Abstract
Rotaviruses are a major cause of acute, often fatal, gastroenteritis in infants and young children world-wide. Virions contain an 11 segment double-stranded RNA genome. Little is known about the cis-acting sequences and structural elements of the viral RNAs. Using a database of 1621 full-length sequences of mammalian group A rotavirus RNA segments, we evaluated the codon, sequence and RNA structural conservation of the complete genome. Codon conservation regions were found in eight ORFs, suggesting the presence of functional RNA elements. Using ConStruct and RNAz programmes, we identified conserved secondary structures in the positive-sense RNAs including long-range interactions (LRIs) at the 5′ and 3′ terminal regions of all segments. In RNA9, two mutually exclusive structures were observed suggesting a switch mechanism between a conserved terminal LRI and an independent 3′ stem–loop structure. In RNA6, a conserved stem–loop was found in a region previously reported to have translation enhancement activity. Biochemical structural analysis of RNA11 confirmed the presence of terminal LRIs and two internal helices with high codon and sequence conservation. These extensive in silico and in vitro analyses provide evidence of the conservation, complexity, multi-functionality and dynamics of rotavirus RNA structures which likely influence RNA replication, translation and genome packaging.
INTRODUCTION
Rotaviruses are responsible for the death from gastroenteritis of over 600 000 children per annum, mainly in developing countries of sub-Saharan Africa, South and South–East Asia (1). Two rotavirus vaccines (2,3) have been licensed since 2006 and are now in use in over 100 countries. Rotaviruses are a genus of the Reoviridae and have a double-stranded RNA (dsRNA) genome comprising 11 segments (4). The segments are monocistronic with the exception of RNA11, which encodes two proteins (NSP5 and NSP6). Rotaviruses replicate in the mature enterocytes at the tip of the villi of the small intestine. Adsorption of infectious virions [triple-layered particles (TLPs)] to specific receptors and interaction with co-receptors are followed by virus particle uptake into the cytoplasm and removal of the outer VP4/VP7 capsid layer. The resulting double-layered particles (DLPs) transcribe and release large numbers of all 11, capped, non-polyadenylated, positive-sense RNAs which can act as mRNA for translation or may be incorporated into progeny viral particles in lipid droplet associated cytoplasmic inclusion bodies termed ‘viroplasms’ (5) where early morphogenesis and viral RNA replication take place. Viroplasms release DLPs into the endoplasmic reticulum where they acquire their outer layer to become TLPs which are released from cells by lysis or other, non-lytic mechanisms (4).
RNA structures are instrumental in RNA–RNA and RNA–protein interactions; in viruses these have many functions including control of transcription, sub-cellular trafficking including nuclear export, translation, packaging, and, in some cases, effector functions such as immune evasion (6) and inhibition of apoptosis (7). Viral RNAs carry multiple overlapping functional cis-acting elements, and it is likely that many of the structures involved from only transiently during passage through the host cell. To date there has been little analysis of the structure function relationships of rotavirus RNAs. Signals essential for negative strand synthesis were identified at the 5′ consensus sequences and 3′ consensus sequences (3′CSs) of viral RNAs (8,9). Structural studies of the individual positive-sense RNAs have been limited to computational analyses of single sequences (9,10).
The genomes of many other RNA viruses have been analyzed much more extensively, and many characteristic motifs have been noted whose structural solution has been critical in establishing their function, such as internal ribosome entry site (IRES) structures to control translation (11), nuclear export signals (12) and RNA packaging signals (13). Packaging in non-segmented RNA viruses relies on specific regions of the RNA genomic strand folding into one or more complex 3D structures which can be recognized by viral structural proteins. Segmented RNA viruses, such as rotaviruses require additional mechanisms to encapsidate 11 separate distinct RNA segments into each particle. Rotaviruses achieve this with remarkable specificity, evidenced by the very low particle-to-pfu ratio (between 2 and 5) in cell culture-adapted rotavirus preparations (14). In addition, substitution of particular segments by their equivalent from a different rotavirus strain (re-assortment) occurs frequently during co-infection, implying a sequentially and/or structurally ordered packaging mechanism. In other segmented RNA viruses such as influenza virus cis-acting sequences involved in genome encapsidation have been identified within coding regions by observing focal restriction of codon usage (15).
To probe RNA structure, there is an increasing variety of physics-based analytical tools relying mostly on prediction of Watson–Crick and wobble base pairs. Free energy minimization is used by many RNA folding programs such as mfold (16,17) and RNAfold (18) for secondary structure prediction of an individual RNA molecule. However, the 2D minimum free energy (MFE) structure derived from these limited parameters calculated under non-physiological conditions, does not truly represent the native biological structure. Its accuracy can be improved by applying constraints derived from biochemical probing experiments. Algorithms which allow pseudoknot prediction (19), and comparative folding methods which consider both thermodynamics and covariation in related RNA sequences can also increase accuracy (20,21). RNAalifold predicts the MFE consensus structure from multiple sequence alignments of a set of related RNA sequences (21), and has been used to search for functional RNA motifs in viral RNAs (22–24). However, covariation analysis using the RNAalifold requires high-quality sequence alignments and careful selection of input sequences to avoid bias towards more closely related sequences within an alignment. The most recent version of the consensus structure prediction programme ConStruct allows the use of the RNAalifold algorithm to measure covariation from sequence alignments; it also has additional features such as a graphical user interface for the optimization of sequence alignments, the assignment of sequence weighting to minimize sampling bias, and the calculation of a thermodynamic score from a superimposed consensus base pairing matrix (25). RNAz is a useful tool to detect non-coding RNAs (ncRNAs) from sequence alignments of genomes (26). Because of its ability to detect conserved and thermodynamically stable secondary structure, it also allows high-throughput screening of conserved structures in viral RNA genomes.
Cis-acting functions in viral RNA are inevitably associated with the presence of structural motifs. For rotavirus genome packaging as well as for other functions of the viral RNA such as replication and translation structural signals should be detectable. Secondary structures may provide clues to the mechanism and the method of maintenance of specificity of packaging, and the capability for and control of re-assortment. To seek evidence for conserved structural elements in the published RNA sequences of rotavirus, we analyzed over 1600 full-length sequences of all 11 segments of group A rotaviruses using a variety of bioinformatic methods.
Conserved long-range interactions (LRIs) were found between the 5′- and 3′-terminal regions, sometimes extending into the coding region. Alternative structures possibly with distinct roles in RNA replication and translation were identified in two segments. Biochemical structure probing of the segment 11 led to a complete secondary structure model and confirmed the in silico prediction of three highly conserved helices. These studies provide a novel first insight into RNA structure function relationships in rotaviruses.
MATERIALS AND METHODS
Rotavirus A sequences
Positive-sense RNA sequences of all segments of group A rotaviruses (Rotavirus A) were obtained from GenBank (27). Sequences with full-length ORFs were used in the codon conservation analysis. Only full-length sequences with complete 5′- and 3′-UTRs were used in the ConStruct and RNAz analyses. Sequences of avian strains and of rearranged RNA segments of mammalian strains were excluded from all analyses. Table 1 summarizes the numbers of sequences evaluated for each RNA segment. Details of the rotavirus strains used in the RNA structural conservation analysis including GenBank accession numbers are listed in Supplementary Table S1.
Table 1.
Numbers of sequences evaluated per RNA segment
| RNA | Length (nt) | Sequences in structural conservation analyses | ORF | Sequences in codon conservation analysis |
|---|---|---|---|---|
| 1 | 3302 | 42a | VP1 | 37 |
| 2 | 2691 | 41a | VP2 | 34 |
| 3 | 2590 | 49a | VP3 | 42 |
| 4 | 2362 | 150a | VP4 | 45 |
| 5 | 1579 | 60 | NSP1 | 29 |
| 6 | 1356 | 164 | VP6 | 219 |
| 7 | 1062 | 687b | VP7 | 1224 |
| 8 | 1059 | 48 | NSP2 | 44 |
| 9 | 1076 | 39 | NSP3 | 46 |
| 10 | 751 | 247 | NSP4 | 237 |
| 11b | 667 | 94 | NSP5 | 64 |
| NSP6 | 87 |
aOnly 5′ 400-nt and 3′ 400-nt terminal sequences were evaluated in the ConStruct analysis for RNA1–RNA4.
bOnly 304 sequences all with <99% difference in sequence identity to each other were used in the prediction of consensus structure.
Sequence variation
The nucleotide variation at each independent position across each segment was calculated as follows:
where N = total number of sequences analyzed at each position and n = number of A,C,G or T nucleotides.
Normalized mean pair-wise distance calculations
The method used for estimating variability inside the rotavirus ORFs was previously successfully applied to influenza A virus RNAs (15). Normalized mean pair-wise distance was calculated for each amino acid position from alignments of rotavirus ORFs as described earlier (15). To identify clusters of low variability, for each site a moving average was computed by taking the average of the normalized MPD (nMPD) scores over an 11 amino acid window: the site in question plus five sites on each side. The statistical significance of low-nMPD scores was determined by computing a bootstrapping value for each site as described earlier (15).
