Abstract
Protein self-organization is essential for the establishment and maintenance of nuclear architecture and for the regulation of gene expression. We have shown previously that the Proline-Rich Homeodomain protein (PRH/Hex) self-assembles to form oligomeric complexes that bind to arrays of PRH binding sites with high affinity and specificity. We have also shown that many PRH target genes contain suitably spaced arrays of PRH sites that allow this protein to bind and regulate transcription. Here, we use analytical ultracentrifugation and electron microscopy to further characterize PRH oligomers. We use the same techniques to show that PRH oligomers bound to long DNA fragments self-associate to form highly ordered assemblies. Electron microscopy and linear dichroism reveal that PRH oligomers can form protein–DNA fibres and that PRH is able to compact DNA in the absence of other proteins. Finally, we show that DNA compaction is not sufficient for the repression of PRH target genes in cells. We conclude that DNA compaction is a consequence of the binding of large PRH oligomers to arrays of binding sites and that PRH is functionally and structurally related to the Lrp/AsnC family of proteins from bacteria and archaea, a group of proteins formerly thought to be without eukaryotic equivalents.
INTRODUCTION
In eukaryotic cells, many chromatin binding proteins compact or loosen chromatin when recruited to DNA by the action of sequence-specific DNA binding transcription factors (1). Some oligomeric transcription factors, such as the homeodomain protein SatB1, have been shown to serve as architectural proteins that provide scaffolds for the recruitment of multiple chromatin binding proteins including co-activators and co-repressors. SatB1 in conjunction with these partner proteins allows specific DNA looping events which form a chromatin landscape that facilitates the activation or repression of specific genes (2,3). However, very few proteins of this type have been characterized in any detail and the relationship between protein oligomerization and the formation of chromatin domains is still poorly understood.
The Proline-Rich Homeodomain protein (PRH, also known as HHex) is an essential transcription factor in vertebrate embryonic development and in the adult (4,5). In the developing embryo, PRH regulates body-axis formation and the formation of multiple tissues including the liver, pancreas, heart and thyroid, the vasculature and the haematopoietic system. In the adult, PRH regulates multiple steps in haematopoiesis and controls cell growth. Mutations that result in the mis-expression or mis-localization of PRH are associated with leukaemia as well as thyroid and breast cancers (6–8). In addition, a fusion protein between nucleoporin protein Nup98 and PRH (Nup-Hex/PRH) that is thought to antagonize the activity of wild-type PRH results in myeloid leukaemia (9).
PRH can repress or activate gene expression but when bound directly to DNA, this protein generally appears to function as a repressor of transcription (10–12). We have shown that PRH recruits members of the TLE family of chromatin binding proteins in order to repress transcription and that it brings about nuclear retention and hyperphosphorylation of TLE proteins (10,13). PRH is a phosphoprotein in cells and phosphorylation by CK2 inhibits the DNA binding activity and transcriptional repression functions of this protein (14). We have used in vivo cross-linking to show that PRH forms oligomeric assemblies in cells (15). The purified recombinant PRH protein also forms oligomeric assemblies and using analytical ultracentrifugation (AUC) and gel filtration chromatography, we have demonstrated that in solution these oligomers appear to be octameric (15). The protein does not appear to form monomers or any other multimers smaller than an octamer (15). The oligomeric complexes formed by the recombinant protein bind to PRH-interacting proteins such as CK2β and the purified oligomer is capable of specific binding to the promoter regions of its target genes in vitro (14,16). Moreover, the recombinant protein can be phosphorylated by CK2 to block DNA binding and then dephosphorylated to restore DNA binding (14). These experiments show that the purified oligomeric PRH protein is functional (14,15). Further, each homeodomain in the octameric complex is capable of binding to DNA and the PRH protein binds with high affinity to arrays of multiple core homeodomain binding sites (16). As might be expected based on these results, several PRH target genes, including the Goosecoid gene (16) and the Vegfr-1 and Vegfr-2 receptor genes (17), contain clustered arrays of sites that mediate PRH binding. When bound to its binding sites in the Goosecoid promoter, PRH oligomers induce significant DNA distortion (16).
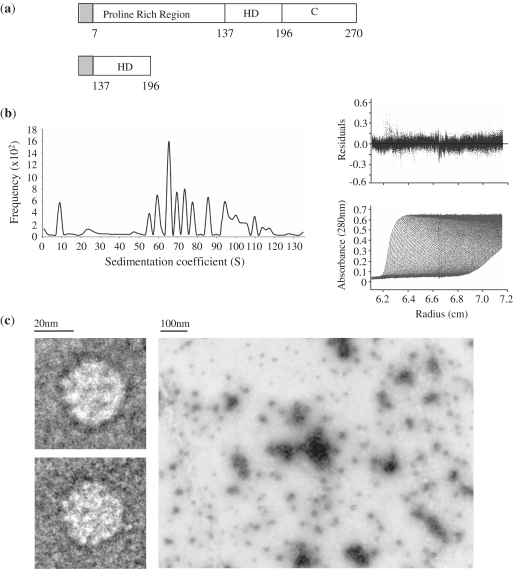
The PRH protein is 270 amino acids in length and consists of three regions: a proline-rich N-terminal transcription repression domain (residues 1–136), a central DNA binding homeodomain (137–197) and an acidic C-terminal domain (198–270) [Figure 1(a)]. The first 46 amino acids of PRH form a novel dimerization motif, while amino acids downstream of residue 46 are required for oligomerization. The region between amino acids 46 and 132 is capable of interacting with the PRH homeodomain and this interaction is probably important during oligomerization (15). Here, we show using AUC that PRH forms discrete disc-shaped octameric complexes and more spherical double octamers (hexadecamers) as well as larger multimers. We show that PRH hexadecamers bind to DNA in an ordered fashion, resulting in PRH–DNA polymers that differ by one repeat unit. We demonstrate using linear dichroism (LD) and electron microscopy (EM) that when PRH oligomers bind to their sites they significantly compact the DNA and form protein arrays or fibres on the DNA.
Figure 1.
Detailed analysis of PRH oligomers. (a) A schematic of the PRH protein and the truncated PRH HD derivative used in this study. The shaded rectangle indicates the 5.4-kDa histidine tag sequence used in protein purification. (b) The results of a sedimentation velocity experiment in which PRH (10 µM) was centrifuged at 16 000 r.p.m. The lower right panel shows a representative analysis of boundary fractions as a function of the centrifugal radius. The main panel shows representative data with best fit (continuous line) C (s) analysis in ULTRASCAN software. The upper panel shows the residuals between the data and the fit. (c) PRH was deposited on a carbon-coated copper EM grid and stained with 1% uranyl acetate before being visualized using transmission EM.
MATERIALS AND METHODS
Protein used in this study
Recombinant full-length PRH protein, the PRH F32E mutant and the truncated PRH homeodomain protein were purified as described previously (18,19). The full-length PRH protein runs as a monomer on non-reducing sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), indicating that there are no interchain disulphide bonds (data not shown). Protein concentrations were determined from the A280 nm using the molar extinction coefficients.
