Abstract
The biological aldehydes, malondialdehyde and base propenal, react with DNA to form a prevalent guanine adduct, M1dG. The exocyclic ring of M1dG opens to the acyclic N2-OPdG structure when paired with C but remains closed in single-stranded DNA or when mispaired with T. M1dG is a target of nucleotide excision repair (NER); however, NER is absent in mitochondria. An in vitro transcription system with purified human mitochondrial RNA polymerase (POLRMT) and transcription factors, mtTFA and mtTFB2, was used to determine the effect of M1dG on POLRMT elongation. DNA templates contained a single adduct opposite either C or T downstream of either the light-strand (LSP) or heavy-strand (HSP1) promoter for POLRMT. M1dG in the transcribed strand arrested 60–90% POLRMT elongation complexes with greater arrest by the adduct when opposite T. POLRMT was more sensitive to N2-OPdG and M1dG after initiation at LSP, which suggests promoter-specific differences in the function of POLRMT complexes. A closed-ring analog of M1dG, PdG, blocked ≥95% of transcripts originating from either promoter regardless of base pairing, and the transcripts remained associated with POLRMT complexes after stalling at the adduct. This work suggests that persistent M1dG adducts in mitochondrial DNA hinder the transcription of mitochondrial genes.
INTRODUCTION
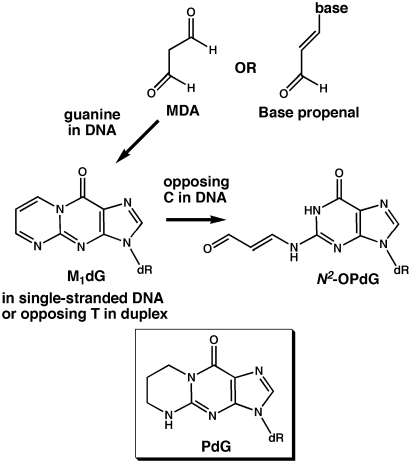
The mitochondria of eukaryotic cells each harbor multiple copies of a 16 569 bp circular chromosome that must be maintained for efficient cellular energy production (1,2). Electrons that escape from the transport chain in the inner membrane react with molecular oxygen and water to produce reactive oxygen species (ROS) in the matrix where mitochondrial DNA (mtDNA) resides (3). The ROS may damage mtDNA directly by oxidation of bases or the sugar backbone. Additionally, ROS oxidation of DNA and inner membrane lipids generates biological aldehydes, such as base propenals, malondialdehyde (MDA) and 4-hydroxynonenal, which react with nucleic acid bases (4). The base propenals generated from DNA sugar backbone oxidation appear to be the major endogenous source of the 3-(2′-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one adduct of guanine known as M1dG (Figure 1) (5–8). M1dG has been reported to occur in human tissues at levels comparable to the most abundant form of base oxidation, 8-oxodeoxyguanine, and to be twice as prevalent in mtDNA than in nuclear chromosomes (4,8–12). In eukaryotic cell nuclei, excision repair pathways remove modified bases and maintain genetic integrity. Base excision repair (BER), which removes oxidized bases such as 8-oxodeoxyguanine and thymine glycol, also functions in mitochondria (13–15). Conversely, nucleotide excision repair (NER), the mechanism implicated in processing of M1dG, is absent in the organelle (16–18). The lack of NER and the presence of ROS in the mitochondrial matrix may contribute synergistically to the persistence of M1dG in mtDNA.
Figure 1.
Formation of M1dG by malondialdehyde (MDA) and base propenal. The exocyclic adduct, M1dG, can react with the amino group of a paired cytosine and form acyclic N2-OPdG. PdG is a stable analog of the closed-ring M1dG.
The chemical properties of M1dG make this form of base damage particularly interesting with respect to its potential detriment to DNA replication and gene expression. As seen in Figure 1, the external ring of M1dG is closed in single-stranded DNA or when mispaired with thymine, but opens to an acyclic N2-(3-oxo-1-propenyl)-dG derivative (N2-OPdG), when correctly paired in DNA with cytosine (19,20). Therefore, a single aldehyde modification of DNA to form M1dG may result in two structurally distinct adducts that must be negotiated by DNA and RNA polymerases and recognized by DNA repair pathways. A chemically stable M1dG analog, 1,N2-propano-2′-deoxyguanosine, called PdG, has been used to elucidate differences in M1dG and N2-OPdG effects on prokaryotic and eukaryotic polymerases.
In bacteria and mammalian cells, M1dG causes a high rate of frameshifts and base substitutions during replication (21–24). M1dG and PdG disrupt nucleotide insertion and translocation by the Klenow fragment of bacterial DNA polymerase I (25), the human BER polymerase, DNA pol β (26,27), and the human translesion DNA polymerase, pol η (28,29). Recent in vitro transcription studies revealed a structure-dependent M1dG arrest of elongation by the T7 bacteriophage RNA polymerase (T7RNAP) and by mammalian RNA polymerase II (RNAP II) that suggests M1dG may also disrupt gene expression (30). Both polymerases exhibited a greater transcriptional arrest at M1dG opposite T; however, RNAP II elongation complexes were more sensitive than T7RNAP to both biological forms of the adduct. PdG, the analog of the closed-ring M1dG, completely blocked RNAP II translocation regardless of the opposing base (30). Human mitochondrial RNA polymerase (POLRMT) is a single subunit enzyme homologous to T7RNAP; therefore, M1dG and its acyclic N2-OPdG conformation may substantially hinder POLRMT transcript elongation. In addition, POLRMT, like RNAP II, requires the assistance of other proteins for promoter initiation, a property that could influence POLRMT behavior upon encountering DNA damage (1,31–34). As POLRMT elongation remains largely uncharacterized, studies of POLRMT interaction with M1dG provide not only information about the biological impact of mtDNA damage on mitochondrial gene expression but also understanding of basic POLRMT function.
