Abstract
DNA topoisomerases manage chromosome supercoiling and organization in all forms of life. Gyrase, a prokaryotic heterotetrameric type IIA topo, introduces negative supercoils into DNA by an ATP-dependent strand passage mechanism. All gyrase orthologs rely on a homologous set of catalytic domains for function; however, these enzymes also can possess species-specific auxiliary regions. The gyrases of many gram-negative bacteria harbor a 170-amino acid insertion of unknown architecture and function in the metal- and DNA-binding TOPRIM domain of the GyrB subunit. We have determined the structure of the 212 kDa Escherichia coli gyrase DNA binding and cleavage core containing this insert to 3.1 Å resolution. We find that the insert adopts a novel, extended fold that braces the GyrB TOPRIM domain against the coiled-coil arms of its partner GyrA subunit. Structure-guided deletion of the insert greatly reduces the DNA binding, supercoiling and DNA-stimulated ATPase activities of gyrase. Mutation of a single amino acid at the contact point between the insert and GyrA more modestly impairs supercoiling and ATP turnover, and does not affect DNA binding. Our data indicate that the insert has two functions, acting as a steric buttress to pre-configure the primary DNA-binding site, and serving as a relay that may help coordinate communication between different functional domains.
INTRODUCTION
DNA topoisomerases (topos) are ubiquitous, essential enzymes that manage supercoiling homeostasis and chromosome topology in all forms of life. Most organisms possess a complement of multiple topoisomerase types, each of which is responsible for a different cellular task (1–3). For example, in Escherichia coli, the type IA topo I relaxes negative supercoils (4–6), whereas its paralog, topo III, helps resolve catenanes and Holliday junctions produced by replication and recombination (7,8). Catenanes and knots are also unlinked by the type IIA enzyme, topo IV (9–11), while its a paralogous counterpart, gyrase, negatively supercoils DNA (5,12).
Type IIA topos, including gyrase, topo IV and eukaryotic topo II, use several conserved catalytic domains to actively pass one duplex DNA through a double-stranded break in another. Strand passage is coordinated by a so-called ‘two-gate’ mechanism, in which the transported DNA duplex (the T-segment) is navigated through the cleaved DNA (the G-segment) by the movement of multiple, dissociable subunit interfaces, or ‘gates’ (13–16). The reaction begins with the binding of G-segment DNA to the central region of the enzyme, the DNA-gate, an interface composed of a pair of metal-binding topo/primase (TOPRIM) domains and two dimer-related winged-helix domains (WHDs). T-segment capture is accomplished upon the nucleotide-dependent dimerization a second interface in the protein, the ATP-gate (17–21). This event stimulates G-segment cleavage, a reaction catalyzed by both a catalytic tyrosine residue present on each WHD and a pair of divalent metal ions liganded by each TOPRIM fold (22–27); ATP hydrolysis and release of inorganic phosphate facilitate passage of the T-segment through the broken DNA (28–31). At the end of the topoisomerase cycle, the T-segment exits the enzyme through a third subunit interface (the C-gate) formed by a pair of small globular domains that each sit at the terminus of a coiled-coil segment (14,16,22,32).
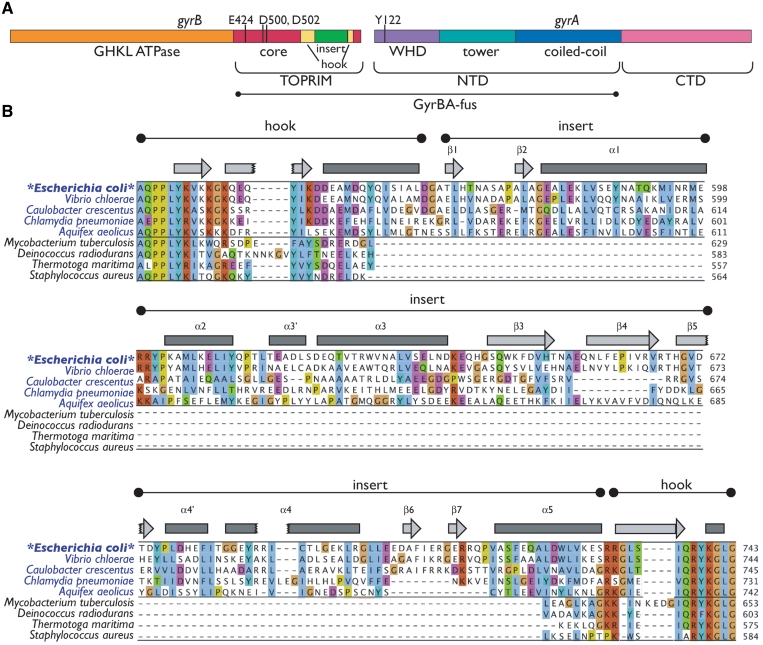
In bacterial type IIA topos, the gyrB and parE genes encode the ATPase and TOPRIM domains of gyrase and topo IV, while the winged-helix and C-gate domains are encoded by gyrA and parC (Figure 1A). The GyrA and ParC subunits also contain a C-terminal, DNA-binding domain that that contributes to the differential substrate specificities of the two paralogs (33,34). Thus far, nearly all of the domains found within type IIA topos have been visualized by X-ray crystallography. One exception is a 170-amino acid region found within the GyrB subunit in a number of bacterial lineages, including proteobacteria, acidobacteria, chlamydia and aquificales (Figure 1B). This insertion, which shares no sequence homology with known proteins, has been proposed to form a distinct structural domain that participates in DNA binding (35,36); however, the overall architecture of the insert and its exact role in gyrase function have remained enigmatic.
Figure 1.
Escherichia coli gyrase primary and secondary structure. (A) GyrA and GyrB domain organization. Catalytic modules of E. coli gyrase are indicated as bars. The regions encompassed by the GyrBA fusion are indicated. (B) Alignment of the GyrB insert region. Selected GyrB genes were aligned using MUSCLE (68,69). Organisms with GyrB genes containing an insert are shown in blue; those with GyrB genes lacking inserts, in black. Regions corresponding to the hook and insert are indicated, as are secondary structure elements as identified in the structure (bars—α-helices; arrows—β-sheets). Insert elements are numbered by secondary structure element as in Figure 3B. Amino acid numbering is at right. The alignment is colored by ClustalX score within the two major subgroups (GyrBs with and without inserts). Note the poor conservation of the insert compared to the hook region. This figure was prepared using Jalview (70).
As a means to understand the role of the insert in gyrase’s mechanism, we determined the 3.1 Å crystal structure of the E. coli GyrB insert in the context of the heterotetrameric GyrB–GyrA DNA binding and cleavage core. This construct encompasses the TOPRIM and insert domains of GyrB and the WHD and C-gate region of GyrA. The insert, which adopts a novel fold, forms an extended element that contacts the coiled-coil arms of its GyrA partner. Using the structure as a guide, we designed precise deletion and point mutants of the insert and found that these variants are defective for a wide range of gyrase activities. Our results indicate that the insert acts as a buttressing element to help pre-orient the DNA gate for efficient G-segment binding, and that it further aids strand passage, possibly by acting as an allosteric relay between distal enzyme gates.
MATERIALS AND METHODS
Construction of a GyrB–GyrA fusion protein for crystallography
To simplify crystallization of a tetrameric GyrB2GyrA2 construct, we created a truncated GyrB–GyrA protein that fuses amino acids 1–525 of GyrA directly to residues 388–804 of GyrB. The coding region for this construct was cloned into a ligation-independent cloning plasmid behind an N-terminal, tobacco etch protease (TEV)-cleavable, hexahistidine-maltose binding protein (His6-MBP) tag, creating the pLIC172-escoGyrBAfus vector.
Expression and purification of selenomethionine-derivatized E. coli GyrBA-fus
For expression, pLIC172-escoGyrBAfus was transformed into E. coli BL21-CodonPlus(DE3)-RIL competent cells (Stratagene). One-Liter cultures were grown in minimal media in 2 l baffled flasks at 37°C to an optical density (A600 nm) of 0.4–0.6. An amino acid cocktail containing leucine, isoleucine, valine, phenylalanine, lysine, threonine and seleno-methionine was added to shut down methionine biosynthesis (37), cultures were transferred to 18–20°C for 30 min, and protein expression was induced by the addition of 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation after ∼16 h, resuspended in buffer A800′ (20 mM HEPES pH 7.5, 800 mM NaCl, 30 mM imidazole pH 8.0, 10% glycerol, 1 mM PMSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 5 mM βME) and frozen dropwise in liquid nitrogen.