DNAs used in this study
The mammalian expression plasmid pMUG1-Myc-PRH expresses Myc-tagged human PRH (amino acids 7–270) and has been described previously (20). pMUG1-Myc-PRH F32E and pMUG1-Myc-PRH N187A express mutated PRH proteins that fail to bind TLE co-repressor proteins and DNA respectively (10). shRNA plasmids shRNAPRH49, shRNAPRH51 and the control shRNA plasmid were obtained from Origene. The pSV-β-galactosidase control reporter (pSV-lacZ) was obtained from Promega. The pTKmin-PRH reporter plasmid has been described previously and contains five PRH binding sites cloned upstream of the minimal thymidine kinase promoter and firefly luciferase gene in pTKmin (20). The Goosecoid reporter plasmid pGL2-GSC contains DNA sequences from −461 to +64 relative to the Goosecoid transcription start point cloned upstream of the luciferase reporter gene in pGL2-basic (Promega) and has been described previously (16). The pGL2-Gsc-PRHx5 plasmid contains five PRH binding sites cloned in the enhancer position around 2 kbp upstream of the Goosecoid promoter. This construct was made by excising five tandemly arranged PRH binding sites from pBS-prh5x (5) using XbaI and HindIII. The fragment containing PRH sites was then treated with Klenow enzyme to produce blunt ends and cloned into the unique BamHI site in pGL2-Gsc plasmid, which had also been treated with Klenow enzyme to produce blunt ends. The pGL2-HS1 reporter contains the human Surf-1/2 bidirectional promoter cloned upstream of the luciferase gene (21,22). The pGL2-HS1-PRHx5 plasmid contains five PRH binding sites cloned in the enhancer position upstream of the Surf-1/2 promoter and was produced exactly as described for pGL2-Gsc-PRHx5. Plasmid pGL2-PRHx5 contains the same five PRH binding sites cloned in the enhancer position with no promoter and was made as above. All constructs were verified by DNA sequencing. DNA concentrations were determined from their absorbance at 260 nM and confirmed by comparison to known DNA samples using agarose gel electrophoresis.
DNA for AUC was purified by CsCl gradient centrifugation. A 267-bp DNA fragment carrying a cluster of PRH binding sites from the Goosecoid promoter was prepared by digesting of 250 mg of pGL2-GSC with 900 U of BanI for 16 h at 37°C yielding around 2.7 mg of the required fragment and several larger digestion products. All of the DNA fragments were applied to a MonoQ column and eluted with a salt gradient. The 267-bp Goosecoid fragment elutes at 750 mM NaCl salt free from the other DNA fragments.
EM
Samples were placed on a carbon-coated copper EM grid and stained with 1% uranyl acetate before examination in a Jeol JEM 2011 transmission EM equipped with an LaB6 filament as an electron source. Magnification ranged between 20 000× and 40 000×.
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments were carried out using a Beckman Optima XL-A Analytical Ultracentrifuge with an An60-Ti rotor and absorbance optics. AUC was performed using two channel centrepieces with 400 µl of sample in dialysis buffer [phosphate-buffered saline (PBS), 10% (v/v) glycerol] loaded into the sample sector against 420 µl of dialysis buffer into the reference sector so that the two menisci were mismatched. Velocity experiments for His-PRH protein were carried out at three loading concentrations (2, 6 and 12 µM). The radial distribution of His-PRH protein at sedimentation was monitored by absorbance as a function of time at either A280 for high protein concentrations or A230 at low (2 µM) concentration. The distribution of sedimentation coefficients was obtained using the program ULTRASCAN employing the enhanced Van Holde–Wischet analysis. Using the calculated values for the molecular weight of His-PRH (34.6 kDa), the partial specific volume of the protein (υ = 0.7216 ml/g), and the buffer density ρ = 1.0317, and taking the relationship S = M (1 – υρ)/Nf, where N is Avagadro’s number, a molecule of this mass will have a sedimentation coefficient of 2S if its frictional ratio is f/fo of 1.3 (fo is the frictional coefficient of an anhydrous sphere with the same mass and a frictional ratio f/fo of 1.3 is typical for proteins with an ellipsoidal shape). Using these values, the 9S sedimentation coefficient PRH species must have a molecular weight of ∼280 kDa. Velocity experiments for DNA and the protein–DNA complex used double-stranded DNA at 4 µM or 4 µM DNA plus 2 µM His-PRH. The protein–DNA complex was formed after incubation of protein with DNA at 4°C for 4 h in sample dialysis buffer. At 3-min intervals, the absorbance of the free DNA or the protein–DNA complex at 260 nm was scanned across the radius of centrifugation. Modelling of protein–DNA complexes was performed using ULTRASCAN software.
LD
LD measurements were carried out in a Jasco J815 spectropolarimeter adapted for LD spectroscopy. Samples were placed in binding buffer (50 mM Tris pH 8, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT) at 20°C and aligned in the light beam using custom made Couette cells exactly as described previously (23).
Cell culture and transcription assays
K562 cells were maintained in Dulbecco’s modified Eagle's medium (DMEM) media supplemented with 10% fetal calf serum and penicillin/ streptomycin at 37°C in 5% CO2. Transiently transfections were performed by electroporation (270 V/950 µF). The cells were co-transfected with the luciferase reporter plasmids described earlier and either the PRH expression vector pMUG1-Myc-PRH or the empty pMUG1 vector. The β-galactosidase reporter plasmid pSV-lacZ was also co-transfected into the cells to act as a control for transfection efficiency. Twenty-four hours post-transfection, luciferase activity was determined using the Promega Luciferase Assay System according to the manufacturer’s instructions (Promega) and β-galactosidase assays were performed using the same lysates. In PRH shRNA knockdown experiments, 5 × 106 K652 cells were transfected by electroporation as described earlier using 10 µg shRNAGFP (control) or 5 µg shRNAPRH49 and 5 µg shRNAPRH51 plasmids in combination. The cells were then grown in the presence of 1 µg/ml puromycin for 10 days to select the transfected cells. Transcription assays were then performed by re-transfecting the cells exactly as described above. Western blotting for PRH and Lamin A/C was carried out as described previously (13).
Chromatin immunoprecipitation and quantitative reverse transcription-polymerase chain reaction
For chromatin immunoprecipitation (ChIP) experiments K562 cells (108 cells per ChIP) were transiently transfected with 5 µg pMUG1-Myc-PRH, pMUG1-Myc-PRH F32E or pMUG1-Myc-PRH N187A as described previously (17). ChIP was carried out exactly as before (16) using primers and polymerase chain reaction (PCR) conditions also described previously (17). Quantitative reverse transcription (RT)-PCR was performed exactly as described previously (17). The amount of Vegfr-1 product was determined relative to product from input chromatin and compared to gapdh as an internal reference.