POLRMT transcription initiates at different promoter sequences on the heavy (H) strand and the light (L) strand of the mitochondrial chromosome and produces polycistronic RNAs (1,31,35–37). The H strand contains genes for 12 of the 13 mtDNA-encoded proteins and the mitochondrial ribosomal RNA (rRNA) and transfer RNA (tRNA) required for translation of the proteins. The heavy strand promoter HSP1 is thought to primarily direct the synthesis of a transcript including 12S and 16S rRNA that terminates in the tRNALeu gene (38), while a second promoter, HSP2, has been proposed to be the site for messenger RNA and tRNA production from the H strand (39). The L strand promoter, LSP, not only permits POLRMT transcription, but also controls POLRMT synthesis of an RNA primer for mtDNA replication at the H strand origin (40–42). The nuclear-encoded proteins, mtTFA, mtTFB1 and mtTFB2, support promoter-directed transcription of mtDNA by POLRMT (1,2,31,43,44). The combination of mtTFA and mtTFB2 maximally stimulates POLRMT activity at LSP and HSP1 in vitro, while the primary role of mtTFB1 appears to be as an rRNA methyltransferase that helps to link mtDNA transcription and translation (1,31,33,34, 45–47). Given the different roles of LSP and HSP1 in directing activities on the mitochondrial chromosome, DNA damage that interferes with POLRMT transcript elongation could hinder both mtDNA gene expression and replication.
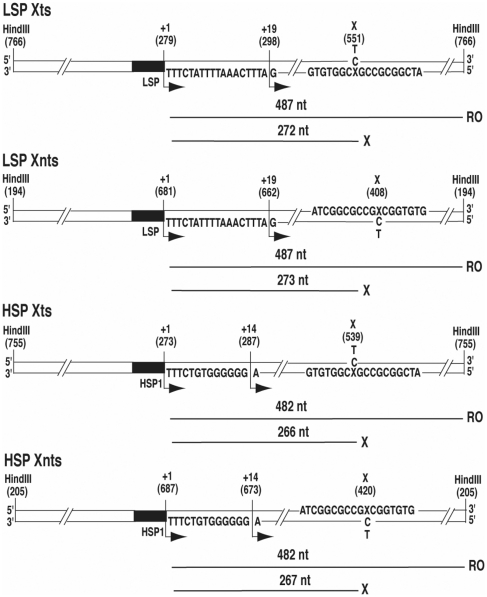
To investigate the consequences of M1dG on transcript elongation by POLRMT and gain insight into POLRMT function, we employed an in vitro transcription system containing purified human POLRMT, transcription factors, mtTFA and mtTFB2, and DNA templates containing a site-specific M1dG or PdG in either the transcribed (Xts) or non-transcribed (Xnts) strand downstream of LSP or HSP1 (Figure 2). M1dG was placed opposite either C or T to elucidate the structural effects of N2-OPdG and M1dG, respectively, on POLRMT. The DNA sequence context of the adduct within the POLRMT templates was identical to that used in the previous studies of RNAP II and T7RNAP, allowing for a comparison of the elongation behavior of these polymerases. In light of the results showing substantial M1dG effects on T7RNAP and RNAP II transcription, we hypothesized that M1dG would impede POLRMT elongation in a structure-dependent manner suggesting that persistent M1dG adducts may disrupt the expression of genes in mtDNA. As POLRMT operates from two distinctly different promoters on the mitochondrial chromosome, we believed that our system also would provide insight into potential promoter-dependent properties of POLRMT elongation, such as the response of POLRMT to mtDNA sequences that govern transcript length.
Figure 2.
DNA transcription templates for POLRMT. The linear Xts and Xnts DNA templates contain dG, M1dG or PdG (X) either in the transcribed or nontranscribed strands, respectively, downstream of promoters for POLRMT transcription (LSP and HSP1) and paired with either C or T. Numbers in parentheses indicate positions in the DNA plasmid construct. Transcript lengths in nucleotides expected from run-off (RO) or from adduct-arrest (X) are depicted below each template. Transcription started at each promoter (+1) and proceeded in the direction of the bent arrows. Transcripts were labeled by [α-32P]-ribonucleotide incorporation within a ‘labeling cassette’ between the bent arrows on each template diagram. Omission of the NTP needed at +19 for LSP templates and +14 for HSP1 templates stalled POLRMT complexes and facilitated the analysis of transcripts from a single-round of POLRMT transcription on each DNA molecule.
MATERIALS AND METHODS
Reagents
T4 polynucleotide kinase, T4 DNA polymerase and M13K07 helper phage were purchased from New England Biolabs (Beverly, MA, USA), and T4 DNA ligase was from Invitrogen (Carlsbad, CA, USA). The F′ Escherichia coli strain MV1184 was developed by J. Messing (Rutgers University, Piscataway, NJ, USA). Non-radioactive ribonucleoside and deoxyribonucleoside triphosphates were purchased from GE Healthcare Bio-sciences Corp (Piscataway, NJ, USA). Radionuclides, [α-32P] UTP, [α-32P] CTP and [γ-32P] ATP, were obtained from Perkin Elmer (Boston, MA, USA). Unmodified oligonucleotides were synthesized by Eurofins MWG Operon (Huntsville, AL, USA). Streptavidin-linked paramagnetic particles were from Promega (Madison, WI, USA).
Preparation of Oligonucleotides
Oligonucleotides of the 18-base sequence 5′-ATC GGC GCC GXC GGT GTG-3′ containing either a single M1dG or PdG at the position X were synthesized as previously described (48), and their purity and presence of the adduct was verified by mass spectrometry. Samples of the oligonucleotides were 32P labeled and resolved by denaturing polyacrylamide gel electrophoresis (PAGE) to check for degradation that could affect construction of the plasmid templates below. The undamaged 18-mer (X = dG) and the oligonucleotides for both strands of LSP and HSP1 were prepared by Eurofins MWG Operon (Huntsville, AL, USA). The LSP and HSP1 sequences were as follows: LSP nontranscribed strand, 5′-AAT TCA TGT GTT AGT TGG GGG GTG ACT GTT AAA AGT GCA TAC CGC CAA AAG ATA AAA TTT GAA ATC T-3′; LSP transcribed strand, 3′-GTA CAC AAT CAA CCC CCC ACT GAC AAT TTT CAC GTA TGG CGG TTT TCT ATT TTA AAC TTT AGA GAT C-5′; HSP1 nontranscribed strand, 5′-AAT TCC GCT GCT AAC CCC ATA CCC CGA ACC AAC CAA ACC CCA AAG ACA CCC CCC T-3′; and HSP1 transcribed strand, 3′-GGC GAC GAT TGG GGT ATG GGG CTT GGT TGG TTT GGG GTT TCT GTG GGG GGA GAT C-5′. These sequences were designed according to studies of human mtDNA promoters by the Clayton group with modifications to facilitate transcription template construction (37)
Construction of transcription templates
The plasmid transcription templates for POLRMT were constructed from plasmids used in previously published studies of RNAP II and T7RNAP transcriptional arrest (30). The identical sequence context for the adduct position (Figure 2, X) was maintained allowing functional comparisons of the three polymerases. In brief, a double-stranded oligonucleotide containing either the mitochondrial LSP or HSP1 sequence modified to create a ‘labeling cassette’ downstream of the promoter was ligated, at a 20:1 molar ratio of oligonucleotide to vector, into the EcoRI–XbaI cloning site replacing the T7RNAP promoter (30) (Figure 2). The constructs were transformed into the F′ E. coli strain MV1184 for replication of the plus strand by the M13K07 phage. Plasmids containing dG, M1dG or PdG in either the transcribed or nontranscribed strand were prepared by T4 DNA polymerase second-strand synthesis on a single-stranded plasmid using the appropriate 18-mer as a primer (49). For each synthesis, a control reaction without the 18-mer was performed to insure that no intact plasmid was generated from aberrant priming by the single-stranded DNA. Products were resolved by electrophoresis in TAE (40 mM Tris–acetate, 2 mM disodium EDTA, pH 8.5) on 1% agarose containing 0.3 µg/ml ethidium bromide. Plasmids were excised, electroeluted from the gel using an Amicon Centrilutor (Millipore, Billerica, MA, USA), and concentrated by centrifugation. The constructs were digested with HindIII, then gel purified and isolated as described earlier.