For protein purification, cell pellets were thawed and sonicated, and the lysate clarified by centrifugation. The lysate was then passed over a Ni2+-affinity column (GE) and washed with buffer A800′, followed by buffer A400′ (buffer A800′ containing 400 mM NaCl). Protein was eluted with buffer B400′ (buffer A400′ containing 500 mM imidazole), concentrated using Amicon centrifugal filter devices, mixed with His6-tagged TEV protease (38) using a ratio of 1:50–1:10 (wt/wt) TEV protease:protein, and dialyzed overnight at 4°C in a 10 000 molecular weight cut-off (MWCO) SlideAlyzer (Pierce) against buffer A400′. The dialyzed mixture was passed over a Ni2+-affinity column, and the flow-through was collected, concentrated and exchanged into buffer C125′ (20 mM HEPES pH 7.5, 125 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM TCEP). The sample was then adsorbed to an anion exchange column (HiTrap HQ, GE) equilibrated in buffer C125′ and eluted using a 125–300 mM NaCl gradient over 60 column volumes. Peak fractions were pooled, concentrated and run over a size exclusion column (Sephacryl S300, GE) equilibrated in buffer SE′ (50 mM HEPES pH 7.5, 500 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM TCEP) to remove aggregated species. Fractions corresponding to the dimer peak were pooled and concentrated, then dialyzed into crystallization buffer (10 mM HEPES pH 7.5, 50 mM KCl and 1 mM TCEP) using a Harvard Apparatus MicroDispoDialyzer.
Crystallization of GyrBA-fus
Crystals of selenomethionine-derivatized GyrBA-fus were grown by hanging-drop vapor diffusion at 18°C by mixing 0.1 µl of protein (1.8 mg/ml) with 0.1 µl of crystallization solution (90 mM imidazole pH 8.0, 11% isopropanol, 18 mM MgCl2, 10 mM spermidine, 3% pentaerythritol ethoxylate 15/4). Crystals grew as 3D trapezoids with two wedge-like ends abutting a thicker, central region. Crystals were cryoprotected by exchanging the drop into harvest solution (2% pentaerythritol ethoxylate 15/4, 6.5 mM spermine, 60 mM imidazole pH 8.0, 12 mM MgCl2, 12.1% isopropanol, 6. mM HEPES pH 7.5, 26.2 mM KCl, 1 mM TCEP, 20% PEG400) prior to harvesting and flash-freezing in liquid nitrogen.
Structure determination
Diffraction data were collected from a single crystal at the Advanced Photon Source microfocus beamline NE-CAT 24-ID-E at the selenium edge (0.9792 Å). Multiple volumes of the crystals were exposed during data collection; regions that yielded data of the highest quality (in terms of mosaicity and I/σ) were merged and processed in HKL2000 (39) (Table 1). A combination of molecular replacement (MR) and single-wavelength anomalous dispersion (SAD) as implemented in PHENIX were used to determine phases (40). A MR solution was obtained using the structure of the E. coli GyrA N-terminal domain (PDBID 1AB4) as a search model (41). These MR phases were used to locate selenium sites, which were then used for SAD phasing (40). The TOPRIM and insert domains, which were not part of the MR search model, were built de novo into SAD-phased maps using COOT (42). Rounds of manual rebuilding and refinement were accomplished using COOT and PHENIX (40,42). TLS groups for refinement were chosen using the TLSMD server (43,44). Stereochemistry and clashes were resolved using MolProbity and COOT (42,45).
Table 1.
Data collection, refinement and stereochemistry
| Data collection | |
|---|---|
| Resolution (Angstroms) (last shell) | 40.0–3.1 (3.2–3.1) |
| Wavelength (Angstroms) | 0.97920 |
| Space group | C2221 |
| Unit cell dimensions (Angstroms, a, b, c) | 108.2,147.4,139.0 |
| Unit cell angles (deg., α, β, γ) | 90,90,90 |
| I/σ (last shell) | 15.0 (3.8) |
| Rsym (last shell) | 0.103 (0.387) |
| Percentage completeness (last shell) | 99.6 (99.6) |
| No. of reflections | 392 313 |
| No. of unique reflections | 38 518 |
| Redundancy (last shell) | 5.0 (4.6) |
| Selenium phasing | |
| No. of sites | 22,27 |
| Figure of merit | 0.39 |
| Refinement | |
| Resolution (Angstroms) | 36–3.1 |
| No. of reflections | 38 485 |
| No. of working reflections | |
| No. of free reflections (% total) | 3778 (10%) |
| Rwork | 23.4% |
| Rfree | 28.2% |
| Structure and Stereochemistry | |
| No. of atoms | 6677 |
| Protein | 6666 |
| Water | 9 |
| Magnesium | 2 |
| R.m.s.d. bond length (Angstroms) | 0.003 |
| R.m.s.d. bond angles (deg.) | 0.618 |
Rsym = ΣΣj | Ij − 〈I〉 / Σ Ij, where Ij is the intensity of the measurement for reflection j and 〈I〉 is the mean intensity for multiply-recorded reflections.
Rwork,free = Σ||Fobs | - |Fcalc || / |Fobs, where the working and free R-factors are calculated using the working and free reflection sets. The free reflections were held aside during refinement.
Cloning and expression of GyrA and GyrB constructs for biochemical studies
The coding region for full-length E. coli GyrA and GyrB, the E. coli GyrB insert (Ala560–Ser735) and CTDless GyrA (Ser2–Thr522, also called GyrAΔCTD) were amplified from either genomic DNA or pre-existing vectors and cloned into either vector pLIC172 (an in-house LIC vector) or pET28b (Novagen). Both vectors include N-terminal hexahistidine (His6) tags upstream of TEV cleavage sites; pLIC172 also contains a maltose-binding protein (MBP) tag between the His6 tag and the TEV site. GyrB point mutants H669A and H669E were created using QuikChange (Stratagene). A GyrB construct lacking the insert (GyrBΔinsert), in which GyrB residues Thr565–Arg731 were replaced with a Gly–Ser–Ser–Gly linker, was created by separately amplifying two segments of the enzyme from the existing GyrB expression vector, then fusing them together in a second PCR step.
Protein expression was carried out in either E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) or E. coli Arctic Express cells (Stratagene). In general, 1 l cultures were grown at 37°C in 2×YT media in 2 l baffled flasks to an A600 nm of 0.4–0.6. Protein expression was induced with 0.25–0.5 mM IPTG. For GyrA and GyrAΔCTD expression, induction was carried out for 4 h at 37°C. All other proteins were expressed for 12–16 h at 18–20°C. Cells were harvested by centrifugation, resuspended in specific buffers (see below), and frozen dropwise in liquid nitrogen before storing at −80°C.
Purification of wild-type E. coli GyrA
Frozen cells resuspended in buffer A1000 (50 mM HEPES pH 7.5, 20 mM imidazole pH 8.0, 1000 mM NaCl, 10% glycerol, 2 mM βME, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mM PMSF) were sonicated and centrifuged, and the clarified lysate passed over a Ni2+-affinity column. The column was washed with buffer A1000 followed by buffer A100 (buffer A1000 containing only 100 mM NaCl). Protein was eluted with buffer B100 (buffer A100 containing 500 mM imidazole). Peak fractions were pooled, concentrated and incubated overnight at 4°C with His6-tagged TEV protease as described for GyrBA-fus. This mixture was then run through a Ni2+-affinity column, and the flow-through concentrated and passed over a Sephacryl S300 gel filtration column equilibrated in 50 mM HEPES pH 7.5, 400 mM KCl, 1 mM EDTA pH 8.0, 2 mM βME, 10% glycerol. Fractions containing GyrA were identified using SDS–PAGE/Coomassie staining, then pooled and concentrated. Glycerol was added to a final concentration of 19%, and protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Final storage conditions were 45 mM HEPES pH 7.5, 360 mM KCl, 0.9 mM EDTA pH 8.0, 1.8 mM βME, 19% glycerol.
Purification of wild-type, Δinsert, H669A and H669E E. coli GyrB
Frozen cells expressing the appropriate proteins and resuspended in buffer A800 (20 mM HEPES pH 7.5, 800 mM NaCl, 30 mM imidazole pH 8.0, 10% glycerol, 1 mM PMSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 2 mM βME) were thawed and sonicated and the lysate clarified by centrifugation. Clarified lysate was passed over a Ni2+-affinity column and washed with buffer A800 followed by buffer A50 (buffer A800 containing 50 mM NaCl). Bound protein was eluted from the Ni2+ column directly to an anion exchange column (HiTrap HQ) using buffer B50 (buffer A50 containing 500 mM imidazole). This column was then washed with buffer C50 (20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, 10% glycerol, 2 mM βME), and bound protein eluted using a 24 column-volume gradient to 100% buffer C500 (buffer C50 containing 500 mM NaCl). Fractions containing the protein of interest were identified by SDS–PAGE/Coomassie staining, pooled and concentrated by centrifugal filtration. Following TEV protease cleavage as per GyrBA-fus, the dialyzed protein was passed over a second Ni2+-affinity column. The flow-through was collected, concentrated by centrifugal filtration, and passed over a Sephacryl S300 gel filtration column equilibrated in buffer SE (50 mM HEPES pH 7.5, 500 mM KCl, 1 mM EDTA, 10% glycerol, 2 mM βME). For initial preps, individual fractions were tested for activity. Fractions corresponding to the dimer and monomer forms of the protein exhibited consistent, high activity, and so only these fractions were pooled and concentrated. Glycerol was added to concentrated fractions to a final concentration of 30%, and protein was aliquoted and snap-frozen in liquid nitrogen and stored at −80°C. Final storage conditions were 50 mM HEPES pH 7.5, 500 mM KCl, 1 mM EDTA pH 8.0, 2 mM βME, 30% glycerol.