RESULTS
PRH octamers are oblate spheroids that self-associate
We have shown previously that the isolated PRH homeodomain is a monomer in solution, whereas the full-length PRH protein forms large oligomers in solution and in cells (15). These oligomers are not the result of non-physiological protein aggregation as they are soluble, capable of binding to PRH-interacting proteins and capable of binding to DNA containing PRH sites (14,15,18). To better characterize PRH oligomeric species, we performed AUC SV experiments at different protein concentrations and centrifugation speeds. Oligomeric PRH comprises species with sedimentation coefficients of 9S and 25S as well as multiple species with sedimentation coefficients larger than 40S [Figure 1(b)]. According to Stokes equation, the Ssphere of a PRH octamer is 15S, which is the maximum sedimentation coefficient value for a protein with a mass of 280 kDa and translational frictional coefficient value, ƒ0, of 8.1411 × 10−8. Thus, the 25S species cannot represent a PRH octamer and as we previously reported, the smallest of these species (9S) most likely corresponds to a PRH octamer (15). Using the Svedberg equation the 9S species are calculated to have a translational frictional coefficient, ƒ, of 1.4447 × 10−7, based on the calculated partial specific volume of PRH (0.7216 ml/g) and the calculated density of water at 20°C (ρ = 0.998). The frictional ratio, ƒ/ƒ0, of proteins provides an estimate of their shape; a ratio of 1 suggests that the protein is completely spherical, whereas ratios >1 suggest that the protein may be more elongated. The frictional ratio, ƒ/ƒ0 of the 9S PRH species in water at 20°C is 1.7. This value corresponds to a highly oblate spheroid with an axial ratio of 19.7, diameters of around 100 Å and 20 Å and a volume of 3.6 × 105 Å3. In contrast, the 25S species shown in Figure 1(b) has a translational frictional coefficient, ƒ, of 1.0257 × 107 and an ƒ/ƒ0 ratio of 1 which suggests that these particles adopt a roughly spherical shape. Using the Svedberg equation, the 25S species molecular weight was determined to be 560 kDa, which correlates precisely with the calculated molecular weight of two 9S PRH octamers. These spherical particles have a calculated diameter of 108 Å and a volume of 6.7 × 105 Å3. This volume is very close to the estimated volume of two PRH octamers (∼6.5 × 105 Å3), which further supports the idea that two 9S oblate particles come together to form these 25S roughly spherical particles. The multiple species larger than 40S have ƒ/ƒ0 ratios, which average at 3.3, suggesting elongated structures. These PRH species have molecular weights in excess of 1 mDa, which suggests polymerized forms of PRH. The association of two or more double octamers would produce complexes with a molecular weights of 1.2, 1.8, 2.4 mDa and higher and these complex would have to be elongated if they associated as rigid bodies. Importantly, we could not detect any differences in the amount of each species when the sample was diluted across a 10-fold range, suggesting that there is a non-reversible association of 9S octameric particles to form the larger assemblies.
This sample of PRH protein was also characterized using negative stain and transmission EM. Under EM, PRH appears to form spherical particles of ∼150–200-Å diameter as well as much larger particles formed by the self-association of the smaller spheres [Figure 1(c)]. It is worth noting that the accumulation of negative stain itself will tend to increase the apparent size of the particles observed under EM. The size of the smallest spherical particles observed under EM (∼150 Å) thus seems to be of the same magnitude as the calculated large diameter of the 9S oblate spheroids (around 100 Å) and the 25S spheres (108 Å), suggesting that both species could be present but indistinguishable when visualized from above. However, it is also possible that the negative stain technique may favour the adherence of some forms of PRH over others on the coated EM grid. We collected several hundred images of individual oligomers and representative views, which are shown in Figure 1(c). Although the majority of single particles are round, some of the oligomers appear to be oval, suggesting that they represent 9S particles that are visualized at an angle. We conclude that the 9S, 25S and larger species characterized above using AUC are representative of the PRH species found under these conditions using EM and that the data are consistent with oblate 9S octameric spheroids coming together to form roughly spherical 25S hexadecameric species.
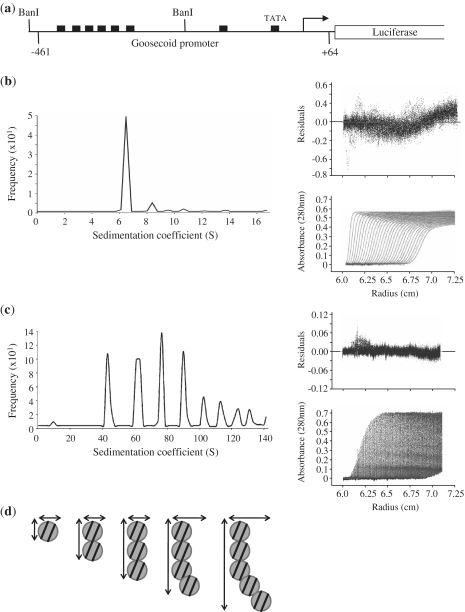
PRH–DNA complexes self-associate in solution
PRH binds to short oligonucleotides carrying a single PRH site with a stoichiometry of 7.3 DNA fragments to 1 PRH octamer, suggesting that each homeodomain within the octamer is capable of binding to DNA (16). PRH binds with high co-operativity to DNA fragments containing multiple PRH binding sites and several PRH responsive genes contain closely spaced arrays of PRH binding sites (16,17). We have previously demonstrated that PRH is able to bind to short oligonucleotides containing PRH binding sites in SV experiments (15). This technique has the potential to provide detailed information on the nature of the PRH–DNA complexes containing longer DNA fragments with multiple PRH binding sites. However, in order to perform these experiments, we require large quantities of a suitable DNA fragment carrying multiple PRH sites. To this end, we chose a 267-bp DNA fragment carrying a cluster of PRH binding sites from the Goosecoid promoter that confers transcriptional regulation by PRH in cells [Figure 2(a)]. Shorter DNA fragments might not contain a sufficient number of PRH sites, while the hydrodynamic properties of longer DNA fragments precludes their use in SV experiments. The 267-bp Goosecoid DNA fragment was obtained by restriction enzyme digestion and purified using ion exchange chromatography. PRH and PRH–DNA complexes were detected by their absorption at 260 nm using SV at centrifugation speeds chosen to best detect the sedimentation of DNA alone as well as the sedimentation of PRH–DNA complexes. Figure 2(b) shows that a single sedimentation boundary is visible for the 267-bp DNA fragment alone, which has a sedimentation coefficient of ∼6S. The frictional ratio ƒ/ƒ0 of this DNA is 3.4, suggesting as expected a highly elongated molecule. The apparent molecular weight is 177 kDa, which is in good agreement with the calculated molecular weight of 166 kDa. When PRH is bound to this DNA, a second sedimentation boundary is detected that corresponds to a PRH–DNA complex with a sedimentation coefficient of 42S. When SV experiments were repeated with slower sedimentation speeds, increased amounts of this 42S complex were observed and in addition a number of additional discrete sedimentation boundaries were also observed having sedimentation coefficients of 63S, 77S, 90S and 102S as well as some larger species [Figure 2(c)]. In order to determine the probable nature of these complexes, we extensively modelled possible protein–DNA complexes and calculated the Ssphere of each complex. The molecular weight was calculated for each potential protein–DNA complex (theoretical mass for 1 PRH octamer plus 1 DNA, 1 PRH octamer plus two DNAs, one PRH octamer plus three DNAs…two PRH octamers plus one DNA, two PRH octamers plus two DNAs, two PRH octamers plus three DNAs, etc.) and used to calculate the maximum Ssphere value for each potential protein–DNA complex. This approach allows us to rule out complexes that could not form the observed species [such as one PRH octamer plus one DNA (Ssphere 26S), two octamers plus one DNA (Ssphere 33S), etc]. The data are summarized in Table 1 for the subset of complexes that are in good agreement with the observed S values (Sobs). The observed and calculated values for a series of double octamers each bound to two DNA fragments are in good agreement assuming that the oligomers deviate somewhat from a linear organization as they grow [as shown in the model in Figure 2(d)]. We conclude that in the presence of DNA, PRH oligomers organize themselves into discrete nucleoprotein complexes that differ in sedimentation coefficient by a discrete repeat unit consistent with a double octamer bound to two 267-bp DNA fragments. Although we could not fit any other models to the data, this analysis cannot exclude the possibility that the complexes do not associate in this manner. It is also not possible to determine whether the regular sedimentation profile for the PRH–DNA complexes is caused by the disruption of pre-existing PRH assemblies that then bind DNA or whether pre-existing PRH assemblies simply bind DNA to form larger complexes.