The 118-bp POLRMT transcription templates were constructed by equimolar annealing of 79-, 18- and 21-base transcribed strand oligonucleotides to a 118-base nontranscribed strand sequence followed by ligation with T4 DNA ligase for 12–16 h at 16°C. The 18-mer was the same PdG-adducted sequence utilized above, and the 21-mer comprising the 5′ end of the transcribed strand contained a 5′ biotin. The ligation reactions were extracted with 25:24:1 phenol/chloroform/isoamyl alcohol and filtered through G-25 then G-100 Sephadex to remove contaminates and unligated transcribed strand 18-mer and biotinylated 21-mer. The 118-bp templates were resolved on 3.5% agarose to ensure purity. The concentration of each template was determined by ultraviolet spectrophotometry with a Shimadzu ND-1000 Nanodrop instrument (Columbia, MD, USA).
POLRMT in vitro transcription
Recombinant human POLRMT, mtTFA and mtTFB2 were purified from E. coli by previously published protocols with modifications submitted separately for publication (43,50,51). Proteins of comparable quality from these preparations can be obtained from Enzymax, LLC (Lexington, KY, USA), which has not provided funding for or influenced the research reported herein. For promoter binding of transcription factors, 100 nM mtTFA and 24 nM mtTFB2 were incubated at 32°C for 5 min with 4 nM LSP or HSP templates in POLRMT transcription buffer (10 mM HEPES, pH 7.9, 10 mM MgCl2, 20 mM NaCl, 0.1 µg/µl bovine serum albumin, 1 mM dithiothreitol). Reactions for LSP template binding contained 100 µM ATP, 20 µM GTP, 10 µM UTP and 0.1 µM [α-32P] UTP (3000 Ci/mmol) and those for HSP template binding contained 100 µM ATP, 20 µM GTP, 10 µM CTP and 0.1 µM [α-32P] CTP (3000 Ci/mmol). Addition of 20 nM POLRMT initiated transcription at each promoter and continued incubation at 32°C allowed POLRMT extension of transcripts to the end of the ‘labeling cassette’ in each template (Figure 2). By excluding the next ribonucleotide required for transcript extension (CTP for the LSP templates and UTP for the HSP1 templates), POLRMT complexes were stalled until ribonucleoside triphosphate (NTP) addition. After 5 min for LSP template reactions and 10 min for HSP template reactions, heparin was added at 50 ng/µl to inactivate uninitiated POLRMT, then elongation by the stalled POLRMT complexes was induced by increasing all four NTP concentrations to 400 µM. The incubation times chosen for POLRMT initiation provided the maximal amount of cassette-arrested transcripts that remained competent for elongation following heparin and NTP addition. Control reactions confirming heparin inhibition of POLRMT re-initiation and an absence of additional 32P incorporation during elongation are shown in Supplementary Data (Figure S1). After elongation times up to 30 min at 32°C, reactions were stopped with 0.3% SDS/22.5 mM EDTA, proteins digested with 0.4 mg/ml proteinase K, and nucleic acids precipitated with ethanol by centrifugation. Samples were dried under vacuum, resuspended in 80% formamide dye, and resolved by denaturing 5% polyacrylamide gel electrophoresis in TBE (89 mM Tris-borate, 1 mM EDTA, pH 8.0). The gels were dried, and then transcripts were detected by phosphorimaging analysis using a Storm 840 system with Image Quant software (GE Healthcare Bio-sciences Corp, Piscataway, NJ, USA).
POLRMT transcription complex stability
The stability of PdG-arrested POLRMT complexes was assessed by isolation of transcripts produced from a biotinylated 118-bp template linked to streptavidin-conjugated paramagnetic particles (SA-PMPs). Briefly, SA-PMPs were washed in cold POLRMT oligonucleotide transcription buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 100 mM NaCl, 0.1 µg/µl bovine serum albumin, 1 mM dithiothreitol) then resuspended at 6 mg/ml. For DNA binding to SA-PMPs, 150 µg of SA-PMPs were mixed with 3.6 pmol biotinylated 118-bp template and incubated at 32°C for 5 min. The buffer was removed from the template-linked SA-PMPs followed immediately by addition of 8 µl of transcription reaction mix containing 360 nM mtTFA, 360 nM mtTFB2, 600 µM ATP, 150 µM GTP, 100 µM UTP, 60 µM CTP and 0.33 µM [α-32P] UTP (3000 Ci/mmol) in POLRMT oligonucleotide transcription buffer. Samples were incubated at 32°C for 2 min to allow transcription factor binding then POLRMT was added at 360 nM to initiate transcription in a total reaction volume of 10 µl. After 2 min, 60 ng/µl heparin was added to prevent further initiation and incubation was continued at 32°C for 1, 5 or 15 min when the SA-PMPs were washed two times with 100 µl 32°C POLRMT transcription buffer. Following buffer aspiration from the SA-PMPs, POLRMT complexes were denatured by heating at 80°C in 80% formamide/50 mM EDTA loading dye. Transcripts associated in stable complexes on the SA-PMPs-linked templates were resolved by 20% polyacrylamide gel electrophoresis in TBE and detected by phosphorimager analysis as above.