Purification of the isolated E. coli GyrB-insert
For protein purification, cells resuspended in buffer A800 were sonicated and centrifuged, and the clarified lysate passed over a Ni2+-affinity column. The column was washed with buffer A800 followed by buffer A400. Protein was then eluted with buffer B400 (buffer A400 containing 500 mM imidazole), and peak fractions were pooled, concentrated and treated with TEV protease as described for GyrBA-fus. This mixture was then passed over a Ni2+-affinity column, and the flow-through collected, concentrated and run over a Sephacryl S200 gel filtration column equilibrated in buffer SE. Fractions containing the insert were identified by SDS–PAGE/Coomassie staining, then pooled and concentrated by centrifugal filtration. Glycerol was added to a final concentration of 30%, and protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Final storage conditions were 50 mM HEPES pH 7.5, 500 mM KCl, 1 mM EDTA pH 8.0, 2 mM βME, 30% glycerol.
Purification of CTD-less E. coli GyrA
Purification was carried out as for the GyrB insert, except that a Sephacryl S300 size exclusion column was used and equilibrated in buffer SE-200 (50 mM HEPES pH 7.5, 200 mM KCl, 1 mM EDTA, 2 mM βME, 10% glycerol). Peak fractions were chosen by SDS–PAGE/Coomassie staining, glycerol was added to a final concentration of 19%, and protein aliquots were snap-frozen in liquid nitrogen and stored at −80°C. Final storage conditions were 45 mM HEPES pH 7.5, 180 mM KCl, 0.9 mM EDTA, 1.8 mM βME, 19% glycerol.
Negative supercoiling assays
Negatively-supercoiled plasmid DNA [pSG483, a derivative of pBluescript (pBSK)] was purified using a Macherey–Nagel maxiprep kit and relaxed with DNA topo I prepared in-house from wheat germ (46). Wild-type, GyrBΔinsert, GyrB–H669A or GyrB–H669E gyrase tetramer at varying concentrations were each incubated with 700 ng (350 fmol) plasmid in 20 µl reactions for 30 min at 37°C. Assay conditions were 30 mM Tris–HCl pH 7.5, 30 mM potassium glutamate, 2 mM MgCl2, 7% glycerol, 1 mM DTT and 1 mM ATP, with an additional ∼50 mM potassium chloride and ∼3% glycerol contributed from the protein storage buffer. Reactions were stopped with the addition of 1% SDS and 10 mM EDTA (final concentrations). Sucrose loading dye was added to samples, and samples were run on a 1% agarose, 0.5 × TBE gel at 1–1.5 V/cm for 14–18 h. Gels were stained with 0.5 μg/ml ethidium bromide (EtBr) in 0.5 × TBE for 30 min, followed by a brief destaining in 0.5 × TBE. Bands were visualized by UV transillumination.
Positive supercoil relaxation assays
Positively-supercoiled plasmid DNA (pSG483) was prepared by treating relaxed plasmid with A. fulgidus reverse gyrase as described by Rodriguez (47). Reactions were carried out as described for negative supercoil introduction.
Negative supercoil relaxation assays
Negatively-supercoiled plasmid DNA (pSG483) was prepared using a maxiprep plasmid purification kit (Macherey–Nagel). Assays were carried out as described earlier for negative supercoiling assays, except ATP was omitted.
Cleavage assays
Negatively-supercoiled plasmid DNA (pBSK) was prepared by CsCl centrifugation. Increasing amounts of gyrase were incubated with 700 ng (350 fmol) pBSK for 30 min in 20 µl reactions at 37°C. Reaction conditions were as follows: 30 mM Tris–HCl pH 7.5, 30 mM potassium glutamate, 2 mM MgCl2, 7% glycerol, 1 mM DTT and 1 mM ciprofloxacin, with an additional ∼50 mM KCl and ∼3% glycerol contributed from the protein storage buffer. Reactions were stopped by the addition of SDS to 1%, followed by the addition of EDTA to 10 mM. Samples were incubated with 0.1 mg/ml proteinase K for 1 h at 37°C, combined with sucrose loading dye, and loaded onto 1% agarose/0.5 × TBE gels. Gels were run at 1.5 V/ cm for 14–18 h and stained with 0.5 μg/ml EtBr in 0.5 × TBE for 30 min. Gels were then destained briefly in 0.5 × TBE and bands visualized by UV transillumination. A linearized pBSK standard was generated by cleaving the negatively-supercoiled DNA with BamHI (New England Biolabs).
DNA-binding assays
DNA binding was visualized by electrophoretic mobility shift assays (EMSAs). The DNA substrate for binding was a 50-bp segment of plasmid pBR322 shown previously to contain a strong gyrase-binding site (48). Two complementary single-stranded oligos (5′-GCG AGA AGA ATC ATA ATG GGG AAG GCC ATC CAG CCT CGC GTC GCG CAA CG-3′; 5′-/6-FAM/ CGT TGC GCG ACG CGA GGC TGG ATG GCC TTC CCC ATT ATG ATT CTT CTC GC-3′,where 6-FAM indicates the position of a fluorescein label for visualization) were obtained from Integrated DNA Technologies and individually resuspended in 100 µl annealing buffer (10 mM Tris–HCl pH 7.5, 50 mM KCl, 5 mM MgCl2) to ∼500 µM final concentration. The two oligos were then combined in equimolar amounts and annealed by boiling in a 1 l water bath for 5 min and allowing the bath to cool to room temperature overnight. Double-stranded DNA was purified by native PAGE and stored in annealing buffer.
Tetrameric gyrase was reconstituted from wild-type, Δinsert, H668A or H668E E. coli GyrB and E. coli GyrAΔCTD at varying concentrations and incubated with 75 nM of the labeled double-stranded 50mer for 30 min at room temperature in 25 µl reactions containing 30 mM Tris–HCl pH 7.5, 2 mM MgCl2, 6% glycerol, 1 mM DTT; the protein storage buffer contributed an additional ∼75 mM KCl and 4.5% glycerol. Reactions were loaded directly onto a 1.8% low-melting temperature agarose gel (SeaPlaque agarose, FMC Bioproducts). Gel running buffer consisted of 45 mM Tris–borate and 2.5 mM MgCl2. Gels were run for ∼3 h at 4.3 V/cm, and DNA was visualized using a Typhoon fluorescent scanner (GE Healthcare), with excitation at 532 nm.
ATPase activity assays
Wild-type and mutant gyrase ATPase activities were measured using an established enzyme-coupled assay (49,50) in which the hydrolysis of ATP to ADP is coupled to the oxidation of NADH to NAD+. To determine standard kinetic parameters (KM, kcat), 0.2 µM tetrameric wild-type, GyrBΔinsert, GyrB–H669A or GyrB-H669E gyrase was incubated with 0–3 mM ATP in 75 µl reactions. Reactions contained 50 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 5 mM βME, 0.1 mg/ml bovine serum albumin, 2 mM phosphoenol pyruvate (PEP), 0.2 mM freshly-prepared β-nicotinamide adenine dinucleotide (NADH), and an excess of pyruvate kinase and lactate dehydrogenase (enzyme mix obtained from Sigma). Assays were performed at 37°C. Oxidation of NADH to NAD+ was monitored by measuring absorbance at 340 nm in a Perkin Elmer Victor 3V 1420 multilabel plate reader. The rate of decrease in absorbance was converted to micromoles of ATP hydrolyzed to ADP using an NADH standard curve and the assumption that one ATP is hydrolyzed for every NADH oxidized. ATPase rate as a function of [ATP] was fit for KM and Vmax using Michaelis–Menten kinetics in KaleidaGraph (Synergy Software).
DNA stimulation of ATPase activity was measured using assays performed as described earlier, except that the concentration of ATP was held constant at 0.5 mM, and reactions were supplemented with varying amounts of sheared, purified salmon sperm DNA (Sigma) resuspended in 10 mM Tris pH 7.5, 50 mM KCl, 5 mM MgCl2.