Figure 2.
The formation of large PRH–DNA complexes. (a) A schematic of the Goosecoid reporter construct used in this study. Goosecoid promoter sequences from –461 to +64 relative to the transcription start point (bent arrow) are located upstream of the luciferase gene. The filled rectangles represent core PRH binding sites and the Goosecoid TATA box sequence. The AUC experiments described below were performed with a 267-bp DNA fragment (96 nM) produced by digestion with BanI. (b) Sedimentation velocity experiments were performed with the BanI fragment shown in (a). Sedimentation was monitored as a function of centrifugal radius using the absorbance at 260 nm (DNA). A single boundary is observed corresponding to the free DNA (bottom right panel) with a sedimentation coefficient of around 7 S (main panel). (c) In the presence of PRH several boundaries are observed corresponding to free and bound DNA (bottom right panel) with sedimentation coefficients of 41 S, 65 S, 86 S, 104 S and 120 S. (d) A model for the formation of PRH–DNA complexes based on the data shown in Table 1. Circles represent PRH double octamers. The lines represent 267-bp DNA fragments.
Table 1.
A summary of the analysis of potential PRH–DNA complexes
| Model | MW (kDa) | V bar | Ssphere | Sobs | f/f0 | Axial ratio |
|---|---|---|---|---|---|---|
| Two PRH octamers + two DNAs | 883.6 | 0.65938 | 41 | 41.2 | 1 | 1 |
| Four PRH octamers + four DNAs | 1767.2 | 0.65938 | 63 | 65.3 | 1.04 | 2 |
| Six PRH octamers + six DNAs | 2650.8 | 0.65938 | 77 | 85.6 | 1.12 | 3 |
| Eight PRH octamers + eight DNAs | 3534.4 | 0.65938 | 90 | 103.7 | 1.16 | 3.5 |
| Ten PRH octamers + ten DNAs | 4418 | 0.65938 | 102 | 120.4 | 1.18 | 4 |
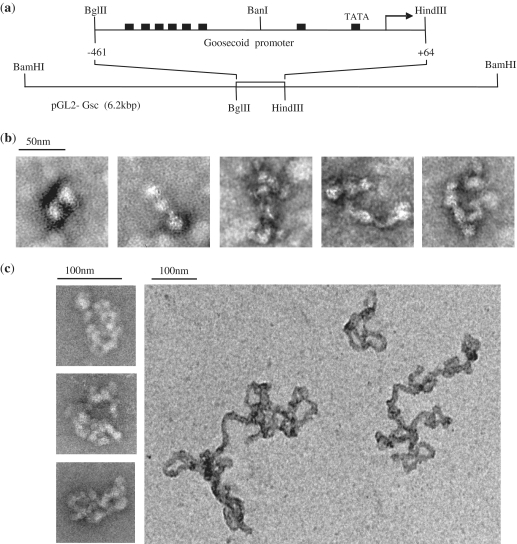
PRH can form protein–DNA fibres
To gain a better understanding of the nature of the higher-order PRH–DNA complexes described earlier, we incubated PRH oligomers with a 525-bp fragment of DNA encompassing the same array of Goosecoid PRH binding sites used in the previous experiments [Figure 3(a)] and viewed negatively stained samples using EM [Figure 3(b)]. In the presence of this DNA fragment, bead-like objects are visible and they associate in a side-by-side manner [Figure 3(b)]. Naked DNA provides very low contrast in transmission EM. Hence, as would be expected, in the absence of PRH protein uranyl acetate stained DNA alone is barely visible. At low numbers of ‘beads’, these assemblies are very similar to the model shown in Figure 2(d). As the number of ‘beads’ increases, complex assemblies are formed that appear mesh like. To determine whether long fragments of DNA are required to form these mesh-like assemblies, the experiment was repeated at the same protein:DNA ratio with small 30-bp oligonucleotides that each contain a single PRH binding site. In the presence of this oligonucleotides, there is very little alteration in the appearance of the protein under negative stain EM (data not shown). This suggests that long DNA fragments carrying arrayed PRH binding sites are required in order to form these assemblies.
Figure 3.
Electron microscopy of PRH with short and long DNA fragments. (a) A schematic of the DNA fragments used in the EM experiments shown here. The top line shows a 525-bp DNA fragment carrying Goosecoid sequences from –461 to +64 relative to the transcription start point obtained by digestion of pGL-Gsc with BglII and HindIII. The bottom line shows the same plasmid linearized by digestion with BamHI to produce a 6.2 kbp DNA fragment with centrally located Goosecoid promoter sequences. (b) The 525-bp DNA fragment shown in (a) was incubated with PRH (1 µM) in binding buffer for 20 min at 4°C. The complexes formed were deposited on a carbon-coated copper EM grid and stained with 1% uranyl acetate for transmission EM. (c) The experiment in (b) was repeated with the 6.2 kbp DNA fragment shown in (a).
To examine the binding of PRH to much longer DNA fragments, the above experiment was repeated using either a linearized 6 kbp plasmid DNA that contains the Goosecoid promoter or the equivalent empty vector [Figure 3(a), line 2]. In the presence of the empty vector, we were unable to visualise either DNA or PRH–DNA complexes using negative stain TEM (data not shown). In contrast, in the presence of the plasmid DNA containing Goosecoid DNA sequences, highly compact PRH–DNA structures are observed [Figure 3(c)]. These complexes form fibres with diameters of around 200 Å that can appear to be highly condensed [Figure 3(c), left panels] or more loosely packaged [Figure 3(c), right panel]. These fibres are very different from the spherical particles formed by PRH alone, although the diameter of the fibre is roughly similar to the diameter of the spheres observed using EM.