To confirm that isolation of the arrested transcripts was dependent upon biotinylated template linkage to the SA-PMPs, identical transcription reactions were performed with 3.6 pmol of non-biotinylated, 118-bp template followed by buffer washes and gel resolution as above. As an additional control for the presence of POLRMT with the arrested transcripts after the SA-PMP washing step, reactions identical to those containing the biotinylated 118-bp substrate, but without [α-32P] UTP, were performed and followed by SA-PMP isolation and resolution of the template-associated proteins by 4–20% SDS-PAGE in Tris–HEPES (Pierce, Thermo Scientific, Rockford, IL, USA). Samples of the purified mitochondrial proteins and a molecular weight marker were resolved alongside the transcription reaction samples, then proteins were detected in the gel by silver staining.
RESULTS
M1dG and PdG adducts block POLRMT transcription from LSP
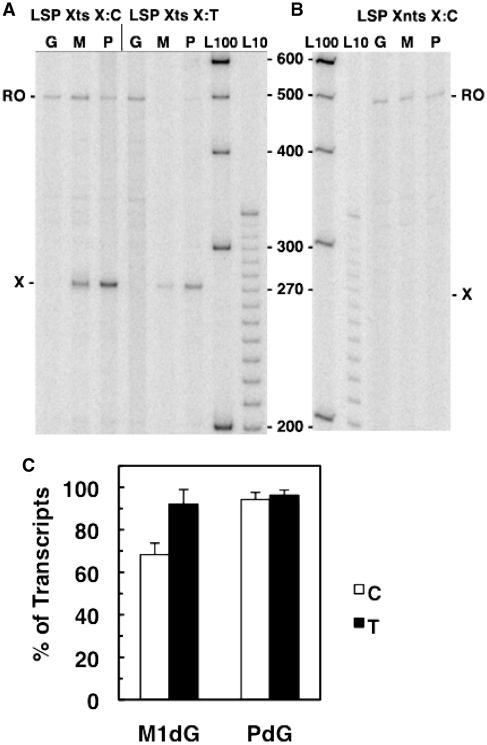
We have employed an experimental design used previously for assessing DNA damage arrest by T7RNAP and RNAP II to analyze the behavior of POLRMT at a site-specific M1dG adduct in the identical sequence context following initiation at either LSP or HSP1 (30,44). Beginning with an analysis of transcripts produced by POLRMT following LSP initiation, we have compared transcript arrest by the adduct and its PdG analog in each template as well as the extent of arrest after initiation at the two promoters. Transcripts from the LSP Xts and Xnts DNA transcription templates were 32P labeled by radionucleotide incorporation along an 18-base sequence cassette downstream of the transcription start site for POLRMT (Figure 2). POLRMT was stalled at the end of the labeling cassette on each DNA molecule by exclusion of CTP. Following heparin addition, all four ribonucleotides were provided for transcript elongation, which minimized further incorporation of 32P due to the presence of the excess cold NTP (see Supplementary Figure S1). By this method, the transcripts produced during POLRMT elongation were the result of a single promoter-initiated, transcription event on each active DNA template molecule. Full-length, run-off transcripts (RO) and transcripts resulting from arrest at the adduct position (X) were identified by comparison with 32P-labeled, single-stranded DNA markers (Figure 3). The percentage of POLRMT transcriptional arrest by the adducts was determined by dividing the intensity of the signal corresponding to adduct arrested transcripts by the sum of the intensities of the arrested and run-off transcripts in each sample lane on the gels.
Figure 3.
POLRMT arrest at M1dG and PdG in LSP templates. The 32P-labeled transcripts from 30 min of elongation by POLRMT on LSP Xts (A) and LSP Xnts (B) DNA templates containing dG (G), M1dG (M) or PdG (P) paired with C (LSP Xts X:C and LSP Xtns X:C) or T (LSP Xts X:T) are shown. Adduct-arrested transcripts are indicated with X and run-off transcripts with RO. Numbers in the center indicate DNA marker lengths in lanes L100 and L10. (C) A graph presenting the averages of the percent of transcripts arrested at M1dG and PdG opposite C (white bars) or T (black bars) in at least four independent experiments.
M1dG present in the transcribed strand of the LSP template arrested ∼70% of transcripts when paired with C and >90% when paired with T (Figure 3A and C). The PdG analog of M1dG blocked around 95% of POLRMT elongation complexes regardless of base pairing, and neither adduct in the nontranscribed strand affected POLRMT transcription (Figure 3B). These results were consistent with the findings from T7RNAP and RNAP II experiments showing that the exocyclic M1dG is a stronger block to elongation than the acyclic N2-OPdG; however, POLRMT arrest by the adducts was more substantial than that observed with the other polymerases (30). The mitochondrial polymerase was more sensitive to M1dG than the nuclear RNAP II (90% versus 70% arrest, respectively), and POLRMT was impeded to a much greater extent than the structurally similar T7 enzyme by both N2-OPdG and M1dG, suggesting that unique amino acid sequences in POLRMT or the association of transcription factors with POLRMT may impart functional differences to the POLRMT elongation complex (1). As POLRMT is capable of transcript elongation past the M1dG adduct, particularly its acyclic form N2-OPdG, a timecourse was performed to ascertain how readily this bypass occurs.
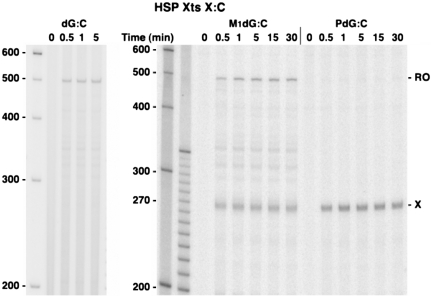
POLRMT is capable of efficient M1dG bypass
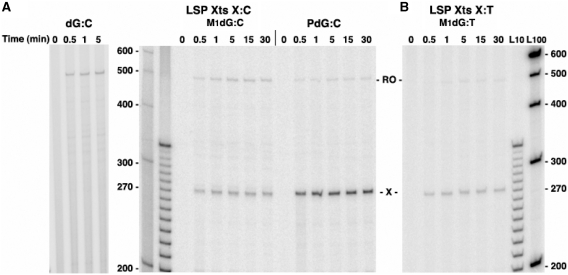
To determine the efficiency of POLRMT elongation past M1dG and the relationship between POLRMT bypass and M1dG structure, transcripts generated from the LSP Xts templates were sampled at times up to 30 min following the restart of POLRMT elongation from the end of the labeling cassette (Figure 4). On templates containing M1dG opposite C, the appearance of POLRMT run-off transcripts coincided with transcripts from arrest at the adduct, indicating that N2-OPdG caused a transient block to POLRMT translocation (Figure 4A). On all templates, the total amount of transcription (arrested plus run-off transcripts) reached a maximum within 1 min, and the proportion of run-off transcripts increased by only 5% over the duration of the time course. Because total transcription did not increase over time, all POLRMT complexes stalled at the labeling cassette apparently restarted transcription simultaneously upon the addition of NTPs, and the majority of POLRMT complexes arrested at N2-OPdG may be unstable. This was supposed as the proportion of run-off transcripts did not significantly increase after the first minute.