RESULTS
The E. coli GyrBA-fusion protein forms a homodimer
To first visualize the GyrB insert in the context of the DNA- and C-gates, we set out to crystallize the entire 212 kDa DNA binding and cleavage core of E. coli gyrase. To prevent subunit dissociation and simplify crystallization, we constructed a fusion protein that couples residues Ala388–Ile804 of E. coli GyrB directly to residues Met1–Ser525 of E. coli GyrA, linking the two proteins through their poorly-conserved C- and N-termini, respectively. Similar subunit fusions have been shown previously to support function of full-length topo IV and gyrase (51,52). The fusion construct, hereafter termed GyrBA-fus, includes the GyrB TOPRIM and insert, along with the WHD, coiled-coil and C-gate domain of GyrA (Figure 1A). Because GyrBA-fus lacks the GyrB ATPase and GyrA C-terminal DNA-binding domains, it is unable to supercoil DNA, but can support DNA cleavage (not shown). We crystallized selenomethionine-derivatized GyrBA-fus in space group C2221 and solved its structure to 3.1 Å resolution. The final model was refined to Rwork/Rfree values of 23.4/28.2%, with good stereochemistry (Table 1).
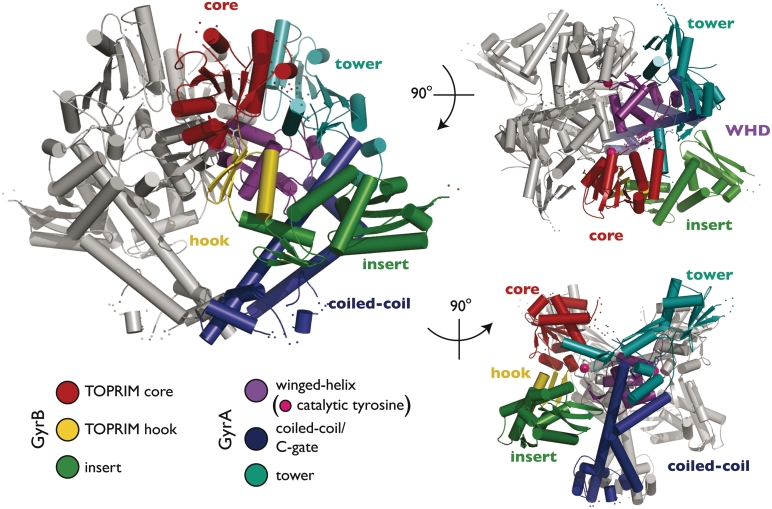
Crystals of GyrBA-fus contain one monomer per asymmetric unit. A crystallographic 2-fold axis recapitulates a dimeric particle, in which the GyrA winged-helix and C-gate domains form homomeric interactions as seen in other structures of this region (Figure 2) (41,53–55, PDBID 2INR). As expected, the E. coli GyrB TOPRIM fold is similar in structure to homologous elements from yeast and bacterial type IIA topos, except for the relative position of a small C-terminal extension that emanates from the central metal-binding domain. This region, sometimes termed the ‘tail’ or ‘hook,’ has been shown previously to be flexible with respect to the TOPRIM core. In GyrBA-fus, the hook adopts an orientation similar to that seen in the isolated Mycobacterium tuberculosis GyrB TOPRIM fold (56), which lies between more extreme conformational states observed for Saccharomyces cerevisiae topo II and Streptococcus pneumoniae topo IV (Supplementary Figure S1) (22,23,57,58). There is no electron density visible for several amino acids at the extreme C- and N-termini of the GyrB and GyrA regions, respectively, indicating that the fusion has not constrained the relative orientations of the two subunits.
Figure 2.
GyrBA-fus structure. The structure of the GyrBA-fus dimer is shown in cartoon. Subdomains are colored as indicated. One protomer is shown in gray, except for the α−carbons of the catalytic tyrosines, both of which are represented as magenta spheres. This and other structure figures were prepared in PyMOL (71).
The insert bears little resemblance to known protein structures
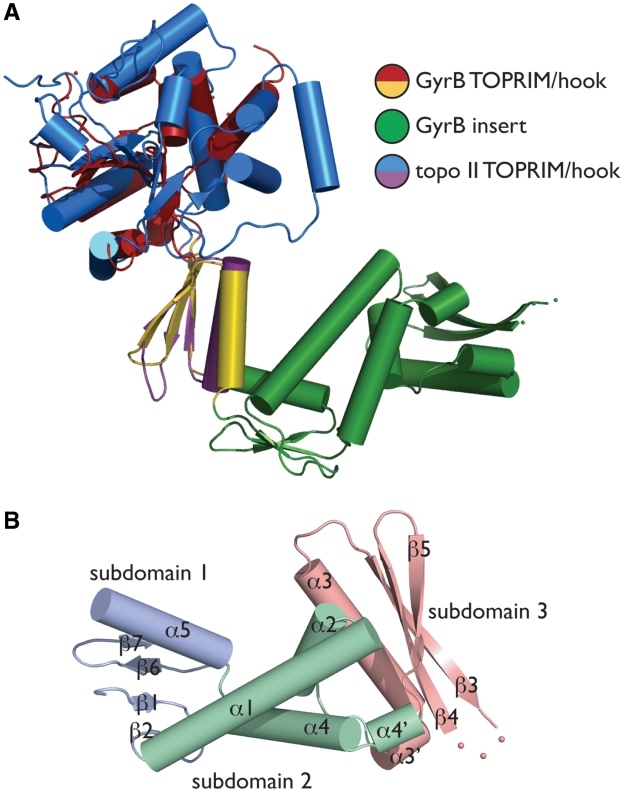
Although the general shape and organization of the GyrB insert has been analyzed by secondary-structure analysis and small-angle x-ray scattering (SAXS) (35,36), our model provides the first high-resolution view of this domain. The insert protrudes from the extreme tip of the TOPRIM hook, away from the central metal-binding fold (Figure 3A). The element is composed of three subdomains: a short, four-stranded β-sheet packed against a single α-helix (subdomain 1, residues Asp562–Gly577 and Asp704–Arg732), a central three-helix bundle (subdomain 2, Glu578–Thr616 and Gly684–Glu703), and a longer three-stranded β-sheet packed against a large helix (subdomain 3, residues Glu617–Thr683). Topologically, subdomain 3 is the only region formed by sequentially contiguous secondary structure elements (Figures 1B and 3B).
Figure 3.
GyrB insert structure and organization. (A) Insert/TOPRIM orientation. The E.coli GyrB TOPRIM is shown aligned with a DNA-bound form of the S. cerevisiae topo II TOPRIM [PDB ID 2RGR (22)]. (B) Insert substructure. The three subdomains of the insert are shown and labeled as per Figure 2.
The mixed α/β fold and extended shape of the insert is largely in agreement with SAXS envelopes for the region, although the fold is distinct from prior predictions (36). Indeed, the overall fold of the GyrB insert appears to be unique, with a DALI search using the isolated insert domain revealing limited structural homology to a small scattering of proteins, nearly all of which appear to be incidental (Z-scores < 5) (59). The best alignment (Z-score 4.4) is between a portion of the insert (subdomain 3) and the C-terminal region of the Desulfitobacterium hafniense tail sheath protein, which adopts a phage sheath 1 superfamily fold. This similarity encompasses only 50 amino acids of the insert with a Cα r.m.s.d. of 2.6 Å (Supplementary Figure S2) (PDBID 3HXL).
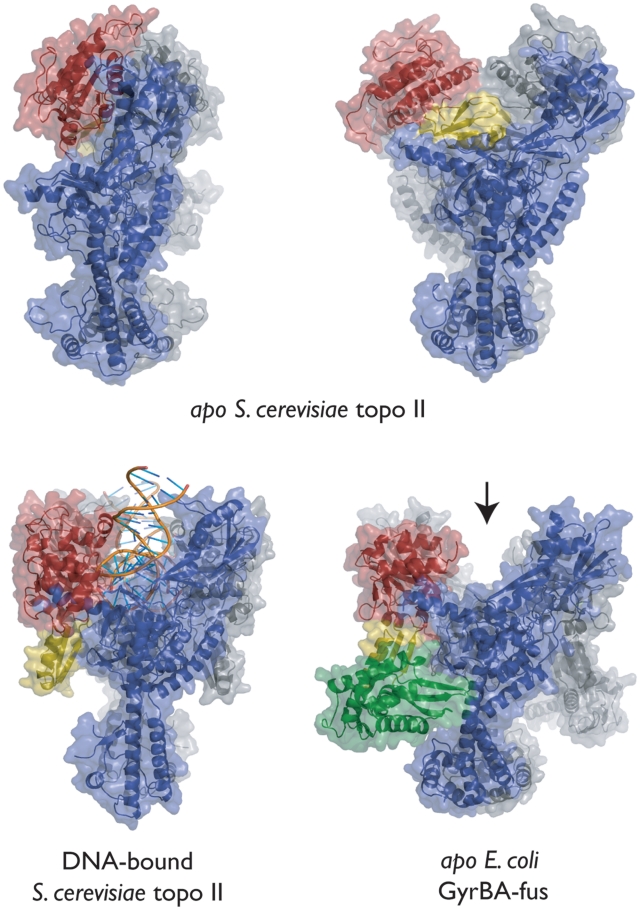
The GyrBA-fus conformation resembles DNA-bound type IIA topo states
There currently exist four models of the type IIA topoisomerase DNA binding and cleavage core, two solved in the absence of DNA and two in its presence (22,23,57,58). These structures have shown that the relative orientation of the TOPRIM domains, which constitute a large portion of the DNA gate, can be extremely plastic. Thus far, the TOPRIM domains have been visualized in three distinct conformations relative to the WHDs. In two of these states, both of which were observed in DNA-free S. cerevisiae topo II, the position of the TOPRIM domains appears to preclude the binding of a G-segment DNA (Figure 4) (57,58). In the third, seen in the DNA-bound complexes of both topo II and S. pneumoniae topo IV (22,23), the TOPRIM domains flank the G-segment-binding site, forming a portion of the long channel that comprises the primary DNA interaction surface. Surprisingly, the TOPRIM domains of DNA-free GyrBA-fus adopt a conformation analogous to that seen in DNA-associated type IIA topo models (Figure 4). This observation demonstrates that the DNA gate of E. coli gyrase can adopt a properly configured DNA-binding conformation even in the absence of nucleic acid substrate.
