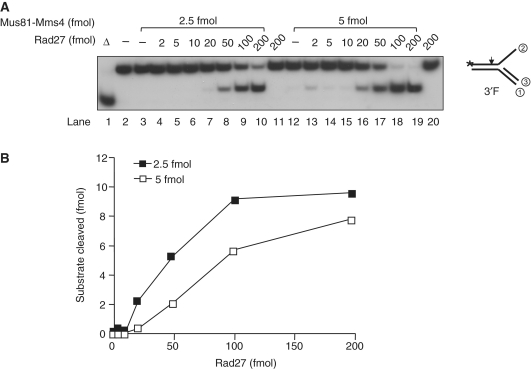
Since genetic analyses revealed that rad27 was synthetically lethal with either mus81 or mms4 (33), we tested the alternative possibility that Rad27 might affect the Mus81–Mms4 endonuclease activity. To confirm this possibility, we incubated the 3′F substrate with two different levels (2.5 and 5 fmol) of Mus81–Mms4 in the presence of increasing amounts (0–200 fmol) of Rad27 (Figure 3A). As expected from substrate specificity, the 3′F substrate was not cleaved by Rad27 (200 fmol; Figure 3A, lanes 11 and 20). In addition, the levels (2.5 and 5 fmol) of Mus81–Mms4 alone hardly produced cleavage products (Figure 3A, lanes 3 and 12) in keeping with the results as shown in Figure 1D. In the presence of Rad27, however, the 3′F substrate was efficiently cleaved by Mus81–Mms4 (Figure 3A and B). In the presence of 2.5 and 5 fmol of Mus81–Mms4, more than 20 fmol of Rad27 was required for detectable levels of cleavage products (Figure 3A, lanes 8 and 16). The cleavage reaction plateaued in the presence of more than 100 fmol of Rad27 (Figure 3A and B). We also examined the Rad27 stimulation of the Mus81–Mms4 cleavage activity using three other substrates, RF, RLe and nHJ. Like the 3′F substrate, cleavage of these substrates by Mus81–Mms4 increased in a Rad27-dependent manner (data not shown). Our data suggest that Rad27 stimulated Mus81–Mms4 regardless of substrates used. In addition, Rad27 did not alter the sites cleaved by Mus81–Mms4. The high-resolution gel analyses showed that identical products were formed in reactions containing Mus81–Mms4 and those also containing Rad27 (data not shown).
Figure 3.
Rad27 stimulates the endonuclease activity of Mus81–Mms4. (A) Standard endonuclease assays for Mus81–Mms4 were carried out with the enzyme levels as indicated. Two levels of Mus81–Mms4 (2.5 and 5 fmol) were used in the presence of increasing amounts of Rad27 as indicated. DNA substrate used was the 3′F substrate (10 fmol) and the mixtures were incubated for 30 min at 30°C. Lanes 11 and 20 are the controls with Rad27 alone carried out independently with the same amounts of enzymes. (B) The amount (fmol) of cleavage products formed in (A) was plotted against the amount of Rad27 used.