Figure 4.
POLRMT transcriptional bypass of M1dG or PdG following initiation at LSP. POLRMT damage-arrested (X) and full-length (RO) transcripts from LSP Xts templates containing dG, M1dG, and PdG paired with C [dG:C, M1dG:C, and PdG:C, (A)] or with T [M1dG:T, (B)] are shown at times following the NTP stimulation of POLRMT elongation. Numbers at the side of the gels indicate DNA marker lengths in the adjacent lanes. The gels shown are one of at least three independent experiments for each template.
Similar elongation kinetics were observed for bypass of M1dG:T (Figure 4B). As predicted from the 30-min elongation reactions above, the exocyclic M1dG was a stronger block than N2-OPdG to transcription in the initial seconds and arrest at the adduct was sustained over time. Regardless of base pairing, PdG immediately halted nearly all POLRMT transcription (Figure 4A, PdG:C; PdG:T not shown), indicating that the chemical dynamics of the exocyclic ring has a significant influence on POLRMT templating and translocation. It is possible that POLRMT complexes that correctly insert cytosine opposite N2-OPdG or M1dG readily extend their transcripts, while complexes that are incapable of insertion or that incorporate another base quickly become unstable and abort transcription. Indeed, if the exocyclic ring is incapable of opening, as with the PdG adduct, base insertion by POLRMT is apparently prevented.
PdG-arrested POLRMT transcription complexes are stable
The arrested transcripts detected in the experiments above may be released during detergent denaturation of stable POLRMT complexes stalled at the adducts or may result from the dissociation of unstable transcription complexes soon after POLRMT encounters an adduct. As PdG induced the strongest POLRMT transcription arrest, biotinylated, oligonucleotide templates containing the LSP and PdG adduct were utilized for in vitro analysis of arrested POLRMT complex stability (Figure 5). Transcripts corresponding to POLRMT arrest at the PdG adduct were observed up to 15 min after heparin addition and appear to reflect insertion up to and possibly opposite the adduct (Figure 5C). The arrested transcripts were absent in reactions containing a biotinylated, undamaged 118-bp template and isolation of the transcripts depended upon biotin linkage of the damaged template to the SA-PMPs (data not shown). The DNA marker (at 40 nt) served only to approximate the migration of the arrested transcripts, expected to be 35 or 36 nt, because mobility discrepancies between short DNA and RNA are known to occur. The transcripts produced by POLRMT complexes were formed prior to heparin addition and were released only when denatured with formamide after washes with copious amounts of buffer, indicating that they are a part of stable transcription complexes on the templates. Further evidence for arrested elongation complex stability was revealed when POLRMT was found in association with the PdG-containing biotinylated template following SA-PMP isolation from 1-min transcription reactions (data not shown).
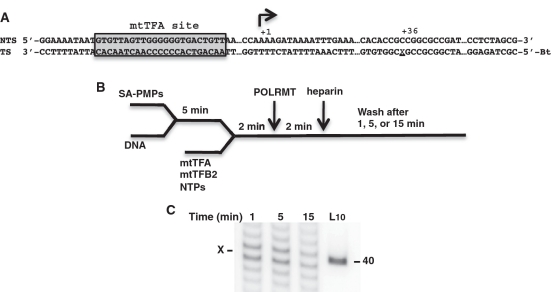
Figure 5.
POLRMT complex stability when arrested by PdG. (A) A schematic of the 118-bp POLRMT template containing LSP and a dG or PdG (position X) 36 bases downstream of LSP is shown. The mtTFA binding site is boxed and shaded, and the start site is indicated by the arrow at +1. (B) The schematic for the reactions to assesses POLRMT complex stability. (C) Transcripts isolated from stable POLRMT complexes arrested at PdG (X) are shown at times following heparin addition. A 40-nt DNA marker is shown in lane L10. The gel shown is one of three independent experiments.
While the time-course studies were conducted with an equimolar excess of NTPs to favor adduct bypass, the reactions to assess stability contained NTPs at a molar ratio of 6 ATP:1.5 GTP:1 UTP:0.6 CTP, which was based on a report of mitochondrial NTP levels in HeLa cells and intended to emulate biological conditions (52). The shorter oligonucleotide templates in this analysis also allowed for the detection of RNA products that may have resulted from templating by the adduct (Figure 5C, X) and suggest that further studies of insertion kinetics by POLRMT at M1dG and PdG are warranted.
POLRMT elongation complexes initiated from HSP1 are less sensitive to M1dG and PdG
POLRMT molecules initiating transcription at LSP and HSP1 in mtDNA are responsive to different signals in the mitochondrial chromosome controlling the length of RNA transcripts (31,36–39). LSP directs transcription of both a polycistronic transcript, nearly the length of the chromosome, and a short RNA primer for mtDNA H strand replication. HSP1 initiates POLRMT transcription to produce an rRNA transcript that terminates at a sequence within the tRNALeu gene (38,39). Previous studies have characterized POLRMT activity at both promoters in the presence of mtTFA and mtTFB2 (33,44). Here, we have compared the response of POLRMT to M1dG and PdG following POLRMT initiation at HSP1 and LSP in our in vitro templates (Figure 2).