MFE structure prediction
RNAfold was used to predict MFE structures of single sequences. RNAcofold was used to predict MFE structures involving two interacting sequences. Both RNAfold and RNAcofold are part of the Vienna RNA package (28).
ConStruct analysis
Full-length sequences were aligned using the multiple sequence alignment program ClustalX2 (29). Due to the large number of closely related full-length sequences of RNA7 available from GenBank, sequences with >99% pair-wise identity were filtered from RNA7 alignments using the WEIGHT utility from the SQUID C function library (Eddy,S. 2005 http://selab.janelia.org/software.html). Full-length alignments were analyzed for RNA5–RNA10. A refined analysis was carried out with an alignment of RNA11 sequences in which the 150-nt long insertion regions in the 3′-UTR of DS-1-like strains were excluded. The extremely high-memory requirement due to the length of RNA1–RNA4 made it impossible to analyze full-length alignments in these four larger segments. Therefore, we analyzed 800-nt hybrid alignments generated by artificially joining the alignment of 400-nt sequences from the 5′-terminus to the alignment of 400-nt sequences from the 3′-terminus in all segments longer that 800-nt (RNA1–9). In order to minimize sampling bias towards closely related strains, a weight value was assigned to each individual sequence from the corresponding ClustalX2 alignment using WEIGHT. The ConStruct package consists of two main programs, the script generator CS_FOLD and the dot plot analysis tool CS_DP (25). Base pairing probability dot-plot and MFE structure were created by RNAfold for each sequence (30). ConStruct project files were either generated by the CS_FOLD, or by custom-made bash scripts to facilitate sequence weight assignment and sequence selection for subgroup analyses (see below). A weighted consensus base pairing probability matrix was computed by CS_DP for each alignment to calculate the thermodynamic scores, while covariation scores were computed using an RNAalifold algorithm which takes stacking into account. Gap positions in some alignments were edited manually. The relative weightings for thermodynamics (wTD) and covariation (wCS) used to calculate the final probability score were wTD = 0.7 and wCS = 0.3, while the minimum thresholds for thermodynamics (tTD) and covariation (tCS) used were tTD = 0.10 and tCS = 0.10 (25). In addition to the probability scores, structural alignments, base frequencies and base pair frequencies (AU, GC and GU) for all predicted conserved base pairs were produced by CS_DP from each sequence alignment (25). Consensus structure and base pairing probability values were mapped to the sequences of the bovine rotavirus UK strain unless specified otherwise. Subgroup analysis was carried out by modifying the project file and sequence alignment to select only a particular group of sequences. Detailed analyses of particular sections of alignment were done by generating project files and alignments using custom-made shell scripts.
RNAz analysis
Short-range RNAz analysis was carried out as previously described earlier (26). Sub-section alignments were generated from full-length alignments using a window size of 60 nt or 120 nt and a sliding distance of 10 or 20 nt. The long-range RNAz analysis involved additional preprocessing of input alignment. All non-overlapping combinations of subsection alignments were joined to create 120-nt hybrid alignments, which were analyzed by RNAz as described earlier. A custom-made shell script was used to extract data from the output file including Z, SCI and P scores. A matrix was generated for each of these values using the OpenOffice Spreadsheet (Supplementary Figure S5). The artificial joining of two alignments may introduce false negatives if one strand of a predicted helix spans across the artificial joining point. Therefore, we manually evaluated the secondary structure predicted for individual strains and for the consensus sequence for all the high scoring alignments (P-score > 0.5). Clusters of high P-scores on the matrix suggest that the conserved structures are true positive results. The calculation of P-score by RNAz 2.0 stems from a support vector machine (SVM) training on functional RNA structures in the Rfam 9.0 database (26). Since ‘hybrid alignments’ were used in the long-range RNAz analysis and the SVM was trained for continuous alignments, the corresponding confidence levels for the 0.9 and 0.5 P-score cut-off points are indicative, but not definitive.
RNA transcription and purification
The cDNA of group A bovine rotavirus UK strain RNA11 has been cloned previously into the multiple cloning site of the TA cloning vector pCR2.1 from Invitrogen (plasmid kindly provided by Malcolm McCrae, University of Warwick, UK). PCR amplification from this plasmid generated cDNA with a T7 promoter attached directly to the 5′-end of the gene. The following primers were used; 5′-TAATACGACTCACTATAGGCTTTTAAAGCGCTACAGTG-3′ (+sense) (T7 promoter sequence underlined) and 5′-GGTCACAAAACGGGAGTGGGG-3′ (−sense) (italicized Gs represent the 5′- and complementary 3′-ends, respectively, of RNA11). Subsequent in vitro transcription from this cDNA using T7 polymerase generated RNA11 transcripts with authentic 5′- and 3′-termini. Viral RNA transcripts were synthesized using the Ambion MEGAscript T7 transcription kits according to the manufacturer’s protocol. Per reaction, 0.5 μg cDNA was used. After 1.5 h incubation at 37°C, 1 μl of DNase Turbo (1 U/μl; Ambion) was added directly to the transcription reaction which was incubated for a further 15 min at 37°C. RNA was then purified by phenol/chloroform extraction, followed by precipitation with 2.5× volumes of ethanol in the presence of 0.5 M ammonium acetate. Polyacrylamide gel electrophoresis was used to confirm the size and cintegrity of the RNA, and the concentration was determined using a NanoDrop® ND-1000 UV-Vis Spectrophotometer.
RNA structure probing
The size of RNA11 (667 nt) made end labeling techniques impractical for probing the majority of the structure, thus the RNA was analyzed by primer extension. To ensure structural uniformity, RNA was heated to 80°C for 10 min and allowed to cool to room temperature before structure mapping was carried out. Enzymatic probing reactions were carried out essentially as described by Ambion using RNases T1, CL3, A, I, V1 (Ambion) and U2 (Thermo Scientific) at limiting dilutions to achieve partial cleavages. RNase A cleaves unpaired cytosine and uracil bases, RNase T1 cleaves unpaired guanine residues, RNase U2 cleaves both unpaired adenines and guanines (but shows a preference for guanines) and RNase I cleaves unpaired bases with no base specificity. RNase V1 cleaves phosphodiester bonds flanked by a helical backbone (31). Reactions were incubated at room temperature for 10 min in 10-μl reactions containing 1 μg carrier yeast tRNA. Each reaction contained 2 μg of RNA11.
For primer extension reactions, 1 μg of partially digested RNA was annealed to 50 pmol primer by heating to 70°C for 1 min in a buffer containing 60 mM Tris–HCl pH 7.5 and 100 mM KCl, followed by slow cooling to 42°C. RNA11-specific primers used in primer extension reactions are listed in Supplementary Table S2. Primer extension reactions were carried out in 10-μl reaction volumes containing 0.5 μg partially digested RNA/primer mix, 16 U RNasin (Promega), 8.8 units AMV RT (Promega) and 0.37 MBq 33P-dATP (Perkin Elmer) in an extension buffer containing 7.5 mM Tris–HCl pH 8.3, 30 mM KCl, 7.5 mM MgCl2, 6 mM DTT, 3 μM dATP and 0.3 mM each of dCTP, dGTP and dUTP. Reaction mixes were incubated at 42°C for 30 min followed by the addition of an equal volume of Ambion Gel Loading buffer II and heating to 80°C for 10 min. Samples were analyzed on 10–12% polyacrylamide 7 M urea gels alongside dideoxy-sequencing ladders of template DNA, primed by the same oligonucleotide primer that had been used for cDNA synthesis.
Due to the nature of the primer extension protocol, it was only possible to obtain structural data up to nucleotide 635 in the 3′ terminal region, and therefore an absence of data beyond nucleotide 635 is not indicative of an absence of cleavage by RNases. Untreated control lanes indicate reverse transcriptase pausing generated by enzyme detachment as a result of homopolymeric runs of nucleotides or the presence of RNA secondary structure ahead of, or behind, the catalytic site (32–34). Cleavage sites were identified by the higher intensity of the cDNA band in the RNase-treated lane compared to the untreated controls. Results represent cleavages in multiple experiments, using the lowest concentration of RNase where a difference between treated and untreated lanes was clearly visible to avoid the small but finite risk of generating artifactual data from secondary cleavages in RNA fragments.