Rad27 stimulates Mus81–Mms4 in a single catalytic cycle
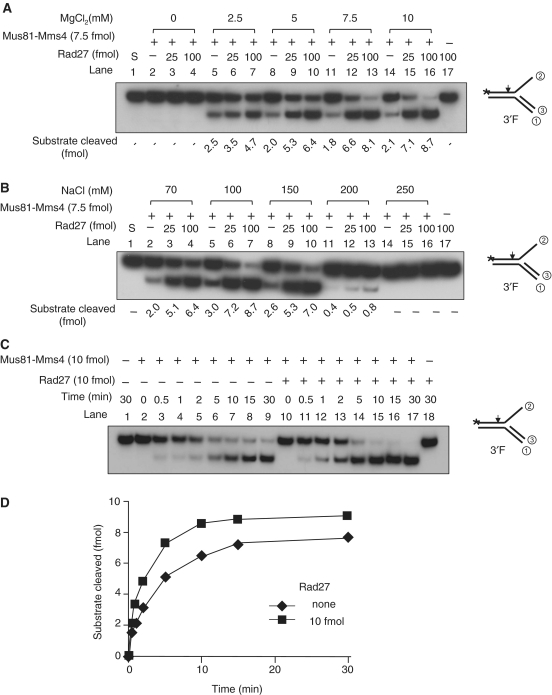
In order to investigate whether Rad27 stimulates Mus81–Mms4 at physiological salt concentrations, we decided to determine concentrations of NaCl for optimal cleavage of the 3′F substrate by Mus81–Mms4. Prior to this, we examined the effect of Mg2+ on the Fen1-mediated stimulation of Mus81–Mms4. In the absence of Mg2+, cleavage did not occur (Figure 4A, lanes 2–4). We noted that increases in Mg2+ concentrations (2.5–10 mM) caused gradual decrease in cleavage efficiencies of 3′F DNA by Mus81–Mms4 alone (Figure 4A, compare lanes 5, 8, 11 and 14). In contrast, the addition of Rad27 (25 or 100 fmol) allowed Mus81–Mms4 to overcome the inhibitory effect of increasing concentrations of Mg2+, resulting in a marked increase in the cleavage of 3′F substrate by Mus81–Mms4. Higher concentrations of MgCl2 caused more robust stimulation of Mus81–Mms4 by Fen1; at 2.5 mM MgCl2, stimulation was weak (<2-fold) with Rad27 added (Figure 4A, lanes 5–7). At 7.5 or 10 mM MgCl2, however, the addition of 100 fmol of Rad27 resulted in more robust cleavage of the substrate (Figure 4A, lanes 11–16). This result demonstrates that effective stimulation of Mus81–Mms4 activity by Rad27 requires higher concentrations (>5 mM) of MgCl2. Thus, we decided to use 10 mM MgCl2 hereafter.
Figure 4.
Rad27 stimulates Mus81–Mms4 at physiological salt concentrations. For all experiments, the 3′F substrate (10 fmol) was used. (A) Mus81–Mms4 stimulation by Rad27 was examined in standard reaction mixtures in increasing concentrations (2.5, 5, 7.5 and 10 mM) of MgCl2. The reactions contained 7.5 fmol of Mus81–Mms4 with two different levels (25 or 100 fmol) of Rad27 as indicated. (B) The standard reactions in (A) were repeated in the presence of increasing concentrations (70, 100, 150, 200 and 250 mM) of NaCl. (C) A time-course experiment was performed with Mus81–Mms4 equimolar with DNA substrate (10 fmol each) in the presence of 100 mM NaCl. The reactions were carried out in the presence (+) and in the absence of Rad27 (10 fmol). The reaction mixtures were incubated at 30°C and aliquots of the reaction mixtures were withdrawn at indicated times. The cleavage products were analyzed as described in Figure 2. (D) The amounts of cleavage products obtained in (C) were plotted against incubation time.
We also investigated whether Rad27 is able to stimulate Mus81–Mms4 at physiological salt concentrations. For this purpose, we tested varying concentrations (70, 100, 150, 200 and 250 mM) of NaCl (Figure 4B) with a fixed concentration (10 mM) of MgCl2. Mus81–Mms4 activity was hardly affected by addition of NaCl up to 150 mM, but significantly inhibited by >200 mM NaCl (Figure 4B, compare lanes 2, 5, 8 and 11). We found that Mus81–Mms4 activity was stimulated by Fen1 under all NaCl concentrations tested. Besides, the extents of Mus81–Mms4 stimulation by Rad27 were similar in the range of 70–150 mM NaCl, confirming that stimulation of Mus81–Mms4 by Rad27 can occur at the physiological salt concentration.