Figure 4.
Relative TOPRIM orientations. Side view of GyrBA-fus (lower-right panel) shown next to models for three other conformations of the type IIA topo DNA binding and cleavage core (top two panels—apo S. cerevisiae topo II, PDB ID BGW and PDB ID 1BJT; lower left panel—DNA-bound S. cerevisiae topo II, PDB ID 2RGR). The arrow marks the open G-segment-binding channel in GyrBA-fus. One protomer of each dimer is colored gray; the other is colored as per Figure 1A.
The insert plays a role in DNA supercoiling and relaxation, but not cleavage
Previous biochemical studies of the E. coli GyrB insert have shown that this region is necessary to support normal gyrase function (35). The effects of deleting this region were severe, resulting in a 50-fold reduction in supercoiling activity and an 80-fold reduction in drug-induced cleavage activity. Based on our present structure, we reexamined the truncation end points used for this analysis (amino acids Leu561–Glu720) and found that they cut within the middle of the subdomain 1 hydrophobic core. To test whether the inclusion of these extra residues might have inadvertently impaired gyrase activity indirectly (e.g. by promoting subunit aggregation, mis-assembly or misfolding), we used the GyrBA-fus model to guide the construction of a more precise insert deletion, replacing residues Thr565-Arg731 of E. coli GyrB with a short, flexible linker (Gly–Ser–Ser–Gly).
To rule out the possibility that preparations of this truncated construct (hereafter termed GyrBΔinsert) might be contaminated with endogenous, wild-type GyrB, we carried out supercoiling assays that varied the relative ratio of GyrB or GyrBΔinsert to GyrA. Briefly, the amount of GyrB or its variants was titrated against a constant amount of GyrA, and the supercoiling activity of the mixture assessed by agarose gel electrophoresis and ethidium bromide staining. Consistent with our estimate of sample purity by SDS–PAGE (>98%), maximal activity was observed at a ratio of one GyrBΔinsert subunit to one GyrA protomer (not shown), indicating that little or no native GyrB copurified with the mutant subunit. We also examined purified GyrB and GyrBΔinsert proteins by circular dichroism and found the spectral signature of the mutant to be virtually identical to wild-type (not shown). This result indicates that removal of the insert did not appreciably alter the secondary structure of the protein, as would be expected if the mutant had severely perturbed proper folding.
To ascertain the functional effects of removing the GyrB insert, we reconstituted tetrameric gyrase from wild-type E. coli GyrA and either wild-type or truncated GyrB. The holoenzyme was pre-formed by first incubating equimolar amounts of GyrB and GyrA at high concentration (∼10 µM) on ice for at least 15 min. Both proteins were then subjected to a battery of gyrase activity assays, including: (i) positive supercoil relaxation and negative supercoil introduction (both ATP-dependent), (ii) the ATP-independent relaxation of negative supercoils, and (iii) drug-induced DNA cleavage.
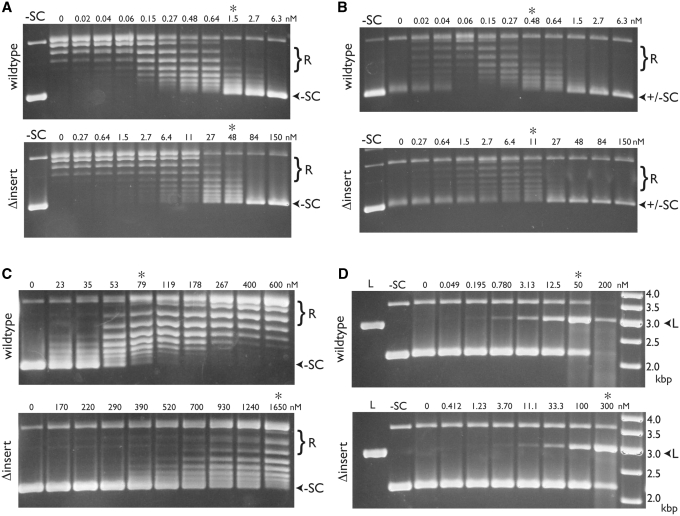
To assess negative supercoiling, we incubated relaxed plasmid DNA with increasing amounts of wild-type or GyrBΔinsert gyrase in the presence of ATP, and visualized the resulting products by gel electrophoresis. Comparison of the relative enzyme concentrations needed to produce comparable levels of supercoil introduction indicates that precise deletion of the insert reduces supercoiling activity ∼20- to 30-fold (Figure 5A). A similar degree of impairment was seen in the ability of GyrBΔinsert gyrase to relax (and subsequently negatively supercoil) a positively-supercoiled plasmid substrate (Figure 5B). In ATP-independent relaxation reactions using negatively-supercoiled substrate, the truncation mutant again displayed an activity ∼25-fold down relative to wild-type gyrase (Figure 5C). Thus, in agreement with Chatterji et al. (35), the GyrB insert appears to be generally important for gyrase reactions that require strand passage.
Figure 5.
Comparison of wild-type and GyrBΔinsert gyrase activities. Negative supercoiling assays (A), positive supercoil relaxation assays (B), negative supercoil relaxation assays (C) and ciprofloxacin-dependent DNA cleavage assays (D) were carried out as described (‘Materials and Methods’ section). The enzyme (wild-type or GyrBΔinsert gyrase) used in each titration is shown at left. Positions of relaxed and supercoiled species are indicated at right. Lanes with negatively supercoiled (–SC) or BamHI-linearized (L) plasmid standards are indicated, as are the concentrations of gyrase tetramer (in nM). Asterisks indicate lanes in each pair of gels with comparable activities. In the cleavage assay (D), the rightmost lane contains a DNA ladder (O’GeneRuler, Fermentas) with the standard sizes (in kb) indicated.
We next set out to define the role of the insert in DNA cleavage. In all type IIA topos, cleavage is disfavored compared to ligation; however, certain drugs, such as fluoroquinolones, can stabilize cleavage complexes (60,61). We incubated increasing amounts of wild-type or GyrBΔinsert gyrase with plasmid DNA and 1 mM of the fluoroquinolone ciprofloxacin. Protein was removed from the reactions using proteinase K, and the resulting products visualized by agarose gel electrophoresis and ethidium bromide staining. In contrast to its strong effects on supercoiling, the GyrBΔinsert gyrase showed a relatively modest cleavage defect (∼6-fold) compared to wild-type gyrase (Figure 5D). This discrepancy may be due to the fact the cleavage assay represents an endpoint reporter that relies on the trapping of a drug-induced cleavage state; hence, the long-term build-up of linear DNA may not accurately reflect differences in activity kinetics. However, the finding also raised the possibility that the observed reduction in supercoiling activity might be linked to one or more specific defects in enzyme function, rather than DNA cleavage per se.