RESULTS
Sequence conservation extends beyond the UTRs of rotavirus RNA
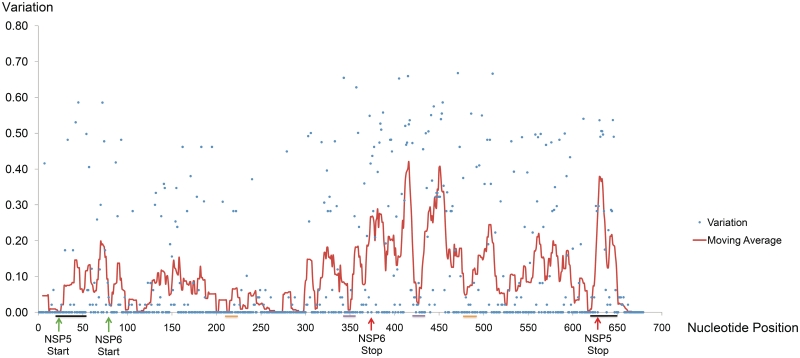
Sequence conservation was identified in many rotavirus RNA segments. For RNA11, conserved regions are present in the 5′- and 3′-UTR, but also as discrete regions within the ORFs, most notably at nucleotides 94–130 and 187–314 (Figure 1). In RNA8 (Supplementary Figure S1), three conserved regions with variation values of <0.1 were found at nucleotides 1–27, 42–60 and 1042–1059. Much of the second conserved region is within the NSP2 ORF. High-nucleotide sequence conservation cannot be wholly explained by the need to maintain protein sequence because alternative codons can be used for most amino acids, suggesting that sequence dependent cis-acting functions contribute to the conservation.
Figure 1.
Nucleotide variation of RNA11. Blue dots represent individual nucleotide variation values. The line graph represents moving average values using a sliding window of 9 nt (from −4 to +4 positions). Locations of start and stop codons are marked by green and red arrows, respectively. Bars indicate locations of the terminal LRIs (black), the H2 helix (orange) and the H3 helix (purple).
Codon conservation analysis reveals potential functional regions in eight rotavirus ORFs
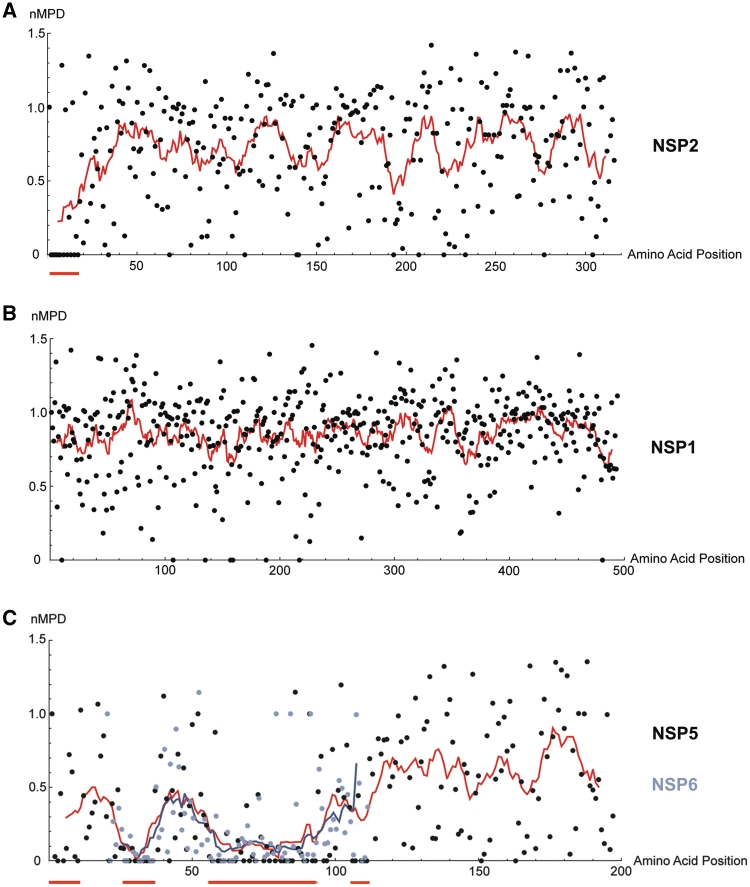
To identify likely cis-acting functional domains within the coding regions of rotavirus RNAs, the nMPD scores for each amino acid codon in the 12 ORFs were calculated. A low-nMPD score represents higher conservation of a particular codon than would be expected from amino acid conservation or codon bias. Examples of nMPD plots for NSP2, NSP1 and NSP5/NSP6 are shown in Figure 2, and the results for these and all the remaining rotavirus ORFs are presented in Supplementary Figure S2. Large areas of low-moving average value were found near the 5′-termini of ORFs at amino acids (aa) 1–12 of NSP2 (Figure 2A), amino acids 1–20 of VP1, amino acids 1–15 of VP3 and amino acids 1–10 of VP7, all with bootstrap values below 0.001. Clusters of low-nMPD scores were also found at amino acids 1–10 of NSP5; amino acids 6–17 of NSP6; amino acids 1–7 of VP4; and amino acids 1–3 and 15–19 of NSP3. Taken together, these observations suggest that functional RNA domains exist in the coding region, possibly as part of a single continuous cis-acting sequence extended from the 5′-UTR. No significant functional constraints were detected in the ORFs for NSP1 (Figure 2B), VP2, VP6 and NSP4. Codon conservation of the two overlapping ORFs on RNA11, NSP5 and NSP6, was found beyond the 5′-terminal regions (Figure 2C). At amino acids 21–112 of the NSP5 ORF where the two ORFs overlap, three distinct regions of low-moving average were found within the overlapping region. The alternating pattern of low (amino acids 25–36, amino acids 59–93 and amino acids 106–111 of NSP5) and high (amino acids 37–58 and amino acids 94–105 of NSP5) moving averages within this region raises two possibilities: (i) the low-nMPD clusters define regions which encode amino acid residues crucial for protein functions in both NSP5 and NSP6 ORFs; or (ii) at least part of the low-nMPD clusters are results of constraints imposed by RNA cis-acting functions. Because NSP6 is not essential for viral replication in vitro (35) and is not expressed in some rotavirus strains (Supplementary Table S1), the second possibility seems more likely.
Figure 2.
nMPD analysis of rotavirus ORFs. Results from the codon variation analysis for (A) NSP2; (B) NSP1; (C) NSP5 and NSP6 are shown. Dots represent individual nMPD scores. Line graphs show moving average values using a sliding window of 11 aa. In (C), data for NSP5 are illustrated by black dots and the red line, while data for NSP6 are illustrated by lilac dots and the purple line. Red bars underneath each plot highlight regions of high codon conservation.
For some ORFs in which we could not identify any low-nMPD regions using alignments of all available sequences, we also analyzed individual or groups of genotypes (Supplementary Figure S3). A small cluster of low-nMPD scores was found at the 5′ terminus of the VP2 ORF (amino acids 1–11) when 42 sequences from the C2 genotype (36) were analyzed alone. A small cluster of conserved codons also appeared at amino acids 387–389 of VP6 when sequences of subgroup II consisting of genotypes I2, I5 and I6 (36) were analyzed separately. Due to insufficient numbers of available sequences this process was not possible for other genotypes of VP2 and NSP4, and for NSP5 from RNA11 sequences which do not express NSP6. The extreme variability including large insertions and deletions in NSP1 precluded meaningful alignment for an analysis based on individual genotypes
Conserved LRIs and local secondary structures in rotavirus RNA segments
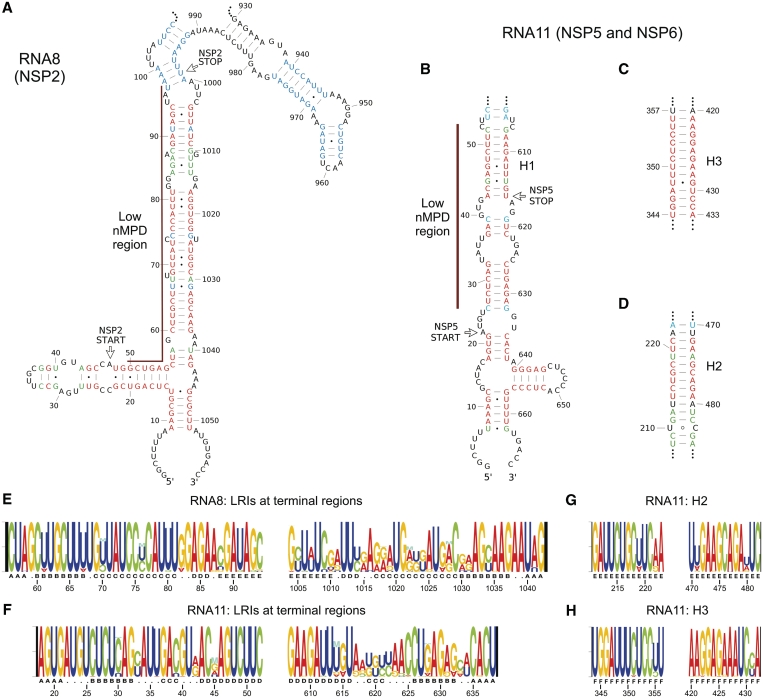
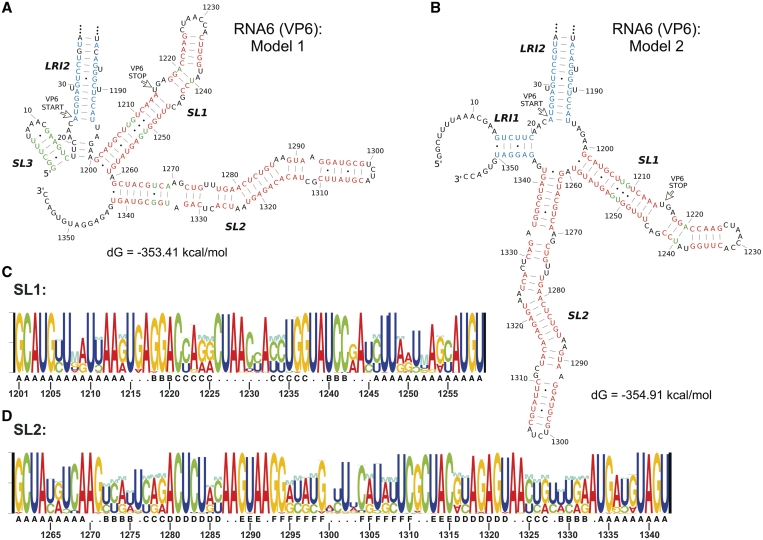
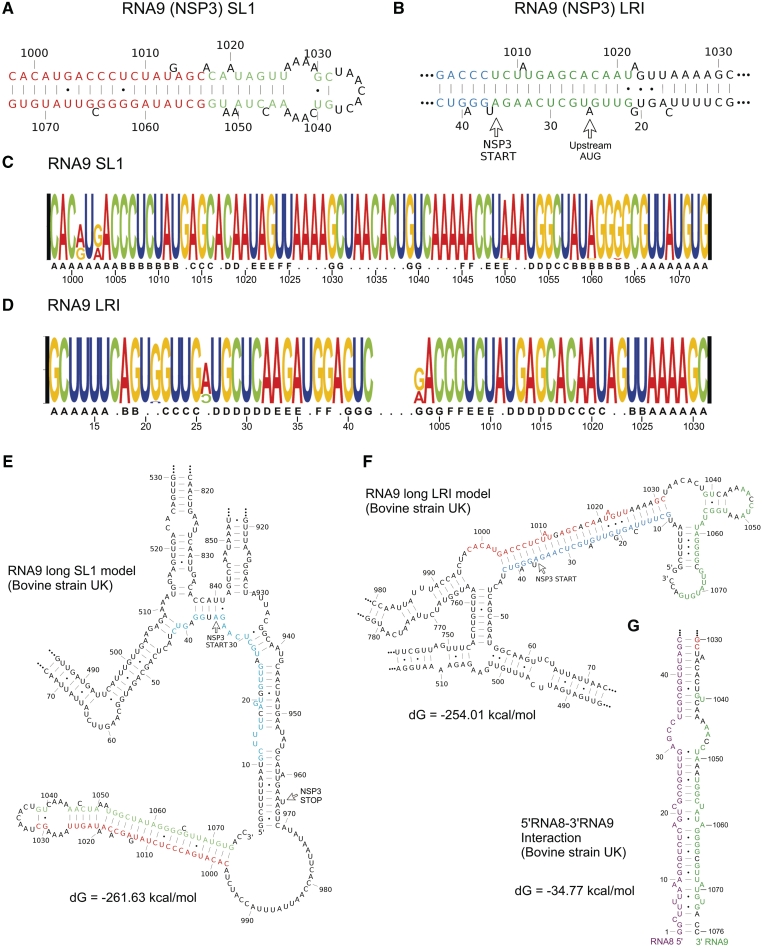