One potential mechanism by which Rad27 stimulates Mus81–Mms4 activity is the nonspecific DNA binding activity of Rad27 that helps dissociate Mus81–Mms4 from cleavage products, thereby rapidly recycling Mus81–Mms4. To exclude this possibility, we examined the effect of Rad27 on Mus81–Mms4 in the presence of a stoichiometric amount with the 3′F substrate DNA. If stimulation were due to the nonspecific DNA binding activity of Rad27, stimulation by recycling of Mus81–Mms4 could be minimized when all the substrates were occupied with Mus81–Mms4. For this reason, we preincubated the 3′F substrate and Mus81–Mms4 (equimolar), followed by addition of Mg2+ (with or without Rad27) to initiate reaction. Under this condition, marked stimulation could not be expected due to the use of a high level of Mus81–Mms4 with respect to the level of substrate. As shown in Figure 4C, we carried out a time-course experiment with reaction mixtures containing equimolar amounts (10 fmol each) of Mus81–Mms4 and 3′F substrate in the absence (Figure 4C, lanes 2–9) and presence (10 fmol; Figure 4C, lanes 10–17) of Rad27. We found that Rad27 was still able to stimulate Mus81–Mms4 activity (Figure 4C and D) more efficiently with shorter incubation period of reaction. Therefore, this result supports the notion that Rad27 stimulates Mus81–Mms4 via a specific protein–protein interaction and the stimulation occurs during a single catalytic cycle (see also below).
When we examined the cleavage products in a high-resolution gel, we found that the substrate specificity as well as the cleavage products formed by both enzymes remained identical, the same as that produced by each enzyme alone. For example, Rad27 stimulated endonuclease activity of Mus81–Mms4 with all DNA substrates tested to a similar extent, indicating that the presence of Rad27 did not alter substrate specificity of Mus81–Mms4 (data not shown). Analyses of cleavage products by a high-resolution sequencing gels revealed that the cleavage sites were not altered (data not shown).
Rad27 interacts directly with Mus81–Mms4
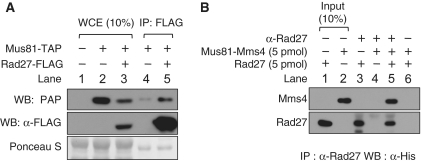
The marked mutual stimulation observed between Rad27 and Mus81–Mms4 prompted us to examine their physical interaction. To this end, we prepared cell-free extracts containing epitope-tagged Mus81-TAP and/or Rad27-FLAG and one of the two tagged proteins was immunoprecipitated with either PAP or anti-FLAG monoclonal antibodies (Figure 5A). Immunoprecipitation of Rad27-FLAG with anti-FLAG beads precipitated Mus81 as well (Figure 5A, lane 5). We noted that expression of Mus81-TAP was inhibited by expression of Rad27-FLAG (Figure 5A, compare lanes 2 and 3). This might account for some background observed in control in the absence of Rad27 (Figure 5A, lane 4). We also attempted to confirm direct interactions between Mus81–Mms4 and Rad27 using purified proteins (Figure 5B). Mms4 and Rad27, tagged with hexahistidine residues at their N- and C-terminus, respectively, were immunoprecipiated with anti-Rad27 polyclonal antibodies. The resulting precipitates, probed with anti-His monoclonal antibodies, revealed the presence of both Mms4 (and thus Mus81) and Rad27 (Figure 5B, lane 5). Controls indicated that the coimmuneprecipitation specifically required the simultaneous presence of Rad27, Mus81–Mms4 and anti-Rad27 antibodies (Figure 5B, lanes 3, 4 and 6). Coimmunoprecipitation in the presence of DNase I did not affect the results above, indicating that the interactions between the two proteins were not due to a DNA contaminant (data not shown). Taken together, these results indicate that Rad27 and Mus81–Mms4 interact directly in vivo and suggest that their mutual activation most likely depends on this property.
Figure 5.
Rad27 interacted physically with Mus81–Mms4. (A) The strains that expressed either Mus81–TAP or Rad27-FLAG or both were constructed as described in ‘Materials and Methods’ section. Crude extracts (5.5 mg), derived from each strain as indicated at the top of figure, were mixed with anti-FLAG antibody-conjugated agarose beads (40 µl), and the mixtures were then incubated at 4°C for 2 h in binding buffer (25 mM HEPES-NaOH/pH7.5, 150 mM NaCl, 10% glycerol, 0.01% NP40). The agarose beads were collected by brief centrifugation, washed, and analyzed by western blotting using either PAP or anti-FLAG (α-FLAG) monoclonal antibodies. WCE, whole cell extract (0.25 mg); Ponceaus S indicates loading controls stained with Ponceasu S. (B) Recombinant Mus81–His6Mms4 and Rad27-His6 (5 pmol each) were mixed and incubated on ice for 1 h in buffer containing 25 mM HEPES-NaOH/pH7.5, 150 mM NaCl, 10% glycerol and 0.01% NP40. After incubation, anti-Rad27 antibodies (α-Rad27, 5 µl) and protein-A agarose (5 µl) were added to reaction mixtures, followed by an additional incubation for 1 h at 4°C with rocking. Western blot analysis was carried out with anti-His monoclonal antibodies (α-His).