Removal of the insert reduces DNA-binding affinity and DNA-stimulated ATPase activity
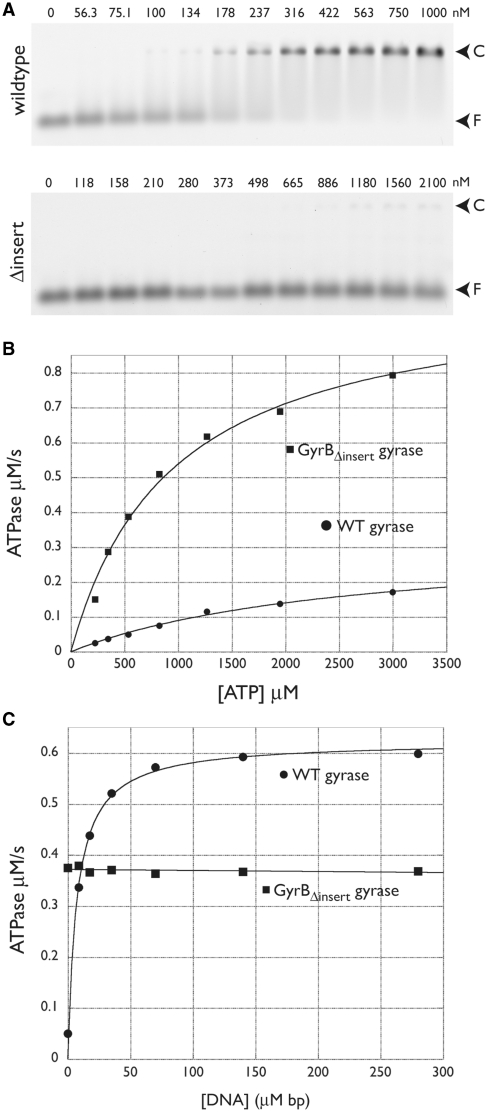
To examine this issue further, we next asked if the insert affects DNA binding. To compare the relative affinities of wild-type and GyrBΔinsert gyrase for DNA, we incubated a fluorescein-labeled 50-bp duplex oligonucleotide corresponding to the pBR322 strong gyrase-binding site with increasing concentrations of gyrase tetramer formed from either wild-type or insert-deleted GyrB. A GyrA construct lacking its C-terminal domain (GyrAΔCTD) was used in this assay to avoid competition from the CTD in G-segment binding. Complexes were visualized by an EMSA in a native agarose gel (Figure 6A). We found that DNA binding by GyrBΔinsert gyrase was substantially (at least 30- to 50-fold) lower than wild-type. This result is consistent with previous studies, which found a complete abrogation of DNA-binding affinity upon removal of the insert (35). It should be noted, however, that these assays were carried out using a small DNA fragment rather than a plasmid substrate, a difference that may accentuate the DNA-binding defect compared than DNA cleavage, relaxation or supercoiling activities.
Figure 6.
DNA binding and ATPase activity of wild-type and GyrBΔinsert gyrase. (A) EMSAs. Titration of wild-type GyrB:GyrAΔCTD is shown at top; the GyrBΔinsert:GyrAΔCTD construct, at bottom. Arrows indicate the positions of free DNA (F) and DNA-protein complex (C). Values above each lane indicate the amount of tetramer in nM. (B) Basal ATPase activity. Wild-type and GyrBΔinsert gyrase activities are plotted as a function of ATP concentration and fit to a standard Michaelis–Menten kinetic model. (C) DNA-stimulated ATPase activity. Wild-type and GyrBΔinsert gyrase activities at a constant ATP concentration are plotted as a function of sheared salmon sperm DNA concentration. Data for wild-type gyrase are fit to a single-site binding equation; GyrBΔinsert gyrase data are fit to a line.
The loss of DNA affinity in the truncation, together with its impaired biochemical functions, suggested that the insert’s predominant role is in facilitating DNA binding. To ascertain whether this function might be due to direct or indirect interactions with nucleic-acid segments, we purified the isolated insert domain and tested its ability to bind DNA by both non-equilibrium (EMSA) and equilibrium (fluorescence polarization) methods. In neither instance was any evidence of DNA binding detectable, even at a 50-fold molar excess of protein (not shown). This result indicates that if the insert does interact with nucleic acid segments, it likely does so only in the context of the holoenzyme.
Finally, we examined the ATPase activity of wild-type and GyrBΔinsert gyrase using a coupled spectrophotometric assay in which ATP hydrolysis is linked to the oxidation of NADH to NAD+ by the sequential action of pyruvate kinase and lactate dehydrogenase (49). Unexpectedly, we found that rather than reducing the basal ATPase activity of gyrase, removal of the GyrB insert actually moderately increased kcat by 2-fold and decreased KM ∼3-fold (Figure 6B and Table 2). By contrast, whereas DNA stimulated wild-type gyrase ATPase activity by 10- to 15-fold (Figure 6C), it completely failed to affect GyrBΔinsert gyrase. Given the observed loss of DNA binding by GyrBΔinsert, it is likely that this behavior arises, at least in part, from an inability of the mutant to properly bind a G-segment duplex.
Table 2.
Basal ATPase activity of wild-type, Δinsert, H669A and H669E gyrase
| Enzyme | KM (µM) | kcat (s−1) |
|---|---|---|
| Wild-type | 3000 ± 500 | 0.97 ± 0.13 |
| GyrB-Δinsert | 1400 ± 500 | 2.6 ± 0.80 |
| GyrB-H669A | 3300 ± 300 | 0.92 ± 0.19 |
| GyrB-H669E | 4600 ± 2300 | 0.56 ± 0.19 |
Errors are standard deviations from at least three measurements.
Structure-based phylogenetic analysis of a coupling point between the insert and GyrA
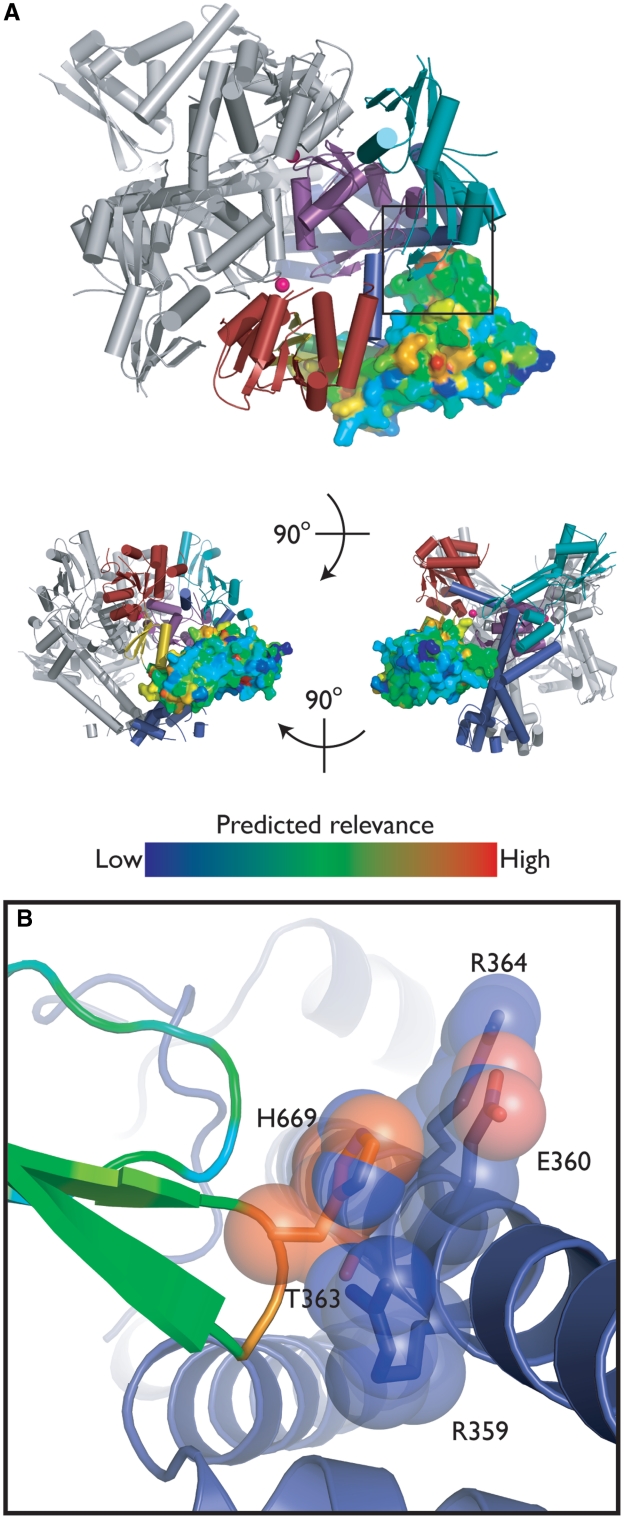
Because ablation of an entire domain is a somewhat extreme physical alteration, we next focused on how specific structural contacts between the insert and the rest of gyrase affect enzyme function. In the context of GyrBA-fus, the insert forms a curved ‘handle’ that protrudes out and away from the core TOPRIM domain (Figure 2). Remarkably, the insert forms essentially no contacts with other parts of gyrase, except between a small loop within insert subdomain 3 (residues 667–669) and the proximal end of the GyrA colied-coil region.