DNA compaction by PRH in solution
In order to follow the formation of the very large PRH–DNA assemblies observed above in solution we made use of ultraviolet (UV) flow-oriented LD (24). This technique can report on the compaction and flexibility of DNA or other macromolecules such as protein fibres, which can be uniformly oriented. Molecules are oriented in solution in one direction by rotation in a Couette. The absorbance of linearly polarized light parallel and perpendicular to an orientation direction, by directionally oriented molecules, is measured and the difference in the absorbances results in an LD signal. In the case of DNA, the difference in absorbance of linearly polarized light (at 260 nm), in parallel and perpendicular directions, by electrons of the DNA bases is the negative minimum. This indicates that the bases lie in a more perpendicular than parallel orientation to the DNA helix axis. Proteins that bind to DNA alter the LD signal in a specific way, resulting in an increased or a decreased negative LD signal at 260 nm together with LD signals arising from the protein itself. Protein fibres containing aromatic residues such as tyrosine, tryptophan and phenylalanine can give LD signals at 280 nm, but the strongest of these signals emanate from absorbance by the peptide backbone chromophore and are in the far UV (190–220 nm).
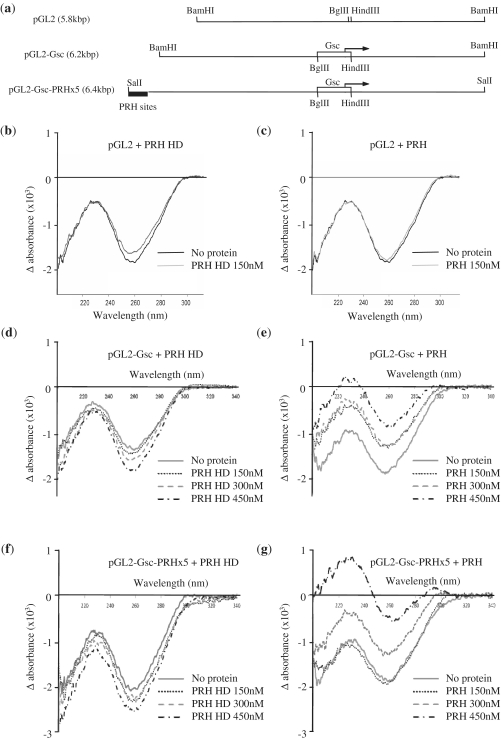
We used LD to examine the binding of PRH and the isolated PRH homeodomain to the linearized 6 kbp plasmid DNA that contains the Goosecoid promoter and the linearized empty vector [Figure 4(a), lines 1 and 2]. LD signals were recorded for DNA alone or DNA in the presence of PRH proteins. Figure 4(b) shows that the 260-nm LD signal for the plasmid lacking Goosecoid sequences becomes less negative upon incubation with the isolated PRH homeodomain (PRH–HD). This LD signal change indicates that the DNA bases are less perpendicular to the helical axis in the presence of PRH–HD and therefore that PRH–HD has increased DNA flexibility. This suggests that PRH–HD binds to ATTA-like sequences in the plasmid and/or possesses high non-specific DNA-binding activity. In contrast, the negative LD signal for this empty vector plasmid DNA does not alter in the presence of full-length PRH [Figure 4(c)]. These data suggest that, as might be expected, the full-length oligomeric PRH protein has increased DNA-binding specificity compared to the isolated homeodomain.
Figure 4.
Linear dichroism indicates DNA compaction by PRH in solution. (a) A schematic of the DNA fragments used in LD experiments. The top line shows pGL2-basic linearized by digestion with BamHI. The middle line shows pGL2-Gsc linearized using the same restriction enzyme. The open rectangle and arrow represent the Goosecoid promoter sequences and Goosecoid transcription start point, respectively. The bottom line shows the pGL2-Gsc-PRHx5 plasmid linearized with SalI. The filled rectangle represents the five PRH binding sites cloned into the BamHI site of pGL2-Gsc to produce this construct. (b–g) LD of the plasmid DNAs shown above (5 nM) with PRH-HD (b, d and f) or the full-length PRH protein (c, e and g).
The negative LD signal for the plasmid carrying Goosecoid DNA sequences becomes more intensely negative in the presence of PRH–HD [Figure 4(d)] in a protein concentration-dependent manner. An increased LD signal indicates that the DNA is aligning to a higher degree. This is the result of the DNA becoming more rigid. This suggests that PRH–HD alters the flexibility/conformation of DNA when bound to its specific binding sites and brings about a more rigid DNA helix. The negative LD signal for this DNA is significantly reduced in intensity by full-length PRH [Figure 4(e)]. This signal is further decreased when more PRH protein is present, that is, at higher PRH:DNA ratios. Given that there is no effect of PRH on the DNA lacking Goosecoid sequences, we interpret the decreased negative LD signal in the presence of PRH sites as resulting from significant bending or compaction of the DNA by PRH in a manner opposite to that produced on specific binding of PRH–HD. It is also observed that the maximum at 230 nm increases until it becomes positive. This is likely to be the result of signals from the peptide backbone of the protein becoming visible as they become aligned as when bound to the aligned DNA. In conclusion, full-length PRH appears to compact/bend DNA when its binding sites are present and shows little non-specific binding to DNA. In contrast, PRH–HD brings about some DNA bending or other conformational change when non-specifically bound to DNA but when bound to specific sites it appears to make the DNA more rigid. We infer that regions outside the homeodomain contribute both to specific DNA binding and to compaction of the DNA associated with specific binding.
Multiple clusters of PRH binding sites increase DNA compaction
To determine whether the presence of additional PRH binding sites in pGL2-Gsc would result in further DNA compaction, we cloned an array of PRH binding sites into this construct and the empty pGL2 vector. Plasmid pGL2-Gsc-PRHx5 contains an array of five PRH binding sites identified in SELEX experiments (5) cloned 3 kb away from the existing array of PRH sites in the Goosecoid promoter [Figure 4(a), line 3]. These sites are well characterized and are known to be bound by PRH in both electrophoretic mobility shift assay (EMSA) and reporter assays (10,20). Plasmid pGL2-PRHx5 contains the same array of five PRH sites but lacks the Goosecoid promoter. LD signals were recorded for DNA alone or DNA in the presence of PRH or PRH HD as described earlier. As seen with the plasmid containing the Goosecoid promoter sequences, the negative LD signal for pGL2-PRHx5 decreases in the presence of full-length PRH, although in this case, the change is only pronounced at 450-nM PRH (data not shown). This confirms that PRH binds to these sites under these conditions [and as shown previously using other methods (5,16)]. The negative LD signal for pGL2-Gsc-PRHx5 is increased in the presence of PRH–HD and the increase depends upon the protein concentration [Figure 4(f)]. The negative LD signal for pGL2-Gsc-PRHx5 is decreased in the presence of full-length PRH [Figure 4(g)]. However, at a low PRH concentration there is little change in the negative signal and possibly even an increase in the negative signal. We attribute this to the fact that the effective PRH:specific site DNA ratio is lower when there are two binding site arrays within the plasmid competing for specific binding by PRH. At higher PRH concentrations, additional LD maxima occur in the region of the far UV (230 nm) and at ∼280 nm and these accompany a significant reduction in the amplitude of the negative LD signal for DNA (260 nm) [Figure 4(g)]. These results suggest that specific DNA binding by PRH compacts pGL2-Gsc-PRHx5 to a greater extent than that seen with pGL-Gsc or pGL2-PRHx5. Furthermore, the LD maximum in the far UV suggests that PRH oligomers associate to form an organized PRH fibre on the DNA, in which proteins become aligned, and hence, produces an overlying LD spectrum containing signals from backbone and side-chain chromophores.