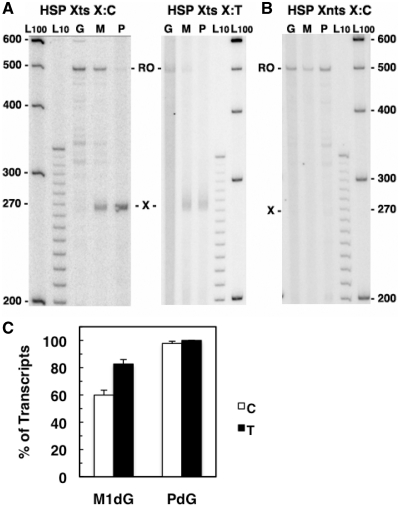
An analysis of the transcripts from POLRMT elongation on the HSP Xts and Xnts templates are shown in Figure 6. As expected, POLRMT translocation was impeded to a greater extent by M1dG, which arrested nearly 80% of HSP1-initiated transcripts, while the acyclic N2-OPdG arrested ∼60% of POLRMT transcripts (lanes M, panel A and panel C). PdG blocked nearly all transcript elongation (lanes P, panel A) regardless of base pairing, and no arrested transcripts were observed when the adducts were in the nontranscribed strand (panel B). Surprisingly, the amount of transcripts arrested by N2-OPdG and M1dG was 10% lower from HSP templates than from LSP templates (Table 1), and POLRMT interaction with the adducts when opposite T resulted in transcripts that appear to arrest immediately and to continue several bases past the adducts. It is unclear whether POLRMT response to the T-paired adducts relates to properties imparted to the polymerase by HSP1 at initiation or to long-range DNA structural effects on HSP1 stemming from a distortion at the mispaired adducts that alter initiation site selection by POLRMT. Two transcripts of ∼315 and 340 bases in length were noted in the lanes for samples with the undamaged and M1dG templates (Figure 6A, lanes G and M). These products likely resulted from stalling of POLRMT at pause sites in the sequence downstream of the adduct position and were not as apparent for POLRMT transcription of the identical sequences in the LSP templates (Figure 3). The distinctive POLRMT pausing after HSP1 and the differences in response to T-paired adducts in the HSP and LSP templates may relate to recently reported differences in POLRMT initiation that could affect POLRMT interaction with the template strand during elongation (44). Taken together, these findings indicate that POLRMT complexes on both strands of the mitochondrial chromosome will respond to M1dG in a structure-dependent manner but that POLRMT transcribing the rRNA genes from HSP1 on the G-rich H strand may more readily bypass both forms of the endogenous adduct.
Figure 6.
POLRMT arrest at M1dG and PdG in HSP templates. The 32P-labeled transcripts from (A) HSP Xts templates (HSP Xts X:C and HSP Xts X:T) or (B) HSP Xnts X:C templates containing dG (G), M1dG (M) or PdG (P). Adduct-arrested (X) and run-off (RO) transcripts are indicated. Numbers at the sides indicate DNA marker lengths in lanes L100 and L10. (C) A graph presenting the averages of the percent of transcripts arrested at M1dG and PdG opposite C (white bars) or T (black bars) for at least four independent experiments.
Table 1.
POLRMT, RNAP II and T7RNAP transcriptional arrest induced by M1dG and PdG (30)
| X:(C/T)a | POLRMTb (%) | RNAP II (%) | T7RNAP (%) |
|---|---|---|---|
| N2-OPdG | LSP 68 ± 5.4 | 60 | 27 |
| (M1dG:C) | HSP1 59.9 ± 3.6 | ||
| M1dG | LSP 92.1 ± 6.8 | 70 | 36 |
| (M1dG:T) | HSP1 82.7 ± 3.3 | ||
| PdG | LSP & HSP1 ≥95 | 100 | 68 |
aX = M1dG shown opposite either C or T; X = PdG results were similar regardless of base pairing.
bPOLRMT transcriptional arrest according to the initiating promoter, LSP or HSP1.
POLRMT elongation complexes initiated from HSP1 also efficiently bypass N2-OPdG
To assess whether the kinetics for adduct bypass are promoter-dependent, POLRMT transcripts initiated at HSP1 were isolated at times up to 30 min following NTP stimulation of elongation (Figure 7). As observed with the LSP templates, all POLRMT molecules appeared to resume transcription concurrently upon NTP addition and to complete elongation within the initial minute. While PdG immediately blocked >95% of POLRMT transcripts, the acyclic N2-OPdG initially arrested around 60% of the transcripts, with run-off transcripts increasing slightly over the first 5 min. While POLRMT was less sensitive to N2-OPdG following HSP1 initiation, the rate at which bypass occurred did not differ significantly from that observed with LSP template. This indicates that neither the stability of POLRMT complexes arrested at these adducts nor the kinetics for POLRMT bypass of N2-OPdG is dependent on the promoter directing transcription. The findings from this in vitro system indicate that M1dG in mtDNA may cause stable arrest of POLRMT complexes thereby interfering with elongation from subsequent initiation events, and in essence, inactivating a polycistronic region of the mitochondrial chromosome.
Figure 7.
POLRMT transcriptional bypass of M1dG or PdG following initiation at HSP1. POLRMT damage-arrested (X) and full-length (RO) transcripts from HSP Xts templates containing dG, M1dG and PdG paired with C (dG:C, M1dG:C or PdG:C) are shown at times following NTP stimulation of POLRMT elongation. Numbers at the left of each gel indicate DNA marker lengths in the adjacent lanes. The gels shown are one of three independent experiments.
DISCUSSION
In the nucleus, RNAP II serves as a sensitive DNA damage recognition factor for NER to increase the efficiency of repair in the transcribed relative to the nontranscribed strand of active genes (53). M1dG is a target of NER and inhibits transcription by mammalian RNAP II and the bacteriophage T7RNAP (16,30). RNAP II arrest by M1dG may facilitate NER recognition and removal the adduct from nuclear DNA, but mitochondria have no known mechanism comparable to NER (16–18). Indeed, our current knowledge of mtDNA repair suggests that the arrest of POLRMT would have only deleterious consequences for mitochondrial function and cellular health. Since M1dG occurs in mitochondria at a 2-fold higher base frequency than in nuclear DNA, the mitochondrial chromosomes are highly susceptible to genetic instability due to the presence of this endogenous form of DNA damage (11).
Our study has provided insight into the ramifications of persistent M1dG for mtDNA gene expression through the use of a POLRMT in vitro transcription system including templates with site-specific M1dG and PdG adducts and the mitochondrial transcription factors, mtTFA and mtTFB2. The DNA template sequence context of the adducts was identical to that in previous work characterizing the effects of M1dG and PdG on RNAP II and T7RNAP transcript elongation; therefore, the behavior of the three polymerases at the adducts could be compared for evident differences in their biochemical function (30).