We investigated whether conserved RNA structures are present in the coding and non-coding regions of the rotavirus RNA segments. Using the ConStruct RNA consensus structure prediction package (25), base pairing probabilities were computed from the alignments of full-length sequences (RNA5–RNA11) and alignments of 800-nt ‘hybrid’ sequences (RNA1–RNA9, ‘Materials and Methods’ section). Most of the conserved RNA structures obtained from the ConStruct analysis were formed by sequences within the 200nt 5′- and-3′ terminal regions of the RNA (Supplementary Figure S4 and Table S2). For RNA5–RNA9, predictions for the 5′- and 3′-terminal 400-nt regions from analyses of full-length and 800-nt ‘hybrid’ alignments were identical (data not shown), validating the ‘hybrid’ alignments as representative for the prediction of conserved secondary structure in the terminal regions of larger segments (RNA1–RNA4). The most common type of conserved secondary structure identified was a long stretch of helical regions held by LRIs between the 5′- and 3′-terminal regions. Structures with a medium to high-average base pairing probability were identified in RNAs1, 2, 3, 5, 7, 8, 10 and 11, and others with a low-average base pairing probability in RNA4 and RNA6. In RNA8, conserved LRI helices were predicted to form between nucleotides 56–94 and 1004–1042 (Figure 3\A). In RNA11, such a structure was predicted with high certainty to form between nucleotides 18–52 and 607–638 including a long-continuous helix H1 (Figure 3A), in agreement with the prediction by Tortorici et al. (9) using the Massively Parallel Genetic Algorithm (MPGAfold). In RNA11, in addition to the terminal LRIs, two remarkably conserved helices with high-base pairing probability, named H2 and H3, were found in the central region of the RNA (Figure 3C and D). Many stem–loop structures were also identified in the 5′- and 3′-terminal regions, including a stem–loop (nt 15–55) in 5′ terminal region of RNA8 (Figure 3A), SL1 (nt 1201–1259) and SL2 (nt 1261–1342) in the 3′ terminal region of RNA6 (Figure 4A) and SL1 (nt 998–1073) within the 3′-UTR of RNA9 (Figure 5A). RNA10 was predicted to have a conserved terminal structure consisting of three conserved stem–loops in the 3′-UTR linked by many LRI helices (Supplementary Figure S4).
Figure 3.
Conserved LRIs in RNA8 and RNA11. Conserved secondary structures predicted by ConStruct and RNAz in RNA8 and RNA11 are shown, including LRIs in the terminal regions of (A) RNA8 and (B) RNA11, and two LRIs in the internal region of RNA11; (C) H3; and (D) H2. Base pairing probabilities calculated by ConStruct are represented by different colors: red (0.500 or higher), green (0.300–0.499) and blue (0.100–0.299). Regions of low-nMPD scores are marked by brown lines. Conserved structures were mapped to the sequence of the bovine rotavirus UK strain. (E and H) Nucleotide compositions of LRIs are illustrated using RNA Structure Logo for the terminal regions of RNA8 (E) and RNA11 (F), and for (G) the H2 helix and (H) the H3 helix. The height of each position indicates nucleotide conservation value. The height ratio between different bases at the same position reflects relative proportion of each base. Different letters under Structure Logos indicate the positions of individual continuous helices.
Figure 4.
Stable 3′-stem–loops in RNA6. (A) Model 1 represents the consensus structure predicted using ConStruct when no constraints were applied. (B) Model 2 represents predicted structure with the 5′-terminal GGC nucleotides unpaired. Consensus structures were mapped to the bovine rotavirus UK strain sequence, and free-energy values were calculated using RNAfold. Base pairing probabilities calculated by ConStruct are represented by different colors: red (≥0.500), green (0.300–0.499) and blue (0.100–0.299). Nucleotide compositions of (C) SL1 and (D) SL2 are illustrated using RNA Structure Logo.
Figure 5.
Dynamic terminal structure of RNA9. The two mutually exclusive conserved structures in RNA9 are (A) a long-stem–loop SL1 and (B) a long-stacked helix formed by LRIs. Base pairing probabilities calculated by ConStruct (A) when no constraints were applied or (B) when the UGUGACC-3′ CS was unpaired are represented by different colors: red (≥0.500), green (0.300–0.499) and blue (0.100–0.299). Nucleotide compositions of (C) SL1 and (D) long LRI are illustrated using RNA Structure Logo. (E and F) MFE structures of bovine rotavirus UK strain predicted by RNAfold using (E) SL1 and (F) long LRI as folding constraints. The 5′-strand and the 3′-strand of SL1 are highlighted in red and green, respectively. The 5′ strand of the long LRI is highlighted in blue. (G) Potential intermolecular interactions between the 5′-terminal region of RNA8 and the 3′-terminal region of RNA9 predicted by RNAcofold. Sequence from RNA8 is highlighted in purple.
In addition to using ConStruct, we evaluated structural conservation using the RNAz programme, which calculates the probability of the presence of thermodynamically stable and conserved secondary structure within a particular region. The short-range-RNAz analysis confirmed the presence of many stem–loops predicted by ConStruct such as 3′SL1 and 3′SL2 in RNA6 (Figure 4B), conserved stem–loops in the 3′-UTR of RNA10 (Supplementary Figure S4) and a stem–loop at nucleotides 939–976 in RNA8 (Figure 3A). The long-range analysis not only confirmed the presence of nearly all LRI helices predicted by ConStruct, but also suggested an additional conserved stacked helix (Figure 5B) formed by LRIs between the 5′-terminal region (nt 11–42) and the 5′-strand of the 3′SL1 (nt 1003–1031) of RNA9.