DNA compaction by PRH does not result in transcriptional repression
The above-described experiments indicate that the presence of an array of PRH binding sites results in DNA compaction and the formation a repetitive PRH–DNA fibre in vitro. As several PRH target promoters contain multiple clusters of PRH binding sites, these findings suggest that DNA compaction could play a role in the repression of transcription by PRH. Alternatively, DNA compaction by PRH could simply be a consequence of the highly oligomeric nature of this protein and be independent of the ability of this protein to repress transcription via, for example, the recruitment of co-repressor proteins. To test these possibilities we transiently transfected a series of reporter plasmids containing varying numbers of PRH sites and different promoter sequences into mammalian cells and examined the effects of PRH on their activity. We made use of K562 cells for these experiments because these cells express PRH endogenously. The luciferase reporter plasmids shown in Figure 5(a) were transiently co-transfected into K562 cells along with a plasmid expressing β-galactosidase that is used as a control for variations in transfection efficiency. Twenty-four hours post-transfection, the cells were harvested and the relative promoter activity calculated in each case by dividing the luciferase activity by the β-galactosidase activity. As expected, based on our previously published results, when an array of five PRH binding sites is present upstream of the minimal thymidine kinase promoter (TKmin-PRH), relative promoter activity is significantly reduced [Figure 5(b), compare columns 1 and 2]. We interpret this reduction in relative promoter activity to be due to endogenous PRH proteins binding to the PRH binding sites in the reporter and bringing about the repression of transcription. In order to confirm, this we repeated this experiment in K562 cells in which endogenous PRH has been knocked down using shRNA [as shown in Figure 5(c)]. In the PRH knock down cells, the relative promoter activities produced by the TKmin and TKmin-PRH constructs are not significantly different [Figure 5(b), columns 3 and 4].
Figure 5.
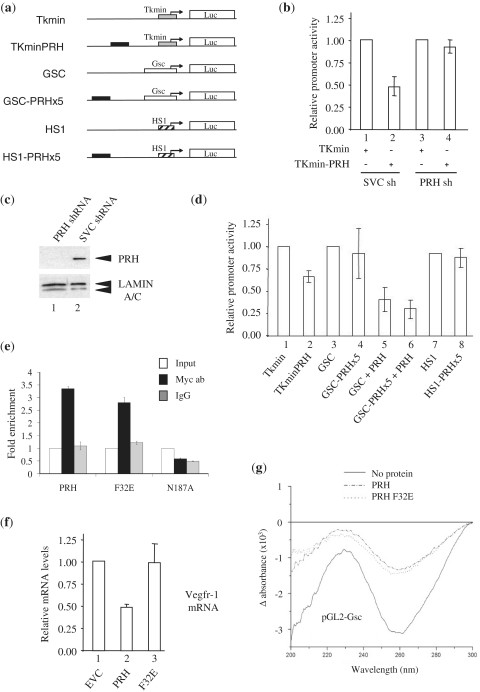
DNA compaction in not sufficient to repress transcription. (a) A schematic representation of the reporter plasmids used in this study. The transcription start point is represented by an arrow, filled rectangles represent a synthetic array of five PRH binding sites and Luc represents the luciferase gene. Not to scale. (b) The relative promoter activity found in extracts prepared from K562 control cells (1 and 2) and K562 cells in which PRH has been knocked down using shRNA (3 and 4) 24 h after transient co-transfection with the 3 ug of the TKmin or TKmin-PRH reporter plasmids shown in (a) and 3 ug of the β-galactosidase expression plasmid (pSV-lacZ). Relative promoter activity is the luciferase activity normalized for transfection efficiency using a co-transfected β-galactosidase expression plasmid. Mean and standard deviation (SD), n = 3. (c) A western blot performed using whole cell extracts prepared from K562 cells in which PRH has been knocked down using shRNA (1) or from control cells (2). PRH was detected using mouse anti-PRH polyclonal antisera. Lamin A/C was detected using a monoclonal anti-Lamin antibody and acts as a loading control. (d) The relative promoter activity found in extracts prepared from K562 cells 24 h after transient co-transfection with the 3 ug of the reporter plasmids shown in (a) and 3 ug of pSV-lacZ. The PRH expression vector pMUG1-Myc-PRH (1 µg) was co-transfected with the GSC reporters in five and six. Mean and SD, n = 4. (e) Quantitative ChIP of Myc-tagged PRH proteins at the Vegfr-1 promoter. The data shown are representative of the results from two independent experiments performed in triplicate. Mean and SD. (f) Vegfr-1 mRNA levels in K562 cells 48-h post-transfection with pMUG1-Myc-PRH (PRH), pMUG1-Myc-PRH F32E (F32E), or pMUG1-Myc-PRH N187A (N187A). mRNA levels were determined by qRT-PCR. Mean and SD, n = 5 (g) LD of linearized pGL2-Gsc (30 nM) with full-length PRH (150 nM) or PRH F32E (150 nM).
Having established that endogenous PRH in K562 cells is able to bind to a reporter plasmid containing an array of PRH sites and repress transcription, we next compared the relative promoter activity of the pGL2-Gsc and pGL2-Gsc-PRHx5 constructs described earlier. These reporter constructs have almost identical relative promoter activity in K562 cells [Figure 5(d), columns 3 and 4] despite the fact that pGL2-Gsc-PRHx5 contains more PRH sites and is more condensed by PRH in vitro as determined using LD. To investigate this further, we repeated this experiment in the presence of over-expressed PRH. Co-transfection of these reporters into K562 cells along with a plasmid that over-expresses PRH results in a significant reduction in relative promoter activity in both cases [Figure 5(d), columns 5 and 6]. However, the reduction in promoter activity brought about by PRH over-expression is very similar for both reporters. We conclude that the presence of additional PRH sites does not result in additional transcriptional repression. To determine whether this is more generally the case, we created reporter constructs in which the array of five PRH sites described earlier is placed upstream of the human Surf-1 (HS1) promoter in a distal position [Figure 5(a), lines 5 and 6]. The HS1 promoter is a well-characterized TATA-less housekeeping promoter that does not contain any sequences that resemble the PRH core binding site (21,25,26). Transient co-transfection of this reporter plasmid and an HS1 reporter plasmid lacking PRH binding sites into K562 cells result in the same amount of relative promoter activity in each case [Figure 5(d), columns 7 and 8]. This demonstrates that the presence of an array of PRH sites does not always result in the repression of transcription and suggests that other factors such as co-repressor recruitment or core promoter structure are important in determining whether PRH is able to represses transcription from a nearby promoter.