To determine whether this interaction or other regions of the insert might be important for gyrase function, we conducted a phylogenetic analysis of the E. coli GyrB insert using INTREPID (INformation-theoretic TREe traversal for Protein functional site IDentification) (62). The INTREPID algorithm weights sequence homology within clusters that are closely related to the target protein more heavily than global homology overall and thus displays improved sensitivity in identifying functional residues that are specific to orthologous subgroups. The mapping of INTREPID scores (Supplementary Table S1) for the GyrB insert onto our structure allowed visualization of potentially important sites. While the majority of these sites corresponded to buried hydrophobic residues, one of the highest-scoring residues, His669, mediates the only direct contact between the insert and the GyrA coiled-coil (Figure 7).
Figure 7.
Insert/GyrA interactions. (A) INTREPID analysis. The structure of GyrBA-fus is shown as in Figure 2, except the insert is displayed as a surface model and colored by INTREPID score (Supplementary Table S1) (62). (B) GyrB His669 contacts GyrA. The GyrB insert is shown colored by INTREPID score; the GyrA coiled-coil is shown in blue. Selected amino acids in close proximity are shown in stick/transparent spheres representation and colored by atom.
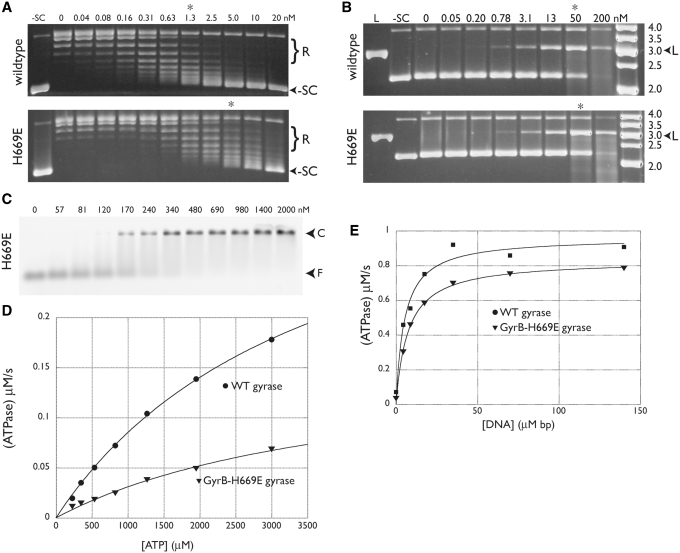
We therefore mutated His669 to alanine and glutamic acid and tested both variants for supercoiling, DNA binding and ATPase activity. Endogenous GyrB contamination was controlled for as described for GyrBΔinsert. The H669A mutant behaved like wild-type gyrase in all respects (not shown); however, the H669E substitution did not. Most notably, theH669E mutation reduced negative supercoiling activity ∼5-fold (Figure 8A). Gyrase-H669E displayed wild-type ciprofloxacin-dependent DNA cleavage (Figure 8B) and DNA binding (Figure 8C), but exhibited a 50% reduction in basal (non-DNA-stimulated) ATPase activity (Figure 8D) and a modest defect in DNA-stimulated ATPase activity (Figure 8E). Taken together, these results suggest that the contact point between the insert and GyrA does not directly participate in the formation of structural or chemical states important for DNA binding or cleavage, but that it does play a role in ATPase and strand passage functions.
Figure 8.
GyrB H669E activities. Negative supercoiling assays (A) and ciprofloxacin-dependent DNA cleavage assays (B) are shown and labeled with the enzyme (wild-type or GyrB-H669E gyrase) used in each titration. Positions of relaxed and supercoiled species are indicated, as are lanes with negatively supercoiled (–SC) or BamHI-linearized (L) plasmid standards. The concentrations of gyrase tetramer (in nM) are also shown. Asterisks indicate lanes with comparable activity for each pair of gels. In the cleavage assay (B), the rightmost lane contains a DNA ladder (O’GeneRuler, Fermentas) with the standard sizes (in kb) indicated. (C) DNA binding. Arrows indicate the positions of free DNA (F) and protein-DNA complex (C) in the EMSA. Values indicate the amount of tetramer in nM. (D) Basal ATPase activity. The ATPase activities of wild-type (circles) and GyrB–H669E (triangles) gyrase are plotted as a function of ATP concentration and fit to a standard Michaelis–Menten kinetic model. (E) The ATPase activities of wild-type (circles) and GyrB–H669E (triangles) gyrase at a constant ATP concentration are plotted as a function of increasing amounts of sheared salmon sperm DNA. Data are fit to a single-site binding equation.
DISCUSSION
The gyrB genes of numerous bacterial species, particularly those of gram-negative organisms, are distinguished by the presence of a moderately-sized insertion embedded within the TOPRIM fold of the subunit (Figure 1). To begin to understand the role of this region in gyrase’s mechanism, we have determined the structure of the insert, together with the accompanying DNA binding and cleavage core of E. coli gyrase. This structure constitutes the first view of the complete DNA gate of a topo from this important model organism, as well of the insert in general. Furthermore, the model reveals that the insert adopts a novel fold that protrudes from the TOPRIM hook subdomain, contacting the associated GyrA subunit at a point near the base of its coiled-coil elements (Figures 2 and 3). Since the poor sequence conservation of the insert and its sporadic presence throughout the bacterial kingdom have hindered efforts to probe its function, we used the structure to guide targeted mutagenesis and biochemical studies. An insert-deletion mutant (GyrBΔinsert) markedly reduced DNA supercoiling and relaxation (∼25-fold), but impacted DNA cleavage less severely (Figure 5). The insert also proved necessary for both DNA binding and DNA-dependent stimulation of ATPase activity (Figure 6); however, the domain in isolation did not appear capable of interacting with a short DNA duplex. Consistent with this result, modeling of a G-segment DNA (as it appears in a co-crystal structure with S. cerevisiae topo II) onto our structure reveals no contacts between the insert and the nucleic acid segment (Supplementary Figure S3).
Together, these results argue that the insert’s contribution to G-segment binding is indirect. One mechanism through which the insert may affect this property is by reducing the conformational space available to the TOPRIM domain, predisposing it to adopt an orientation that, until now, has been observed exclusively in DNA-bound forms of type IIA topos (Figure 4) (22,23). In support of this premise, modeling of the TOPRIM and insert from our present structure onto DNA-free models of topo II, neither of which appear capable of binding an intact G-segment DNA, results in significant clashes between the insert and the rest of the protein (Supplementary Figure S4). Thus, the insert appears to function in part as a steric buttress, stabilizing the DNA-gate of gyrase in a conformation that favors G-segment binding (Figure 9). It is interesting to note that this role appears to specific to those topos that bear the insert (such as E. coli), as otherwise ortholologous enzymes lacking this element are not obviously impaired functionally. We hypothesize that these ‘insert-less’ topos, which comprise the bulk of type IIA topo homologs, possess sequence variations or structural differences that stabilize the formation of a competent DNA-binding state without the need for this domain.
Figure 9.
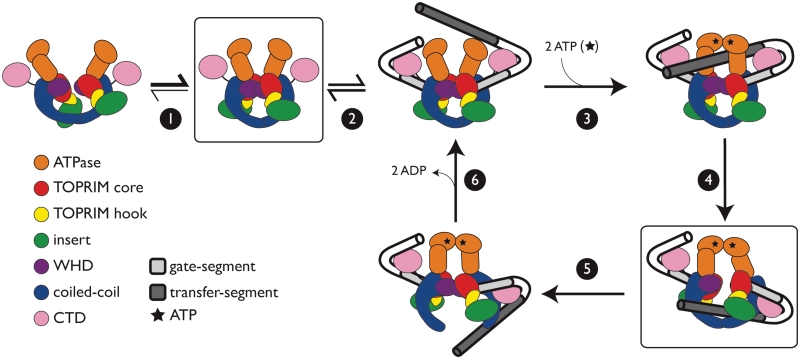
Model for the role of the insert in strand passage. An abbreviated schematic of the gyrase catalytic cycle is shown with protein domains colored as indicated. Individual steps of the reaction are depicted as follows: (1) Prior to G-segment binding, the DNA-gate is oriented in a state competent for DNA binding, (2) A G-segment is bound and wrapped, (3) ATP binding facilitates T-segment capture and G-segment cleavage, (4) G-segment opening permits T-segment transport, (5) G-segment closure and T-segment release, and (6) ADP release resets the enzyme for another round of strand passage. The insert (green) is proposed to favor formation of a state competent to bind G-segment DNA (step 1) and to aid T-segment transport (step 4).
The poor sequence conservation of the insert likewise supports the notion of a primarily structural role for this domain; however, we were able to uncover one functionally significant, surface-exposed residue in the region (His669) using INTREPID, a phylogenetic tree-based algorithm for scoring amino acid sequence conservation. In the GyrBA-fus structure, His669 sits at the apex of a loop that forms the only contact between the insert and GyrA (Figure 7). When His669 is mutated to glutamic acid, the resultant gyrase lost ∼50% of its ATPase activity and ∼80% of its negative supercoiling activity (Figure 8). In contrast, the H669E mutant was normal for DNA binding and cleavage and retained its DNA-stimulated ATPase activity.