Enzymatic stimulation depends on specific protein–protein interactions
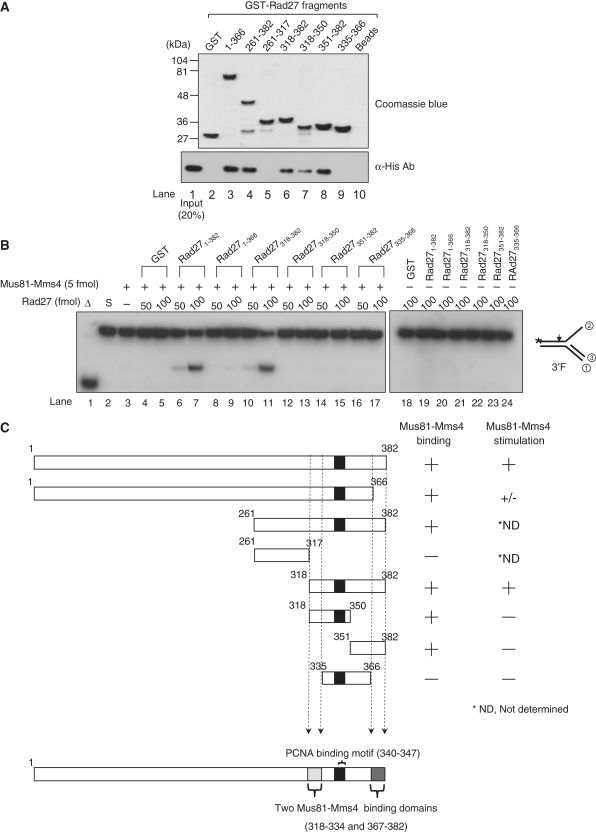
The regions in Rad27 that contribute to its direct physical binding to Mus81–Mms4 were examined. For this purpose, a series of GST-fusion Rad27 fragments were prepared as described in ‘Materials and Methods’ section (Figure 6A, top panel, Coomassie blue), and examined for their ability to bind Mus81–Mms4 using GST-pull-down assays. The presence of Mms4-His6 in the precipitated complexes was examined by western blot analysis with anti-His monoclonal antibodies (Figure 6A, bottom panel, α-His Ab). All of the GST-fusion Rad27 derivatives, except Rad27261–317 and Rad27335–366 (Figure 6A, bottom panel, lanes 5 and 9, respectively), interacted with Mus81–Mms4. GST or GST-beads alone failed to do so (Figure 6A, bottom panel, lanes 2 and 10), indicating that specific binding occurred between Rad27 fragments and Mus81–Mms4. We noted that two regions (between aa 318–334 and 367–382), flanking the known PCNA binding motif (aa 340–347), bound Mus81–Mms4 (Figure 6C). Each region alone was sufficient to bind Mus81–Mms4, since Rad27318–350 or Rad27351–382 interacted with Mus81–Mms4 as efficiently as Rad27318–382 (Figure 6A, compare lanes 6–8; see also Figure 6C). It appears that the amounts of Mus81–Mms4 bound to GST-Rad27318–350, GST-Rad27318–382 and GST-Rad27351–382 varied, but this is most likely due to varying levels of GST-Rad27 fragments used for the GST pull-down assays (compares lanes 6–8 in Figure 6A, bottom panel). Next, we tested whether the GST-fusion Rad27 derivatives that bound to Mus81–Mms4 were able to stimulate the enzymatic activity of Mus81–Mms4 using the 3′F substrate as shown in Figure 6B (see also Figure 6C). We found that Rad27 fragments containing only one of the two binding regions (Rad271–366, lanes 8 and 9; Rad27318–350, lanes 12 and 13; Rad27351–382, lanes 14 and 15) or none (Rad27335–366, lanes 16 and 17) failed to stimulate the activity of Mus81–Mms4, whereas Rad27318–382 containing both regions stimulated Mus81–Mms4 as efficiently as Rad271–382 (full-length) (Figure 6B, compare lanes 6and 7 to 10 and 11). Each GST-fusion Rad27 derivative did not cleave the 3′F substrate, excluding the possibility that the cleavage was due to contaminating activity derived from E. coli (data not shown). Thus, simultaneous presence of at least two binding motifs in Rad27 (that support the binding of Mus81–Mms4) appears to be essential for the stimulation of Mus81–Mms4 activity.