Conserved LRIs in RNA8 and RNA11
Data from nMPD, nucleotide variation and structural conservation analyses can be used to study how conserved RNA structures are maintained. Both sequence conservation and covariation are involved in maintaining the terminal structures of RNA8 and RNA11. Three types of base pairing can be found within the long-terminal LRIs of RNA8 (Figure 3): (i) between two conserved nucleotides (e.g. base pairs G69:C1029); (ii) between a conserved nucleotide (e.g. U81 in base pairs 81:1017) and a varying 3′ nt (e.g. swapping between A1017 and G1017); and (iii) between two co-varying nucleotides (e.g. U70:A1028 or C70:G1028). In both RNA8 and RNA11, the long-terminal LRIs are formed between highly conserved sequences on the 5′-strand overlapping the low-nMPD regions, and highly variable sequences within the 3′-UTRs (e.g. nucleotides 1005–1034 of RNA11) on the 3′-strand. The much higher sequence conservation on the 5′-strands may be due to protein-coding constraints in addition to the formation of LRIs, but also to other cis-acting functions. Sequence conservation seems to play a key role in maintaining some conserved LRIs. In H1, H2 and H3 helices of RNA11, most base pairs are formed between conserved nucleotides within clusters of low-nMPD scores (Supplementary Text S1), indicating strong selection pressure to maintain these helical structures. Individual and clusters of low-nMPD scores allowed us to evaluate the functional importance of many predicted conserved structures, but due to the mechanism of covariation the lack of codon conservation does not necessarily indicate a lack of functional constraints.
Conserved long stem–loop structures in RNA6 including a translation enhancer SL1
In RNA6, both SL1 and SL2 are largely maintained by covariation (Figure 4). In contrast to the high level of nucleotide variation at the stem region, most nucleotides in the loop region (nt 1226–1232) are highly conserved and may be important for tertiary interactions, intermolecular interactions or protein binding. The 3′-UTR of RNA6 had previously been shown to possess translation enhancement activity, attached to either the RNA6 sequence or a reporter gene (37). This activity is contained within nucleotides 1218–1276, therefore it seems very likely that the stacked stem–loop at nucleotides 1218–1238 of SL1 functions as a translation enhancer when placed after the stop codon of an ORF. A conserved stem–loop (SL3) was predicted at the 5′-terminus but with only low-base pairing probabilities (Figure 4A). The SL3 structure is maintained mostly by weak A:U and G:U wobble base pairs. Evaluation of individual MFE structures shows that within all groups of similar sequences, SL3 is predicted in some sequences while a short-LRI helix (LRI-1) is predicted in others. Using SL3 or LRI-1 as folding constraints, RNAfold predicted two MFE structure models for the bovine rotavirus UK strain with a very small free energy difference of 1.50 kcal/mol. The major conserved LRI-2 was extended with the formation of a 3-bp-short helix in model 1 (Figure 4A), while a 4-helix junction is formed between LRI-1, LRI-2, SL1 and SL2 in Model 2 (Figure 4B). Because the 5′-terminus of all rotavirus RNAs contains a cap structure (38), we carried out an additional ConStruct analysis with the first three bases 5′-GGC unpaired to mimic interaction with a cap-binding protein. With this small change the consensus structure prediction switches to the formation of LRI-1 at low base pairing probability.
Alternative conformations of RNA9
In RNA9, both SL1 (Figure 5A) and the alternative LRI (Figure 5B) structures can be formed involving extremely conserved sequences (Figure 5C and D). The low-nMPD region in the N-terminal of NSP3 ORF (amino acids 1–3, nucleotides 35–43) was also found to be completely within the 5′-strand of the LRI. In the bovine rotavirus UK strain, both the long- and short-range interactions involve a total of 27 bp with identical numbers of 11 G:C, 13 A:U and three G:U pairs. The prediction of these two mutually exclusive conformations with equal thermodynamic stability suggests the presence of a molecular switch in RNA9.
Two rotavirus proteins, VP1 and NSP3, are known to specifically bind to the 3′CS of all rotavirus positive strand ssRNAs. VP1 interacts with the 3′CS UGUGACC-3′ (39,40), while NSP3 interacts with the four base sequence GACC-3' at the 3′-termini of rotavirus RNAs (41,42). Because the 3′-strand of SL1 involves the first 4 nt of the 3′CS, we speculated that the switch from the SL1 to the LRI structure may be induced by VP1 binding, causing the destabilization of the lower helix of SL1, while NSP3-binding to GACC-3′ may not be sufficient to cause the destabilization of SL1. To test the hypothesis in silico, alignments of RNA9 sequences with a deletion of the entire 3′CS (mimicking VP1-binding) or GACC-3′ (mimicking NSP3-binding) were analyzed by ConStruct. When UGUGACC-3′ was deleted a large part of the LRI structure (base pairs from 22:1021 to 42:1003) was predicted with medium to low-base pairing probability (Figure 5B), while the SL1 structure was predicted when GACC-3′ was deleted. To evaluate the effect of the formation of SL1 or LRI on the overall structure of RNA9, MFE structures for the RNA9 of the bovine rotavirus UK strain were predicted using RNAfold with folding constraints forcing the formation of SL1 or LRI. In the SL1 model (Figure 5E), the 5′-terminal sequence was found to form another long-LRI structure, and a long-single-stranded region is formed at nucleotides 970–997. In the long-LRI model (Figure 5F), weak helices were formed at the 5′-terminal region. Interestingly, no ‘hinge region’ was found to isolate the switch region from sequences in the central region of the RNA. Instead, the central region is predicted to undergo extensive structural rearrangement.
As a consequence of the switch from SL1 to the long-LRI conformation, the accessibility of the 3′-terminal region is much higher in the LRI model. To test if there are possibilities of intermolecular RNA–RNA interactions between this 3′-accessible region and 5′-regions of other RNA segments, RNAcofold was used to predict structures between nucleotides 1026–1076 of RNA9 and nucleotides 1–80 of all RNA segments of the bovine rotavirus UK strain. The highest sequence complementarity and lowest free energy were found between nucleotides 1–45 of RNA8 and nucleotides 1030–1076 of RNA9 (Figure 5G). A large part of this potential interaction is formed between the loop region (nucleotide 22–48) of the conserved stem–loop in RNA8 (Figure 3A) and the discontinuous loop region of SL1 (nucleotide 1017–1054) in RNA9, raising the possibility of a kissing-loop interaction between the two RNA segments which may trigger the switch in conformation in RNA9. However, the RNAcofold algorithm does not allow the prediction of kissing loop interactions.
Diverse terminal structures of RNA4
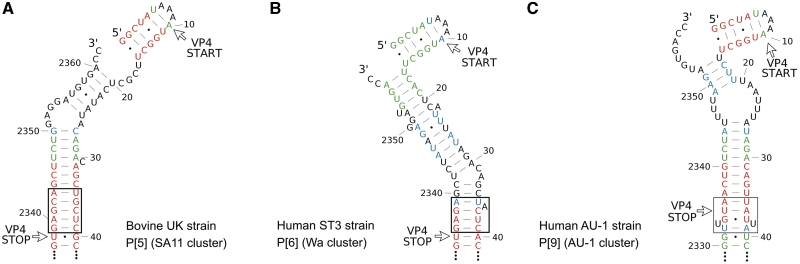
The nMPD analysis identified a short-low-scoring region at the 5′-terminus of the VP4 (amino acids 1–7). However, prediction of the terminal structure using ConStruct yielded a consensus structure with a 5′ terminal stem–loop and two short helices with a lower base pairing probability than in other segments (Supplementary Figure S4). It is possible that different strains adopt different secondary structures due to high-nucleotide variability at the 3′-terminal region. We divided the RNA4 sequences into three large clusters of genotypes based on their phylogenetic distances and analyzed them separately using ConStruct. The ‘Wa’ cluster contains the most common human P genotypes P[8], P[4], P[19] and P[6], the ‘AU-1’ cluster contains P[14], P[9] and P[25]; while the ‘SA11’ cluster contains P[1], P[3] and P[5], mostly animal rotavirus strains. By mapping the base pairing probability to the MFE structure of representative strains in each cluster (the Wa cluster: human P[6] ST3 strain; the SA11 cluster: bovine P[5] UK strain; the AU-1 cluster: human P[9] AU-1 strain), it became obvious that different RNA4 clusters adopt different LRI structures at the terminal regions (Figure 6). In each cluster, the conserved LRI was found to extend beyond the 5-bp consensus LRI, and the 5′-terminal stem–loop is always predicted. Despite the difference in positions of bulges, most nucleotides in the 3′-UTR are base paired with the 5′-terminal region in all clusters to form a similar terminal structure. The low-structural conservation and high-codon conservation at the terminal regions of RNA4 suggest that the much more conserved 5′-strand may be involved in other functions.
Figure 6.