Thus, while the simple physical presence of the insert aids DNA binding by gyrase, the insert-mediated connection between the TOPRIM and the GyrA coiled coil regions appears important for the strand passage reaction overall. One means by which the insert could assist this event is by binding T-segment DNA; however, we see no particular attractive (e.g. electropositive) surface that might mediate such a function (Supplementary Figure S3), nor did the insert show any propensity to engage nucleic acids in vitro. Alternatively, the insert might play a role in strand passage by aiding communication between holoenzyme gates. The promotion of unidirectional strand passage by type IIA topos is known to require the physical and temporal coordination of structurally disparate functional modules (2). For example, in the homologous ATPase regions of topo II and topo VI (a type IIB enzyme), a conserved lysine (Lys337 in E. coli GyrB) has been suggested to mediate communication between the ATP- and DNA-gates, contacting the γ-phosphate of ATP and inducing conformational changes in response to ATP hydrolysis (63–66). The GyrB insert may constitute a similar, albeit non-catalytic, structural relay between the DNA-gate and either the ATP- or C-gate to communicate conformational signals between these structurally distinct protein interfaces (Figure 9).
As there have been relatively few systematic comparisons of gyrases from different organisms, it is difficult to assign a conserved role for the insert among the many species in which it appears. The presence of the domain in the deeply-rooted hyperthermophilic genus Aquifex (67) suggests that the insert may have been present in early GyrB genes and subsequently lost in the majority of bacterial lineages. Where it has been retained, species-specific variations, such as residue His669 in the proteobacterial GyrBs, may have contributed to the biochemical functionality of this element, promoting its retention despite the cost of maintaining such a relatively large protein component. Alternatively, it is possible that the insert may have been acquired through horizontal gene transfer to serve some as yet undefined role in the cell. In the future, it will be important to determine whether gyrases lacking this region have acquired compensatory mutations that stabilize the TOPRIM domain orientation or promote DNA binding. Moreover, it will be interesting to see if the GyrB insert serves as a platform for associating with specific regulatory proteins. Additional structural information, preferably in the presence of DNA and larger constituent pieces of the holoenzyme, should help further elucidate the precise mechanism through which the insert fine-tunes gyrase function.
ACCESSION NUMBER
3NUH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation graduate research fellowship (to A.J.S.); National Cancer Institutes (CA077373 to J.M.B.); the NIGMS (GM071326-02 to A.P.M.). Funding for open access charge: National Cancer Institute (grant no. CA077373).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors extend special thanks to Kanagallagatha Rajashankar and the Advanced Photon Source beamline NE-CAT 24IDE for assistance with X-ray data collection. The authors would also like to thank Hiroshi Hiasa for helpful suggestions in the early stages of our structural studies. Coordinates have been deposited in the RCSB PDB 3NUH.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Schoeffler AJ, Berger JM. DNA Topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 3.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Drlica K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992;6:452–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 5.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 7.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 8.Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, DiGate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol. Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 9.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 10.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 11.Peng H, Marians KJ. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl Acad. Sci. USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns in DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 14.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 16.Williams NL, Maxwell A. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999;38:13502–13511. doi: 10.1021/bi9912488. [DOI] [PubMed] [Google Scholar]
- 17.Bjergbaek L, Jensen S, Westergaard O, Andersen AH. Using a biochemical approach to identify the primary dimerization regions in human DNA topoisomerase IIalpha. J. Biol. Chem. 1999;274:26529–26536. doi: 10.1074/jbc.274.37.26529. [DOI] [PubMed] [Google Scholar]
- 18.Olland S, Wang JC. Catalysis of ATP hydrolysis by two NH(2)-terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem. 1999;274:21688–21694. doi: 10.1074/jbc.274.31.21688. [DOI] [PubMed] [Google Scholar]
- 19.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key state in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 20.Ali JA, Orphanides G, Maxwell A. Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry. 1995;34:9801–9808. doi: 10.1021/bi00030a018. [DOI] [PubMed] [Google Scholar]
- 21.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 22.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 23.Laponogov I, Sohi MK, Veselkov DA, Pan X, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 24.Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 1983;258:15365–15370. [PubMed] [Google Scholar]
- 25.Morrison A, Cozzarelli NR. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979;17:903–906. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- 26.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 27.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 28.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 29.Harkins TT, Lewis TJ, Lindsley JE. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998;37:7299–7312. doi: 10.1021/bi9729108. [DOI] [PubMed] [Google Scholar]
- 30.Harkins TT, Lindsley JE. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry. 1998;37:7292–7298. doi: 10.1021/bi9729099. [DOI] [PubMed] [Google Scholar]
- 31.Tingey AP, Maxwell A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996;24:4868–4873. doi: 10.1093/nar/24.24.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl Acad. Sci. USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 34.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl Acad. Sci. USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterji M, Unniraman S, Maxwell A, Nagaraja V. The additional 165 amino acids in the B protein of Escherichia coli DNA gyrase have an important role in DNA binding. J. Biol. Chem. 2000;275:22888–22894. doi: 10.1074/jbc.M001047200. [DOI] [PubMed] [Google Scholar]
- 36.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure. 2007;15:329–339. doi: 10.1016/j.str.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 acomplexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 38.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides wi which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Procesing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 41.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 44.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl. Cryst. 2006;39:109–111. [Google Scholar]
- 45.Davis ID, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall B, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W388. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dynan WS, Jendrisak JJ, Hager DA, Burgess RR. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme) J. Biol. Chem. 1981;256:5860–5865. [PubMed] [Google Scholar]
- 47.Rodriguez AC. Studies of a positive supercoiling machine. Nucleotide hydrolysis and a multifunctional “latch” in the mechanism of reverse gyrase. J. Biol. Chem. 2002;277:29865–29873. doi: 10.1074/jbc.M202853200. [DOI] [PubMed] [Google Scholar]
- 48.Cove ME, Tingey AP, Maxwell A. DNA gyrase can cleave short DNA fragments in the presence of quinolone drugs. Nucleic Acids Res. 1997;25:2716–2722. doi: 10.1093/nar/25.14.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J. Biol. Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- 50.Tamura JK, Gellert M. Characterization of the ATP bidning site on Escherichia coli DNA gyrase. Affinity labeling of Lys-103 and Lys-110 of the B subunit by pyridoxal 5’-diphospho-5’-adenosine. J. Biol. Chem. 1990;265:21342–21349. [PubMed] [Google Scholar]
- 51.Lavasani LS, Hiasa H. A ParE-ParC fusion protein is a functional topoisomerase. Biochemistry. 2001;40:8438–8443. doi: 10.1021/bi0155201. [DOI] [PubMed] [Google Scholar]
- 52.Trigueros S, Roca J. A GyrB-GyrA fusion protein expressed in yeast cells is able to remove DNA supercoils but cannot substitute eukaryotic topoisomerase II. Genes Cells. 2002;7:249–257. doi: 10.1046/j.1365-2443.2002.00516.x. [DOI] [PubMed] [Google Scholar]
- 53.Laponogov I, Veselkov DA, Sohi MK, Pan X, Achari A, Yang C, Ferrara JD, Fisher LM, Sanderson MR. Breakage-reunion domain of Streptococcus pneumoniae Topoisomerase IV: Crystal structure of a gram-positive quinolone target. PLoS ONE. 2007;2:e301. doi: 10.1371/journal.pone.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards MJ, Flatman RH, Mitchenall LA, Stevenson CE, Le TB, Clarke TA, McKay AR, Fiedler HP, Buttner MJ, Lawson DM, et al. A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science. 2009;326:1415–1418. doi: 10.1126/science.1179123. [DOI] [PubMed] [Google Scholar]
- 55.Tretter EM, Schoeffler AJ, Weisfield SR, Berger JM. Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis. Proteins. 2010;78:492–495. doi: 10.1002/prot.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng JY, Zhang XE, Bi L, Wang D. Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 58.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 59.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hooper DC, Rubinstein E. Quinolone Antimicrobial Agents. 3rd edn. Washington DC: ASM Press; 2003. [Google Scholar]
- 62.Sankararaman S, Sjolander K. INTREPID–INformation-theoretic TREe traversal for Protein functional site IDentification. Bioinformatics. 2008;24:2445–2452. doi: 10.1093/bioinformatics/btn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005;13:873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 66.Bjergbaek L, Kingma P, Nielsen IS, Wang Y, Westergaard O, Osheroff N, Andersen AH. Communication between the ATPase and cleavage/religation domains of human topoisomerase IIalpha. J. Biol. Chem. 2000;275:13041–13048. doi: 10.1074/jbc.275.17.13041. [DOI] [PubMed] [Google Scholar]
- 67.Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl Acad. Sci. USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high througput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeLano WL. USA: Palo Alto, CA; 2002. DeLano Scientific. [Google Scholar]
- 72.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.