Figure 6.
Mapping of specific Rad27 domains required for binding and stimulation of Mus81–Mms4 activity. (A) The numbers above the gel indicate amino acid positions present in the GST-fusion Rad27 derivatives. GST indicates glutathione-S transferase. Beads, Mus81–Mms4 incubated with glutathione-agarose beads in the absence of Rad27. GST-fusion Rad27 fragments bound to glutathione-agarose beads were analyzed in a 10% SDS-polyacrylamide gel and stained with Coomassie blue (top gel, Commassie blue). For GST-pull-down assays (bottom gel), 1 pmol of Mus81–Mms4 was mixed with 100 pmol of GST (lane 2) or GST-fused Rad27 fragments (lanes 2–9) in binding buffer (25 mM HEPES-NaOH/pH7.5, 150 mM NaCl, 10% glycerol, and 0.01% NP40). The reaction mixtures were incubated for 1 h at 4°C with rocking. The complex formed was collected by centrifugation and washed. The presence of Mus81–Mms4 in the precipitated materials was determined by western blotting using anti-His monoclonal antibodies against His6-Mms4 (α-His Ab, bottom panel). Lane 1 contained 0.2 pmol of Mus81–Mms4 as an input control. (B) The reaction mixtures contained a fixed amount (5 fmol) of Mus81–Mms4, and the reactions were carried out with 10 fmol of the 3′F substrate and two levels (50 or 100 fmol) of GST-fused Rad27 fragments using standard reaction condition as described in ‘Materials and Methods’ section. The cleavage products formed from the 3′ F substrates were analyzed as described in Figure 2B. (C) A schematic summary of the results obtained in (A) and (B). The numbers in each fragment indicate the positions of amino acids present in full-length Rad27.
DISCUSSION
In this report, data are presented showing that the three endonucleases, Rad27, Mus81–Mms4 and Dna2, interacted genetically and functionally in vivo and in vitro. Our experimental results suggest that MUS81–MMS4 is involved in the processing of Okazaki fragments by virtue of its genetic and functional interaction with DNA2 and RAD27, both of which play critical roles in lagging strand processing. In support of this notion: (i) Overexpression of Mus81–Mms4 rescued growth defects associated with dna2K1080E. Furthermore, we found that dna2-2 and dna2-4, the two other dna2 mutant alleles isolated by Formosa and Nittis (44) were also suppressed by overexpression of Mus81–Mms4 (data not shown). (ii) Mus81–Mms4 and Rad27 mutually stimulated the structure-specific endonuclease activity intrinsic to each protein. (iii) Finally, the functional interactions between Rad27 and Mus81–Mms4 were shown to be mediated by specific protein–protein interactions.
We believe that suppression of dna2 mutants by Mus81–Mms4 occurred via stimulation of Rad27. A number of previous results have shown that elevated levels of Rad27 can substitute for a functionally impaired Dna2. These include: (i) Overexpression of Rad27 suppressed several dna2 mutant alleles, including dna2-1 and dna2Δ405N (31,45). (ii) We also have shown that the RAD27 gene can suppress dna2K1080E mutant (data not shown). (iii) Several suppressors of dna2 mutations were found to stimulate the nuclease activity of Rad27. For example, in vivo the nonessential yeast MGS1 gene could suppress dna2 mutations, but in vitro the purified Mgs1 protein stimulated Rad27, but not Dna2 endonuclease activity (39). The observed suppression was absolutely dependent on the presence of a functional copy of RAD27. Thus, enhancement of Rad27 activity (or its abundance) could lead to rapid removal of flaps generated by pol δ-catalyzed displacement DNA synthesis. The net effect of this activation blocks formation of long flaps. Normally, when long flaps are formed, they could bind RPA and are efficiently cleaved by Dna2, but resistant to cleavage by Fen1 (31). Thus, enhanced or elevated levels of Rad27 activity could make cells less dependent on Dna2 activity, allowing cells to grow with impaired Dna2 activity.