As observed with T7RNAP and RNAP II, POLRMT arrest by M1dG was dependent upon the structure of the adduct such that the polymerase more readily bypassed the ring-open N2-OPdG (formed when M1dG is opposite C) than the exocyclic M1dG (when M1dG is opposite T) (Table 1). PdG, the closed-ring analog of M1dG, halted nearly all POLRMT elongation, suggesting that chemical dynamics of M1dG may allow the active site of POLRMT to accommodate templating and insertion of a ribonucleotide for bypass of the endogenous adduct (54). Nuclear magnetic resonance (NMR) studies have shown that the oxopropenyl group of N2-OPdG lies within the DNA minor groove and causes little distortion at the base pair (19), while PdG, the planar analog of the M1dG, participates in Hoogsteen pairing and rotates to a syn conformation at the glycosylic bond placing the exocyclic ring in the major groove (55). M1dG is predicted to cause base pair distortions similar to those observed in DNA containing PdG; however, the chemistry of the M1dG ring appears to determine the behavior of RNA polymerases attempting to ‘read’ the adduct during transcription. The observed differences in the arrest of T7RNAP, RNAP II and POLRMT by M1dG within an identical sequence context suggest that the polymerases have unique active site interactions with the transcribed strand and with the adduct itself during elongation.
The initiation and elongation mechanisms of T7RNAP and RNAP II have been characterized through biochemical and structural studies, but information about POLRMT is limited. Based on its sequence homology to T7RNAP, POLRMT has been presumed to operate much like the single-subunit bacteriophage enzyme (1,31). However, our results suggest that POLRMT elongation complexes have properties that cannot be predicted from structural alignments with the T7 enzyme. POLRMT arrest at N2-OPdG and M1dG was 2-fold greater than that observed for T7RNAP, indicating that nonhomologous active site or amino terminal sequence in POLRMT may affect function (Table 1). The amino terminal domain, which is not found in T7RNAP, may also help establish promoter-specific conformations of the POLRMT transcription complex through associations with transcription factors and the promoter. POLRMT displayed greater sensitivity to M1dG and N2-OPdG after initiation at LSP than at HSP1 (promoter differences for N2-OPdG, P = 0.002 and for M1dG, P = 0.018, in a two-tailed, unpaired t-test), and transcripts from possible pause sites were more apparent from transcription of the HSP templates (Figure 6). This finding indicates that promoter sequence may play a significant role in determining POLRMT interaction with the template during transcript elongation.
While the extent of POLRMT arrest at M1dG and N2-OPdG was promoter dependent, the ability of the enzyme to transcribe past the adducts was not. On both LSP and HSP1 templates, POLRMT transcription reached maximal levels (assessed as the sum of all transcripts) within the first minute of elongation (Figures 4 and 7). The majority of N2-OPdG and M1dG bypass occurred during the first minute, with only a ≤5% increase in the proportion of run-off transcripts thereafter. This indicates that POLRMT base insertion opposite both ring structures of the adduct occurs readily in the first seconds of elongation, but POLRMT extension of transcripts may depend on the base inserted by enzyme. If arrested POLRMT complexes remained stable over the time course, a gradual increase in the run-off transcripts would have been expected due to delayed nucleotide insertion opposite the adduct and continued elongation by POLRMT. However, the arrested transcripts did not appear to lengthen past the adducts after the initial 5 min, suggesting that POLRMT dissociated from the template and released the truncated RNAs. These findings indicate that N2-OPdG and M1dG stall POLRMT and induce elongation complex instability if nucleotide insertion does not occur within a short time frame after POLRMT encounters the damage. Further studies are necessary to determine properties of POLRMT base insertion at M1dG and whether the resulting transcripts are extended with different efficiencies by the enzyme.
Under conditions that approximate the NTP ratios present in mitochondria (52), POLRMT elongation complexes arrested at PdG remain stable for several minutes in vitro and possibly contain transcripts that include a nucleotide insertion opposite the adduct (Figure 5C). A detailed investigation of POLRMT insertion kinetics opposite M1dG will be necessary to assess the potential impact of the transcriptional arrest on mtDNA gene expression. Mitochondria may contain factors resembling nuclear elongation factor SII that could intervene to rescue stable complexes; however, such a factor or proteins constituting a repair mechanism to remove the guanine adducts have yet to be identified. Whether POLRMT complexes continue past M1dG or become unstable after damage arrest, essential transcripts will not be produced in a timely fashion resulting in a deficiency of mitochondrial RNAs and proteins, an imbalance in the transcription of the mtDNA strands, and perhaps a failure of mtDNA replication and mitochondrial biogenesis.
Only a few human mitochondrial proteins involved in POLRMT transcript initiation and elongation have been identified (31,56). A recent study identified several proteins associating with the yeast polymerase, Rpo41p, that may have human orthologs (57). The human POLRMT contains a unique pentatricopeptide repeat sequence in its amino terminal domain that could mediate specific interactions with the RNA transcript or with proteins that couple transcription and translation (56,58). Yet to be discovered factors in the organelle also could enhance POLRMT elongation complex stability, enable transcriptional bypass of DNA damage or facilitate mtDNA damage repair. While T7RNAP lacks associated proteins, RNAP II carries several factors during elongation and recruits additional proteins when arrested at pause sites or DNA damage (59,60). The previous study of M1dG arrest of RNAP II revealed that RNAP II complexes remain stable at PdG and are responsive to the elongation factor SII (59,61). While SII stimulates an RNAP II nuclease activity, an activity not associated with POLRMT, it is possible that a mitochondrial factor may associate with arrested POLRMT elongation complexes to prevent dissociation or to promote backward translocation of POLRMT from a damaged base to permit mtDNA repair.
As no NER-like mechanism for M1dG repair has been identified in mitochondria, the findings presented here suggest that this endogenous form of DNA damage may significantly impair mitochondrial gene expression by arrest of POLRMT or by induction of transcriptional mutagenesis. Studies are underway to characterize M1dG effects on POLRMT nucleotide insertion kinetics and fidelity to better understand the biological impact of our findings. Certainly, this work has led us to consider whether the inability of mitochondria to repair M1dG and M1dG effects on mtDNA transcription contribute to the decline in cellular energy production observed in aging and diseased tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM087681 to S.D.C. and CA087819 to L.J.M); Mercer University Seed Grant (to S.D.C.); Berg Endowment of the Eberly College of Science (to C.E.C.). Funding for open access charge: National Institutes of Health and National Institute of General Medical Sciences (grant GM087681).
Conflict of interest statement. Co-authors C.E.C. and J.J.A. receive royalties in connection with the distribution of human mitochondrial transcription proteins by Enzymax, Inc.