To investigate this in a setting in which the relative contribution to repression from co-repressor recruitment and DNA compaction can be determined, we made use of a PRH mutant that fails to bind TLE co-repressor proteins. PRH F32E carries a mutation that blocks binding to TLE proteins and blocks co-repression in cells (10,13). We first compared the ability of wild-type PRH and PRH F32E to bind to a PRH target gene in cells using quantitative ChIP. The Vegfr-1 gene is directly repressed by PRH in K562 cells (17). Both PRH and PRH F32E bind to Vegfr-1 promoter sequences in a ChIP assay [Figure 5(e)]. In contrast, a DNA binding defective PRH protein (PRH N187A) fails to bind to the same Vegfr-1 sequences. Quantitative RT-PCR shows that PRH represses Vegfr-1 mRNA levels in these cells, whereas PRH F32E has little or no effect [Figure 5(f)]. To examine whether PRH F32E brings about DNA compaction, we purified the protein and performed LD experiments. Figure 5(g) shows that the F32E mutation has no effect on the ability of the mutant protein to bring about DNA compaction. We conclude that DNA compaction by PRH and transcriptional repression are separable events.
DISCUSSION
In prokaryotes, DNA is packed into the nucleoid by a variety of DNA binding proteins including H-NS, HU and the Lrp proteins (27). In many cases, these architectural proteins can also act as gene-specific transcription factors regulating the expression of one or more genes. In eukaryotes, architectural proteins include the histone proteins, the nuclear lamins and scaffolding factors such as SATB1 (28,29). However, the distinction between architectural proteins and gene-specific transcription factors is again not clear-cut, as some proteins that influence the architecture of the genome also have effects on the expression of specific genes. For example, mutations in the lamin genes disrupt both nuclear organization and the expression of individual genes (28). Conversely, proteins that have gene-specific regulatory properties, such as the homeodomain protein SatB1, can also have more global roles in the architecture of chromatin (2). Architectural DNA binding proteins can be divided into those that wrap DNA around themselves to form nucleosome-like structures, those that bind to distantly spaced sites on DNA to form bridges and those that introduce DNA bending (27). The nucleosome is the paradigm for DNA wrapping and consists of around 150 bp of DNA wrapped around an octamer of histone proteins (30). Several gene-specific transcription factors are also thought to wrap DNA including the GAGA factor and the Polycomb nucleoprotein complex (31,32). However, these proteins are not thought to form stable homo-oligomers but rather act as monomers or other species that can self-associate and/or form heteromeric complexes with other proteins and thereby produce larger assemblies.
PRH is a gene-specific transcription factor that self-associates to form large oligomeric complexes in vitro and in cells (15). We have shown previously that these oligomers bind co-operatively to DNA sequences containing arrays of core PRH binding sites inducing a significant degree of DNA distortion (16). Here, we have shown that PRH oligomers compact DNA and form regular higher-order structures that likely correspond to protein hexadecamers bound to ∼500 bp of DNA. EM images of the PRH–DNA complexes suggest that PRH particles bind to specific sites resulting in further protein self-association and the formation of a protein–DNA fibre. DNA compaction therefore appears to be a consequence of the binding of large PRH oligomers to tandem arrays of binding sites. In these respects, PRH seems to be very different from the gene-specific transcription factors described earlier. Whilst PRH forms closed oligomers that bind DNA and then further self-associate, several well-characterized gene-specific transcription factors such as GAGA factor (32) and the bacterial MalT protein (33) appear to form open oligomers. These open oligomers can self-associate to build complexes which spread along DNA. In contrast, PRH appears to be able to spread along DNA by the association of hexadecameric protein–DNA complexes. In this respect, PRH is similar to some members of the oligomeric Lrp/AsnC family of proteins (34,35). These DNA binding proteins from bacteria and archaea form octamers and hexadecamers and are involved in both gene-specific transcriptional regulation and the global control of genome architecture. In this protein family, DNA binding is mediated by an N-terminal helix–turn–helix (H–T–H) motif (which is preceded by an α-helix) and the proteins bind co-operatively to arrays of suitably spaced recognition sites inducing DNA wrapping (35). PRH contains a central homeodomain (H–T–H–T–H) and also forms octameric and hexadecameric oligomers. Furthermore, the leucine-responsive regulatory protein (Lrp), the best characterized member of the family, forms disc-shaped oligomers with a diameter of ∼120 Å (34,35). DNA is thought to wrap around the edges of two stacked LRP discs to form a nucleosome-like structure. This is again highly reminiscent of the octameric discs and hexadecameric spheroids formed by PRH. Although the Lrp/AsnC proteins do not have known eukaryotic homologues and PRH does not show sequence similarity to these proteins, at least at low resolution, these proteins appear to be structural as well as functional homologues.
Once bound to DNA, PRH can bring about the repression of transcription (10,20). Several PRH target genes contain arrays of PRH core binding sites and the protein binds tightly these arrays (16). Our data suggest that PRH oligomers either (i) wrap DNA around themselves in a manner similar to the nucleosome or (ii) spread along DNA in a fibre-like fashion. In either case, the PRH–DNA assemblies can further associate to form regular repeating units. In both cases, this might be expected to contribute to the repression of transcription by steric hinderance (12). However, we have shown that the presence of an array of PRH binding sites is not in itself sufficient to bring about the repression of an adjacent promoter. Although an array of five PRH sites placed upstream of the minimal thymidine kinase promoter is able to bring about transcriptional repression, the same array of sites is unable to repress transcription when it is placed upstream of the HS1 promoter. As PRH-induced DNA condensation is brought about in vitro in both cases, this suggests that DNA condensation alone is not sufficient to bring about repression. In agreement with this conclusion, we have shown that increased DNA condensation brought about by increasing the number of PRH binding sites at a PRH repressible promoter does not result in increased transcriptional repression. Furthermore, we have shown that a mutated PRH protein that is unable to recruit co-repressor proteins (PRH F32E) brings about DNA condensation in vitro, although it fails to repress transcription when bound to a PRH target gene in cells. In this regard, PRH seems to be acting in a similar fashion to some of the components of the Sir protein complexes that repress gene expression at yeast telomeres and silent mating-type loci (36). Sir2-3-4 heterotrimers bind chromatin and naked DNA and bring about DNA condensation but the enzymatic activity of Sir2 is required for repression (37,38). Similarly, once it is bound to DNA, PRH must recruit TLE co-repressor proteins that can in turn recruit histone deacetylases in order to repress transcription (10). TLE proteins also oligomerize and condense chromatin and their oligomerization appears to be necessary for repression (39–41). Although the binding of PRH and the associated DNA condensation does not repress transcription, recruited TLE proteins might spread along the PRH–DNA structures at PRH target genes resulting in repression. Further experiments will be required to determine whether like the Sir complex, PRH is itself able to bind nucleosomes or whether this protein replaces nucleosomes when it binds to PRH binding site arrays.