Diverse terminal structure of RNA4. The terminal structures of three large clusters of RNA4 genotypes are represented by the MFE structure of (A) the bovine UK strain for the SA11 cluster; (B) the human ST3 strain for the Wa cluster; and (C) the human AU-1 strain for the AU-1 cluster. Base pairing probabilities calculated by ConStruct for each cluster of genotypes are represented by different colors: red (≥0.500), green (0.300–0.499) and blue (0.100–0.299). Consensus structures predicted by ConStruct using a full set of sequences with all genotypes include the 5′-terminal stem–loop and the short helix marked by the black rectangles.
Biochemical secondary structure probing of the complete RNA11 confirmed the terminal structure and the presence of H1, H2 and H3
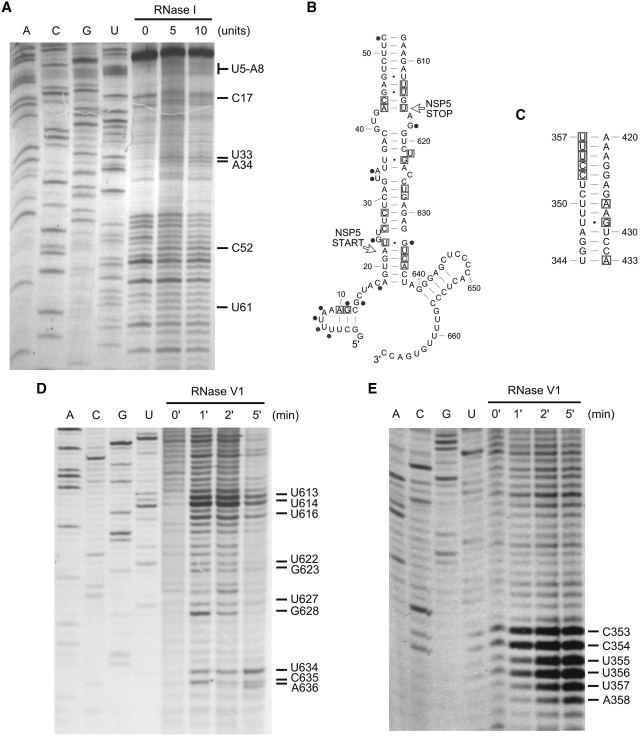
We analyzed the in vitro secondary structure of the shortest segment, RNA11, by biochemical structure probing followed by primer extension (Figure 7). The presence of H1, H2 and H3 LRIs are confirmed by the presence of many cleavage sites on both 5′- and 3′-strands by RNase V1, which cleaves specifically within regions with helical backbone conformation (31). Because helical backbone conformation may extend to several nucleotides adjacent to a helix, RNase V1 cleavages are observed in nucleotides between helices or nucleotides flanked by long helices (31). The presence of the 5′-terminal stem–loop is confirmed by RNase I data.
Figure 7.
Biochemical structure probing for RNA11. Representative auto-radiographs of primer extended products showing cleavage sites (A) by RNase I at the 5′-terminal region; (D) by RNase V1 at the 3′-terminal region; and (E) by RNase V1 close to the 5′-strand of the H3 helix. Cycle sequencing ladders are shown in the A, C, G and U lanes. Combined results from multiple experiments (B) for the terminal regions and (C) for the H3 helix are also shown, where boxes represent RNase V1 cleavages (indicating the presence of helical conformation at flanking nucleotides) and dots represent cleavages by RNases with single-strand specificities (RNases T1, U2, A, I and CL3).
Complete secondary structure model of RNA11
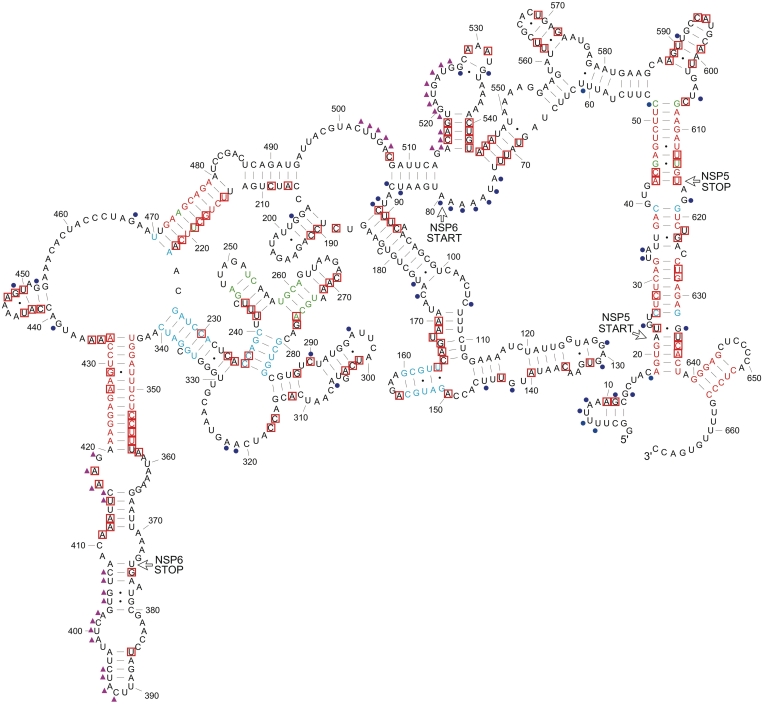
Based on both, biochemical structure mapping data and consensus structure models derived from ConStruct and RNAz analyses, we built a complete secondary structure model for the RNA11 of the bovine rotavirus UK strain. The first step was to map the RNase cleavage sites to different models including the consensus model from ConStruct, MFE predictions from high-scoring windows in RNAz analysis, and the MFE structures predicted by RNAfold and Mfold. This led to a selection of helices and stem–loops which are compatible with the structure mapping data. By joining different combinations of these substructures together, a model with maximal agreement of the biochemical data, conservation data and thermodynamics was derived (Figure 8). Many of the structures from the ConStruct model are compatible with the biochemical data and are also present in this strain-specific model. In the H2 helix, only 11 bp predicted with high probabilities by ConStruct is included in this model (base pairs from 213:480 to 223:470), because the MFE structure by RNAfold predicts the formation of a 7-bp helix adjacent to H2 which gives an overall higher thermodynamic stability and agrees with the RNase V1 cleavages at A207 and C209.
Figure 8.
Complete secondary structure model of RNA11. Base pairing probabilities calculated by ConStruct are represented by different colors: red (0.500 or higher), green (0.300–0.499) and blue (0.100–0.299). Red boxes represent RNase V1 cleavages (indicating the presence of helical conformation at flanking nucleotides), and blue dots represent cleavages by RNases with single-strand specificities (RNases T1, U2, A, I and CL3). Purple triangles represent potential tertiary interactions predicted by RNAz.
Many interesting features are found in the complete structure of RNA11. First, the sequence immediately before the start codon of NSP6 is highly accessible, strongly supported by RNase I cleavages at nucleotide 74–80. Second, there are many repeat sequence motifs. For example, 5 CUUC motifs are found at nucleotide 49–52 (within H1), 53–56, 62–65, 89–92 and 218–221 (within H2); while 5 GAAG motifs are found at 183–186, 472–475 (within H2), 555–558, 582–585 and 607–610 (within H1). Interestingly, all the CUUC and GAAG motifs are base paired with each other and the CUUC motif is always on the 5′-strand of the resulting helix. Third, a highly conserved palindromic sequence motif with extremely high-GC content 5′-G640GGAGCUCCC649-3′ is found in the small stem–loop at nucleotide 640–656 in the 3′-terminal region. In retroviruses autocomplementary motifs, underpin intermolecular RNA pairing (43) and Hepatitis C virus may have a similar mechanism (44). Fourth, the region between the 3′-strands of H2 and H3 helices is highly conserved and is highly accessible in all MFE and consensus models. Lastly, a tertiary interaction between regions nucleotides 391–419 and nucleotides 502–526 (triangles in Figure 8) was suggested by RNAz from two overlapping hybrid alignments with P = 0.47 and 0.39 (Supplementary Figure S5). This interaction was evaluated using ConStruct by joining subsection alignments of the two regions and large regions of sequence complementarity can be found between the two regions in most strains. However, a single consensus structure cannot be determined because of the varying position of interaction sites in different strains. We notice that many base pairs in this proposed tertiary interaction are formed between regions of high accessibility in our secondary-structure model with no conserved structure present. The long single-stranded region between 5′-strands of H2 and H3 may allow enough flexibility for H3 to flip towards H2 to form this tertiary interaction, bringing the start and stop codons of NSP6 into close proximity. More structure mapping data will be needed to prove whether this proposed tertiary structure can be formed.