Although Rad27 and Mus81–Mms4 are involved in two seemingly separate processes, their mutual stimulation described in this study suggest a more direct interfunctional role between the two structure-specific endonucleases. Based upon the genetic and biochemical interactions between Rad27 and Mus81–Mms4, a model can be proposed that may account for how they collaborate in repairing unprocessed flaps. All flaps may not be correctly processed for a variety of reasons. For example, the two critical processing enzymes, Dna2 and Fen1, may not be able to process efficiently flaps that form a structure. Failure in processing such flaps could result in formation of double strand breaks in replicated lagging strand, which are supposed to be resected by the MRX complex to generate 3′ ssDNA overhang. The 3′ overhang then would start homologous recombination by invading replicated sister chromatid DNA, resulting in formation of recombination intermediates that are toxic to cells unless resolved. They can be normally removed by Mus81–Mms4 or Sgs1–Top3. The choice between the two pathways is governed by the availability of nicks in the recombination intermediates. Thus, processing of resilient Okazaki fragments could be ultimately assisted by Mus81–Mms4 when nicks are present in the recombination intermediates or by Sgs1–Top3 when all nicks are sealed (46).
The joint role of Rad27 and Mus81–Mms4 could come into effect via the interconversion between DNA structures specific for each endonuclease that arise from reannealing of flaps to the template DNA. It should be noted that numerous structural intermediates, called ‘equilibrating flaps’ (double-flap DNA with both 5′ and 3′ flaps of varying length) can result from the reannealing of flaps. These intermediates are not cleaved by either Rad27 or Mus81–Mms4 until they convert into a structural form susceptible to cleavage by each enzyme. The 5′ or 3′ flap can be converted into a 3′ or 5′ flap, respectively, in a manner similar to that seen in Holliday junction migration. If the equilibrating flaps are trapped in a structure suitable for the action of each endonuclease, they could be processed more effectively and rapidly if the two enzymes involved could stimulate each other’s activity. This could constitute a feedforward and feedback stimulatory mechanism, increasing the rate of flap removal. Although BLM (47) and Mph1 (34) could directly stimulate Fen1, they are able to migrate Holliday junctions. Therefore, it is conceivable that the two helicases could contribute to Okazaki fragment processing by facilitating conversion of the equilibrating flaps into a specific structure that can be cleaved by one of the two enzymes. One example is that Sgs1 drives the annealing process to completion by virtue of its ability to displace the downstream strand. The product formed by this reaction would contain a 5′ ssDNA flap that is a substrate for Fen1. Other helicase may reverse this process to generate 3′ flap, structure that can be processed by Mus81–Mms4. Recently, the human BLM helicase was shown to stimulate nuclease activity of the Mus81–Eme1 complex (47). In addition, Rad54 was found to strongly stimulate Mus81–Mms4 in an ATP-independent fashion in humans and yeasts (48). This is interesting, considering that both BLM and Rad54 are capable of mediating the migration of branched DNA structures. Alternatively, a nuclease(s) that can simultaneously process both 5′ and 3′ double flaps could reduce the length of both flaps, and generate more rapidly a DNA substrate that can be processed by either Rad27 or Mus81–Mms4.
Although Rad27 and Mus81–Mms4 differ in their structural substrate specificity, therefore, they can act jointly to remove flap structures more efficiently particularly with aid of other protein factors that facilitate inter-conversion of substrate DNA.