Supplementary Material
REFERENCES
- 1.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 3.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 4.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- 5.Plastaras JP, Riggins JN, Otteneder M, Marnett LJ. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: base propenals, malondialdehyde, and N(epsilon)-oxopropenyllysine. Chem. Res. Toxicol. 2000;13:1235–1242. doi: 10.1021/tx0001631. [DOI] [PubMed] [Google Scholar]
- 6.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc. Natl Acad. Sci. USA. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Taghizadeh K, Dedon PC. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J. Biol. Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 8.Jeong YC, Swenberg JA. Formation of M1G-dR from endogenous and exogenous ROS-inducing chemicals. Free Radic. Biol. Med. 2005;39:1021–1029. doi: 10.1016/j.freeradbiomed.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Jeong YC, Sangaiah R, Nakamura J, Pachkowski BF, Ranasinghe A, Gold A, Ball LM, Swenberg JA. Analysis of M1G-dR in DNA by aldehyde reactive probe labeling and liquid chromatography tandem mass spectrometry. Chem. Res. Toxicol. 2005;18:51–60. doi: 10.1021/tx049853l. [DOI] [PubMed] [Google Scholar]
- 10.Kadlubar FF, Anderson KE, Haussermann S, Lang NP, Barone GW, Thompson PA, MacLeod SL, Chou MW, Mikhailova M, Plastaras J, et al. Comparison of DNA adduct levels associated with oxidative stress in human pancreas. Mutat. Res. 1998;405:125–133. doi: 10.1016/s0027-5107(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 11.Jeong YC, Nakamura J, Upton PB, Swenberg JA. Pyrimido[1,2-a]-purin-10(3H)-one, M1G, is less prone to artifact than base oxidation. Nucleic Acids Res. 2005;33:6426–6434. doi: 10.1093/nar/gki944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Analysis of 7,8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 14.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson KA, Fink SP, Marnett LJ. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J. Biol. Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 17.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascucci B, Versteegh A, van Hoffen A, van Zeeland AA, Mullenders LH, Dogliotti E. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J. Mol. Biol. 1997;273:417–427. doi: 10.1006/jmbi.1997.1268. [DOI] [PubMed] [Google Scholar]
- 19.Mao H, Schnetz-Boutaud NC, Weisenseel JP, Marnett LJ, Stone MP. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl Acad. Sci. USA. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szekely J, Rizzo CJ, Marnett LJ. Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J. Am. Chem. Soc. 2008;130:2195–2201. doi: 10.1021/ja074506u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benamira M, Johnson K, Chaudhary A, Bruner K, Tibbetts C, Marnett LJ. Induction of mutations by replication of malondialdehyde-modified M13 DNA in Escherichia coli: determination of the extent of DNA modification, genetic requirements for mutagenesis, and types of mutations induced. Carcinogenesis. 1995;16:93–99. doi: 10.1093/carcin/16.1.93. [DOI] [PubMed] [Google Scholar]
- 22.Riggins JN, Marnett LJ. Mutagenicity of the malondialdehyde oligomerization products 2-(3′-oxo-1′-propenyl)-malondialdehyde and 2,4-dihydroxymethylene-3-(2,2-dimethoxyethyl)glutaraldehyde in Salmonella. Mutat. Res. 2001;497:153–157. doi: 10.1016/s1383-5718(01)00253-4. [DOI] [PubMed] [Google Scholar]
- 23.VanderVeen LA, Hashim MF, Shyr Y, Marnett LJ. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl Acad. Sci. USA. 2003;100:14247–14252. doi: 10.1073/pnas.2332176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J. Biol. Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 25.Hashim MF, Riggins JN, Schnetz-Boutaud N, Voehler M, Stone MP, Marnett LJ. In vitro bypass of malondialdehyde-deoxyguanosine adducts: differential base selection during extension by the Klenow fragment of DNA polymerase I is the critical determinant of replication outcome. Biochemistry. 2004;43:11828–11835. doi: 10.1021/bi049360f. [DOI] [PubMed] [Google Scholar]
- 26.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 27.Hashim MF, Schnetz-Boutaud N, Marnett LJ. Replication of template-primers containing propanodeoxyguanosine by DNA polymerase beta. Induction of base pair substitution and frameshift mutations by template slippage and deoxynucleoside triphosphate stabilization. J. Biol. Chem. 1997;272:20205–20212. doi: 10.1074/jbc.272.32.20205. [DOI] [PubMed] [Google Scholar]
- 28.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc. Natl Acad. Sci. USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stafford JB, Eoff RL, Kozekova A, Rizzo CJ, Guengerich FP, Marnett LJ. Translesion DNA synthesis by human DNA polymerase eta on templates containing a pyrimidopurinone deoxyguanosine adduct, 3-(2′-deoxy-beta-d-erythro-pentofuranosyl)pyrimido-[1,2-a]purin-10(3H)-one. Biochemistry. 2009;48:471–480. doi: 10.1021/bi801591a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc. Natl Acad. Sci. USA. 2004;101:7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 34.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 35.Shuey DJ, Attardi G. Characterization of an RNA polymerase activity from HeLa cell mitochondria, which initiates transcription at the heavy strand rRNA promoter and the light strand promoter in human mitochondrial DNA. J. Biol. Chem. 1985;260:1952–1958. [PubMed] [Google Scholar]
- 36.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- 38.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 39.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 40.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4:1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 43.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J. Biol. Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 46.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Schnetz-Boutaud NC, Mao H, Stone MP, Marnett LJ. Synthesis of oligonucleotides containing the alkali-labile pyrimidopurinone adduct, M(1)G. Chem. Res. Toxicol. 2000;13:90–95. doi: 10.1021/tx990141i. [DOI] [PubMed] [Google Scholar]
- 49.Bregeon D, Doetsch PW. Reliable method for generating double-stranded DNA vectors containing site-specific base modifications. Biotechniques. 2004;37:760–762, 764, 766. doi: 10.2144/04375ST01. [DOI] [PubMed] [Google Scholar]
- 50.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 51.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bestwick RK, Moffett GL, Mathews CK. Selective expansion of mitochondrial nucleoside triphosphate pools in antimetabolite-treated HeLa cells. J. Biol. Chem. 1982;257:9300–9304. [PubMed] [Google Scholar]
- 53.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 54.Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. Kinetics and mechanism of the general-acid-catalyzed ring-closure of the malondialdehyde-DNA adduct, N2-(3-oxo-1-propenyl)deoxyguanosine (N2OPdG-), to 3-(2′-Deoxy-beta-D-erythro-pentofuranosyl)pyrimido[1,2-alpha]purin- 10(3H)-one (M1dG) J. Am. Chem. Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- 55.Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem. Res. Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–440. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reines D. RNA polymerase II elongation complex. Elongation complexes purified using an anti-RNA antibody do not contain initiation factor alpha. J. Biol. Chem. 1991;266:10510–10517. [PMC free article] [PubMed] [Google Scholar]
- 60.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reines D, Chamberlin MJ, Kane CM. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.