DISCUSSION
This detailed and extensive study has evaluated sequence, codon and structural conservation in all 11 rotavirus RNA segments using all the available complete published sequences. All segments show structurally conserved elements and in some we have identified different alternative stable conformations. We also derived a complete in vitro secondary-structure model for RNA11 from a combination of in silico data and biochemical structure mapping. These data not only suggest the presence of cis-acting elements within the highly structured rotavirus (+)RNAs, but also illustrate the potential dynamics and multifunctionality of these RNA structures. Both sequence conservation and covariation are important in maintaining these conserved structures. Where strict sequence conservation is observed it is likely to be the result of multiple functional constraints. We were able to use knowledge of the functions of certain rotavirus proteins (VP1, VP6, NSP2 and NSP3) along with the RNA structures we identified to develop hypotheses on the association of these latter with particular functions.
For RNA conservation analyses, the ConStruct and RNAz programmes both have advantages and disadvantages. ConStruct allows the detailed analysis of conservation, stability and dynamics of structures within intact RNA, and enables analyses of a large number of sequences while applying sequence weight to minimize sampling bias. However, ConStruct is impractical for in silico analyses of larger RNAs and does not easily identify tertiary interactions and alternative conformations involving mutually exclusive structures. In contrast, RNAz allows high-throughput genomic screening of regions containing conserved local structures. By using hybrid alignments, we can detect potential LRIs, intermolecular interactions, mutually exclusive structures and tertiary interactions. However, the SVM model in RNAz was not designed for such a purpose, and the incorporated folding algorithm cannot directly predict some tertiary interactions such as kissing hairpins due to their pseudoknot character. Our analysis showed that RNAz is still highly useful in suggesting candidate functional structures because nearly all of the conserved interactions predicted by ConStruct with high-base pairing probabilities lead to a high P-score of ≥0.5 (and mostly >0.9) in the long-range RNAz analysis. Further developments of the RNAz programme may allow more accurate predictions of LRI and other RNA–RNA interactions. We showed that the two in silico tools when used together are very powerful in detecting potential local structures and LRIs. This highlights the importance of cross-validation of results obtained by different methodologies.
Previous studies of two single rotavirus sequences from separate strains have suggested that a terminally unpaired long-helical ‘panhandle’ may link the 5′- and 3′-termini in RNA8 and RNA11 (9,10). We found such structures, with evidence of evolutionary conservation, in all rotavirus RNA segments. Predicted conserved LRIs in the terminal regions of all RNA segments do not result from bias within the folding algorithms as they are powerfully supported by sequence and codon conservation data. These LRIs may facilitate RNA circularization which has been observed in many other viruses including HIV (45), orthomyxoviruses (46–52), enteroviruses (53) and hepatitis C virus (54). Circularization may be important for a number of steps in the rotavirus life cycle such as RNA replication (10,55), translation (9,10,55) or genome assortment. NSP3 interacts with the GACC-3′ 4-nt sequence at the 3′-termini of rotavirus as well as with the cap-binding eIF4GI (56), however these interactions may be insufficient for efficient circularization of the RNA molecule and the terminal LRIs may approximate the 5′- and 3′-terminal regions before the NSP3–eIF4GI interaction can occur. Furthermore, several terminal LRIs including those in RNA4, RNA8 and RNA11 link the stop codon to the start codon via a long-stacked helix and may have additional roles in translation reinitiation. LRI structures may also function to prevent 5′- and 3′-terminal sequences from base pairing with sequences in the central region of the RNA and thus maintain the structural integrity of functional terminal stem–loops and accessibility of sequences at the termini for intermolecular interactions. For example, replication of rotavirus (+)RNAs requires the binding of UGUG from the 3′CS UGUGACC-3′ (39,40) and possibly the 5′-cap structure to the viral RdRP VP1 (57,58).
The in vitro structure mapping data for RNA11 (Figure 7) support a previously proposed stem–loop formed by 12 nt at the 5′-terminus (9). Similar 5′-terminal structures were predicted by ConStruct in RNA1, 2, 4 and 6 (Supplementary Figure S4). However, alternative terminal conformations involving a short-LRI helix was predicted by ConStruct for RNA6 and RNA11. The small difference in free energy between the 5′-stem–loop and the short-LRI conformations suggests a flexible terminal structure which may adopt different conformations in vivo at different stages of the life cycle such as translation and replication. The 3′CS is likely unpaired in the conformation adopted during replication (10,40,59).
The proposed structural rearrangement between two alternative conformations of RNA9 is striking. Despite the extremely high-thermodynamic stability and sequence conservation, SL1 is incompatible with RNA replication since the UGUG nucloetides of the 3′CS form part of a highly stable helix. This is good evidence for the existence of distinct, function specific, structural conformations at different stages of the viral life cycle. Such a model may explain the presence of an extensive predicted molecular switch in RNA9. SL1 formation may be important for translation of NSP3, supported from the compatibility of SL1 with NSP3-binding and the shortened distance between the start and stop codon in the SL1 conformation. Replication requires switching from the SL1 to the LRI conformation triggered by VP1 binding to the 3′CS. Biochemical analyses will be needed to address the validity of such models.
The biochemical structure mapping analysis of RNA11 validated the presence of all conserved structures predicted by ConStruct at high probability and many others at medium to low probability. The presence of two highly conserved internal LRI helices, H2 and H3, is unique to RNA11 and may be particularly important to ensure correct folding of RNA11 and to isolate the intervening sequence between them, such that it folds independently of the remainder of the structure. This is most obvious between H2 and H3 where the formation of the two LRIs creates a highly structured region on the 5′-side and a highly unstructured region on the opposite side. The co-localization of conserved LRIs and isolated codon-conservation regions suggest that RNA sequence is important beyond the maintenance of two ORFs.
Several structures suggest translational control mechanisms. In RNA9, a second upstream initiation codon (AUG/CUG) exists at nucleotide 26 in frame with the probable authentic AUG nucleotide 35. This, together with the presence of strong structure, raises the possibility that translation initiation of NSP3 is mediated through an IRES (60,61). The identification of the conserved SL1 in RNA6 has provided us with structural insights to the previously described translation enhancement activity of the 3′-UTR of this RNA segment (37). IRESs are usually found in viral 5′-UTRs but there are well characterized examples of translational elements in the 3′-UTR such as the cap-independent translation elements found in many plant viruses (62). Because of the abundance of G:C base pairs, a possible function of the small stem–loop at the tip of SL1 may be to force ribosomal disassembly and facilitate the recycling of 40S ribosomal subunits at the 5′-cap. The conserved internal sequences and structures within RNA11 may be linked with the expression of the second ORF encoding NSP6. Currently, the translation mechanism used to express this protein is not known. Pulse/chase radio-labeling of virus infected cells using antiserum against NSP6 showed that it is expressed at a low but steady rate throughout the replication cycle and localizes to viroplasms within the cytoplasm (63). Expression in Rabbit Reticulocyte Lysate found the NSP5 and the NSP6 ORFs to be translated into proteins of the expected size and to be expressed in equimolar quantities (data not shown). Neither the NSP5 nor the NSP6 start codons are found in the most optimal Kozak start codon context (GCC(A/G)CCAUGG) (64,65). Expression of NSP6 is likely to be initiated through leaky scanning, with a proportion of ribosomes passing through the NSP5 start codon and starting translation at the NSP6 start codon. The potential tertiary interactions bringing the start and the stop codons of NSP6 in close spatial proximity and the high accessibility of sequence immediately before the NSP6 start codon suggest a ribosomal shunting reinitiation mechanism (66), as proposed for cauliflower mosaic virus (67), prototype foamy virus (68), Sendai virus (69) and adenovirus (70).
Packaging and replication of rotavirus (+)RNA are most likely controlled by the interaction between VP1 and the 3′CS (57), however currently there is no mechanism to explain the precision of genome assortment which ensures that one copy of each RNA segment is packaged per virion. In bacteriophage Phi6 which has a genome of 3 dsRNA segments, the availability of an in vitro assembly system (71,72) led to the proposal of a mechanism involving capsid expansion, exposure of RNA-binding sites on capsid, and sequential segment packaging (73,74). In rotavirus, the requirement of 11 different RNA-binding sites makes this model unlikely. It has been proposed that rotavirus RNA segments form inactive prereplication complexes with VP1 (57), and it is possible that these complexes are linked to each other through RNA–RNA interactions. Because many RNAs have strong intra-molecular structures, the helix destabilizing activity (75) of NSP2 in viroplasms may assist strand exchange reactions by reducing the kinetic barrier between intra- and intermolecular interactions to form inter-segmental linkages. Our data from RNA9 suggest that the formation of a prereplication complex and establishment of RNA–RNA interactions are likely to involve structural changes. Further in silico and in vitro analyses are needed to test these hypotheses.
The consistent finding of conserved structures at the segment termini suggests that they may contribute to packaging specificity. Reoviruses efficiently package internally deleted genomic RNA segments indicating that the packaging signals are located within the 200 nt at the 5′- and 3′-terminal regions (76,77). For influenza virus, another virus with a segmented RNA genome, specific packaging signals regions of 20–200 nt in length were identified in segments encoding NA (78), HA (79,80), NS (81), PB2, PB1 and PA (82–84). Codon variability studies suggested that the signals extended into the coding region from both 5′- and 3′-ends (15) and segment specificity of the signals was shown to influence influenza A virus segment re-assortment (85). Codon variability studies revealed 30–90 nt sequences of low variability at and near most of the segment termini where mapping had suggested packaging signals and in addition, previously unidentified conserved areas in the ORFs for PA, MA and PB2 (15).