The mutual stimulatory effects of Rad27 and Mus81–Mms4 also suggest that that Rad27 can play critical roles in the resolution of toxic recombination intermediates and the re-initiation of damaged RFs by activating the Mus81–Mms4 complex (see ref. 46 for details). Mus81–Mms4 is known to produce collapsed RFs, when RFs encounter damaged DNA (16,22). Then, cells attempt to repair collapsed RFs (produced by Mus81–Mms4 action) by synthesis-dependent strand annealing initiated by double-strand breaks. Therefore, the mutual stimulatory effects of Rad27 and Mus81–Mms4 provide with cells more effective means to restore damaged RFs. The collaboration between Rad27 and Mus81–Mms4/Sgs1-Top3 appears to be physiologically relevant since the synthetic-lethal interaction was detected in cells harboring mutations in MUS81 or SGS1 in combination with rad27Δ (33,49,50). This view is also in keeping with the finding that rad27Δ mutant was sensitive to HU, which induces replication fork arrest (51).
FUNDING
Funding for open access charge: National Research Foundation of Korea (Grant No. 20100000009) funded by the Ministry of Education, Science and Technology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Jerard Hurwitz for critical reading of the manuscript.
REFERENCES
- 1.Cox MM. Historical overview: searching for replication help in all of the rec places. Proc. Natl Acad. Sci. USA. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 4.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 5.Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. BioEssays. 1999;21:286–94. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. (review) [DOI] [PubMed] [Google Scholar]
- 6.Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 7.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Whitby MC. Junctions on the road to cancer. Nat. Struct. Mol. Biol. 2004;11:693–695. doi: 10.1038/nsmb0804-693. [DOI] [PubMed] [Google Scholar]
- 9.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;16:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 12.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre F, Chan A, Heyer W-D, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2001;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 16.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 17.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 18.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 19.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein MUS81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Esser J, Hanada K, Chahwan R, Khaw AK, et al. EME1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cell. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, Xiao W. Functional domains required for the Saccharomyces cerevisiae Mus81-Mms4 endonuclease complex formation and nuclear localization. DNA Repair. 2003;2:1435–1447. doi: 10.1016/j.dnarep.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rgh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 23.McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Kanaar KR, Jasin M, et al. Involvement of mammalian MUS81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 24.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 25.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 27.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 29.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 31.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 32.Ii M, Brill SJ. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr. Genet. 2005;48:213–225. doi: 10.1007/s00294-005-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 34.Kang YH, Kang MJ, Kim JH, Lee CH, Cho IT, Hurwitz J, Seo YS. The MPH1 gene of Saccharomyces cerevisiae functions in Okazaki fragments processing. J. Biol. Chem. 2009;284:10376–10386. doi: 10.1074/jbc.M808894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 36.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang KY, Baek K, Kim HY, Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 38.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JH, Kang YH, Kang HJ, Kim DH, Ryu GH, Kang MJ, Seo YS. In vivo and in vitro studies of Mgs1 suggest a link between genome instability and Okazaki fragment processing. Nucleic Acids Res. 2005;33:6137–6150. doi: 10.1093/nar/gki900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CH, Shin YK, Phung TT, Bae JS, Kang YH, Nguyen TA, Kim JH, Kim DH, Kang MJ, Bae SH. Involvement of Vts1, a structure-specific RNA-binding protein, in Okazaki fragment processing in yeast. Nucleic Acids Res. 2010;38:1583–1595. doi: 10.1093/nar/gkp1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 42.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 44.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 46.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 47.Zhang R, Sengupta S, Yang Q, Linke SP, Yanaihara N, Bradsher J, Blais V, McGowan CH, Harris CC. BLM helicase facilitates Mus81 endonuclease activity in human cells. Cancer Res. 2005;65:2526–2531. doi: 10.1158/0008-5472.CAN-04-2421. [DOI] [PubMed] [Google Scholar]
- 48.Matulova P, Marini V, Burgess RC, Sisakova A, Kwon Y, Rothstein R, Sung P, Krejci L. Cooperativity of Mus81·Mms4 with Rad54 in the resolution of recombination and replication intermediates J. Biol. Chem. 2009;284:7733–7745. doi: 10.1074/jbc.M806192200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair. 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 51.Lopes J, Ribeyre C, Nicolas A. Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol. Cell Biol. 2006;26:6675–6689. doi: 10.1128/MCB.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]