FUNDING
The BBSRC and Wellcome Trust. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ian Portman and the Electron Microscopy Facility, Department of Biological Sciences, University of Warwick (Wellcome Trust grant reference: 055663/Z/98/Z) for instrument use and technical support. A.S. is grateful to the Royal Thai Government for a Ph.D. studentship. P.N. is grateful to the University of Birmingham for a Ph.D. Studentship.
REFERENCES
- 1.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 2.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 3.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 4.Soufi A, Jayaraman PS. PRH/Hex: an oligomeric transcription factor and multifunctional regulator of cell fate. Biochem. J. 2008;412:399–413. doi: 10.1042/BJ20080035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crompton MR, Bartlett TJ, MacGregor AD, Manfioletti G, Buratti E, Giancotti V, Goodwin GH. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 1992;20:5661–5667. doi: 10.1093/nar/20.21.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, Jordan CT, Borden KL. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell Biol. 2003;23:8992–9002. doi: 10.1128/MCB.23.24.8992-9002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puppin C, Puglisi F, Pellizzari L, Manfioletti G, Pestrin M, Pandolfi M, Piga A, Di LC, Damante G. HEX expression and localization in normal mammary gland and breast carcinoma. BMC Cancer. 2006;6:192. doi: 10.1186/1471-2407-6-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Elia AV, Tell G, Russo D, Arturi F, Puglisi F, Manfioletti G, Gattei V, Mack DL, Cataldi P, Filetti S, et al. Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J. Clin. Endocrinol. Metab. 2002;87:1376–1383. doi: 10.1210/jcem.87.3.8344. [DOI] [PubMed] [Google Scholar]
- 9.Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, Baty F, Brutsche M, Jayaraman PS, Santoro A, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111:5672–5682. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 10.Swingler TE, Bess KL, Yao J, Stifani S, Jayaraman PS. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J. Biol. Chem. 2004;279:34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 11.Bess KL, Swingler TE, Rivett AJ, Gaston K, Jayaraman PS. The transcriptional repressor protein PRH interacts with the proteasome. Biochem. J. 2003;374:667–675. doi: 10.1042/BJ20030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol. Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjobert C, Noy P, Swingler T, Williams H, Gaston K, Jayaraman PS. The PRH/Hex repressor protein causes nuclear retention of Groucho/TLE co-repressors. Biochem. J. 2009;417:121–132. doi: 10.1042/BJ20080872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soufi A, Noy P, Buckle M, Sawasdichai A, Gaston K, Jayaraman PS. CK2 phosphorylation of the PRH/Hex homeodomain functions as a reversible switch for DNA binding. Nucleic Acids Res. 2009;37:3288–3300. doi: 10.1093/nar/gkp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soufi A, Smith C, Clarke AR, Gaston K, Jayaraman PS. Oligomerisation of the developmental regulator proline rich homeodomain (PRH/Hex) is mediated by a novel proline-rich dimerisation domain. J. Mol. Biol. 2006;358:943–962. doi: 10.1016/j.jmb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Williams H, Jayaraman PS, Gaston K. DNA wrapping and distortion by an oligomeric homeodomain protein. J. Mol. Biol. 2008;383:10–23. doi: 10.1016/j.jmb.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Noy P, Williams H, Sawasdichai A, Gaston K, Jayaraman PS. PRH/HHex controls cell survival through coordinate transcriptional regulation of VEGF signalling. Mol. Cell Biol. 2010;30:2120–2134. doi: 10.1128/MCB.01511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butcher AJ, Gaston K, Jayaraman PS. Purification of the proline-rich homeodomain protein. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003;786:3–6. doi: 10.1016/s1570-0232(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 19.Soufi A, Gaston K, Jayaraman PS. Purification and characterisation of the PRH homeodomain: removal of the N-terminal domain of PRH increases the PRH homeodomain-DNA interaction. Int. J. Biol. Macromol. 2006;39:45–50. doi: 10.1016/j.ijbiomac.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Guiral M, Bess K, Goodwin G, Jayaraman PS. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J. Biol. Chem. 2001;276:2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 21.Lennard A, Gaston K, Fried M. The Surf-1 and Surf-2 genes and their essential bidirectional promoter elements are conserved between mouse and human. DNA Cell Biol. 1994;13:1117–1126. doi: 10.1089/dna.1994.13.1117. [DOI] [PubMed] [Google Scholar]
- 22.Gaston K, Fried M. CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 1995;23:901–909. doi: 10.1093/nar/23.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dafforn TR, Rajendra J, Halsall DJ, Serpell LC, Rodger A. Protein fiber linear dichroism for structure determination and kinetics in a low-volume, low-wavelength couette flow cell. Biophys. J. 2004;86:404–410. doi: 10.1016/S0006-3495(04)74116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dafforn TR, Rodger A. Linear dichroism of biomolecules: which way is up? Curr. Opin. Struct. Biol. 2004;14:541–546. doi: 10.1016/j.sbi.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Gaston K, Fried M. YY1 is involved in the regulation of the bi-directional promoter of the Surf-1 and Surf-2 genes. FEBS Lett. 1994;347:289–294. doi: 10.1016/0014-5793(94)00567-2. [DOI] [PubMed] [Google Scholar]
- 26.Vernon EG, Gaston K. Myc and YY1 mediate activation of the Surf-1 promoter in response to serum growth factors. Biochim. Biophys. Acta. 2000;1492:172–179. doi: 10.1016/s0167-4781(00)00116-0. [DOI] [PubMed] [Google Scholar]
- 27.Luijsterburg MS, White MF, van DR, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 28.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 29.Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 31.Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Mol. Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Larquet E, Schreiber V, Boisset N, Richet E. Oligomeric assemblies of the Escherichia coli MalT transcriptional activator revealed by cryo-electron microscopy and image processing. J. Mol. Biol. 2004;343:1159–1169. doi: 10.1016/j.jmb.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der OJ, Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de los Rios S, Perona JJ. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 2007;366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- 37.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 38.Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song H, Hasson P, Paroush Z, Courey AJ. Groucho oligomerization is required for repression in vivo. Mol. Cell Biol. 2004;24:4341–4350. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jennings BH, Wainwright SM, Ish-Horowicz D. Differential in vivo requirements for oligomerization during Groucho-mediated repression. EMBO Rep. 2008;9:76–83. doi: 10.1038/sj.embor.7401122. [DOI] [PMC free article] [PubMed] [Google Scholar]