This first comprehensive combined computational and biochemical analysis of rotavirus RNA has generated novel and unexpected insights into the pervasiveness, complexity and dynamic nature of RNA structures within the genome. A number of the questions raised by the data are currently under investigation, others await development of a universally applicable reverse genetics system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust (grant WT082031MA to A.M.L. and U.D.); Biomedical Research Centre (grant RG52162 to A.M.L); Studentship from the Medical Research Council (to J.C.vK.); Royal Society University Research Fellowship (to J.R.G.). Funding for open access charge: Wellcome Trust (grant WT082031MA to A.M.L. and U.D.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Suzanne Diston for secretarial assistance; Malcolm McCrae, University of Warwick, UK, for kindly providing the plasmid containing the RNA11 sequence of the bovine rotavirus UK strain; and Gerhard Steger, University of Düsseldorf, Germany, for advice on installing and using the ConStruct RNA consensus structure prediction package.
REFERENCES
- 1.Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 2006;12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- 3.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 4.Estes MK, Kapikian AZ. In: Fields’ Virology. 5th edn. Knipe DM, Howley PM, editors. Vol. II. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. pp. 1917–1974. [Google Scholar]
- 5.Cheung W, Gill M, Esposito A, Kaminski C, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gale M, Jr, Katze MG. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 7.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 8.Wentz MJ, Patton JT, Ramig RF. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J. Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortorici MA, Shapiro BA, Patton JT. A base-specific recognition signal in the 5′ consensus sequence of rotavirus plus-strand RNAs promotes replication of the double-stranded RNA genome segments. RNA. 2006;12:133–146. doi: 10.1261/rna.2122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Patton JT. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J. Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen HS, Nelbock P, Cochrane AW, Rosen CA. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990;247:845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- 13.Lever AM. HIV-1 RNA packaging. Adv. Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 14.Hundley F, Biryahwaho B, Gow M, Desselberger U. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology. 1985;143:88–103. doi: 10.1016/0042-6822(85)90099-6. [DOI] [PubMed] [Google Scholar]
- 15.Gog JR, Afonso Edos S, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruan J, Stormo GD, Zhang W. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics. 2004;20:58–66. doi: 10.1093/bioinformatics/btg373. [DOI] [PubMed] [Google Scholar]
- 20.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004;85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 23.Witwer C, Rauscher S, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 2001;29:5079–5089. doi: 10.1093/nar/29.24.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofacker IL, Stadler PF, Stocsits RR. Conserved RNA secondary structures in viral genomes: a survey. Bioinformatics. 2004;20:1495–1499. doi: 10.1093/bioinformatics/bth108. [DOI] [PubMed] [Google Scholar]
- 25.Wilm A, Linnenbrink K, Steger G. ConStruct: improved construction of RNA consensus structures. BMC Bioinformatics. 2008;9:219. doi: 10.1186/1471-2105-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber AR, Findeiss S, Washietl S, Hofacker IL, Stadler PF. RNAz 2.0: Improved Noncoding RNA Detection. Pac. Symp. Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]
- 27.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinformatics. 2009;26:12.2.1–12.2.16. doi: 10.1002/0471250953.bi1202s26. [DOI] [PubMed] [Google Scholar]
- 29.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowman HB, Draper DE. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- 32.Klarmann GJ, Schauber CA, Preston BD. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 1993;268:9793–9802. [PubMed] [Google Scholar]
- 33.Harrison GP, Mayo MS, Hunter E, Lever AM. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klasens BI, Huthoff HT, Das AT, Jeeninga RE, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 35.Lopez T, Rojas M, Ayala-Breton C, Lopez S, Arias CF. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 2005;86:1609–1617. doi: 10.1099/vir.0.80827-0. [DOI] [PubMed] [Google Scholar]
- 36.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008;153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang AD, Barro M, Gorziglia MI, Patton JT. Translation enhancer in the 3′-untranslated region of rotavirus gene 6 mRNA promotes expression of the major capsid protein VP6. Arch. Virol. 2004;149:303–321. doi: 10.1007/s00705-003-0211-9. [DOI] [PubMed] [Google Scholar]
- 38.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J. Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortorici MA, Broering TJ, Nibert ML, Patton JT. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 2003;278:32673–32682. doi: 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]
- 40.Chen D, Barros M, Spencer E, Patton JT. Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology. 2001;282:221–229. doi: 10.1006/viro.2001.0825. [DOI] [PubMed] [Google Scholar]
- 41.Poncet D, Aponte C, Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J. Virol. 1993;67:3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poncet D, Laurent S, Cohen J. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greatorex JS, Laisse V, Dockhelar MC, Lever AM. Sequences involved in the dimerisation of human T cell leukaemia virus type-1 RNA. Nucleic Acids Res. 1996;24:2919–2923. doi: 10.1093/nar/24.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shetty S, Kim S, Shimakami T, Lemon SM, Mihailescu MR. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–925. doi: 10.1261/rna.1960410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35:5253–5261. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desselberger U, Racaniello VR, Zazra JJ, Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 47.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leahy MB, Dessens JT, Pritlove DC, Nuttall PA. The Thogoto orthomyxovirus cRNA promoter functions as a panhandle but does not stimulate cap snatching in vitro. J. Gen. Virol. 1998;79:457–460. doi: 10.1099/0022-1317-79-3-457. [DOI] [PubMed] [Google Scholar]
- 50.Lee YS, Seong BL. Nucleotides in the panhandle structure of the influenza B virus virion RNA are involved in the specificity between influenza A and B viruses. J. Gen. Virol. 1998;79:673–681. doi: 10.1099/0022-1317-79-4-673. [DOI] [PubMed] [Google Scholar]
- 51.Pritlove DC, Fodor E, Seong BL, Brownlee GG. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 1995;76:2205–2213. doi: 10.1099/0022-1317-76-9-2205. [DOI] [PubMed] [Google Scholar]
- 52.Weber F, Haller O, Kochs G. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch. Virol. 1997;142:1029–1033. doi: 10.1007/s007050050138. [DOI] [PubMed] [Google Scholar]
- 53.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 55.Patton JT. Rotavirus RNA replication and gene expression. Novartis Found Symp. 2001;238:64–77. doi: 10.1002/0470846534.ch5. [DOI] [PubMed] [Google Scholar]
- 56.Vende P, Piron M, Castagne N, Poncet D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 2000;74:7064–7071. doi: 10.1128/jvi.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guglielmi KM, McDonald SM, Patton JT. Mechanism of intraparticle synthesis of the rotavirus double-stranded RNA genome. J. Biol. Chem. 2010;285:18123–18128. doi: 10.1074/jbc.R110.117671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barro M, Mandiola P, Chen D, Patton JT, Spencer E. Identification of sequences in rotavirus mRNAs important for minus strand synthesis using antisense oligonucleotides. Virology. 2001;288:71–80. doi: 10.1006/viro.2001.1054. [DOI] [PubMed] [Google Scholar]
- 60.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 61.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem. Soc. Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rainsford EW, McCrae MA. Characterization of the NSP6 protein product of rotavirus gene 11. Virus Res. 2007;130:193–201. doi: 10.1016/j.virusres.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schepetilnikov M, Schott G, Katsarou K, Thiebeauld O, Keller M, Ryabova LA. Molecular dissection of the prototype foamy virus (PFV) RNA 5′-UTR identifies essential elements of a ribosomal shunt. Nucleic Acids Res. 2009;37:5838–5847. doi: 10.1093/nar/gkp609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yueh A, Schneider RJ. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 71.Olkkonen VM, Gottlieb P, Strassman J, Qiao XY, Bamford DH, Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage phi 6: formation of a recombinant double-stranded RNA virus. Proc. Natl Acad. Sci. USA. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poranen MM, Paatero AO, Tuma R, Bamford DH. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell. 2001;7:845–854. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- 73.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol. Mol. Biol. Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 75.Taraporewala ZF, Patton JT. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou S, Brown EG. Identification of sequence elements containing signals for replication and encapsidation of the reovirus M1 genome segment. Virology. 1992;186:377–388. doi: 10.1016/0042-6822(92)90003-8. [DOI] [PubMed] [Google Scholar]
- 77.Ni Y, Kemp MC. Subgenomic S1 segments are packaged by avian reovirus defective interfering particles having an S1 segment deletion. Virus Res. 1994;32:329–342. doi: 10.1016/0168-1702(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 78.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl Acad. Sci. USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 2003;77:10575–10583. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 2005;79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marsh GA, Rabadan R, Levine AJ, Palese P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 2008;82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao Q, Palese P. Rewiring the RNAs of influenza virus to prevent reassortment. Proc. Natl Acad. Sci. USA. 2009;106:15891–15896. doi: 10.1073/pnas.0908897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.