Abstract
Using numerical simulations, we investigate the underlying physical effects responsible for the overall organization of chromosomal territories in interphase nuclei. In particular, we address the following three questions: (i) why are chromosomal territories with relatively high transcriptional activity on average, closer to the centre of cell's nucleus than those with the lower activity? (ii) Why are actively transcribed genes usually located at the periphery of their chromosomal territories? (iii) Why are pair-wise contacts between active and inactive genes less frequent than those involving only active or only inactive genes? We show that transcription factories-mediated contacts between active genes belonging to different chromosomal territories are instrumental for all these features of nuclear organization to emerge spontaneously due to entropic effects arising when chromatin fibres are highly crowded.
INTRODUCTION
During the typical life cycle of eukaryotic cells the stage of division is followed by the G1 stage. In the G1 stage, cells grow, synthesize RNA and proteins, and prepare for DNA replication (1). Chromosomes that were highly condensed during cell division decondense in the G1 stage (2,3). Interestingly, in higher eukaryotes the decondensed chromatin fibres of individual chromosomes that are long enough to span the entire nucleus do not spread out but remain confined within rather compact chromosome territories (4–8). Several recent papers revealed physical reasons why highly crowded, long, polymer molecules undergoing thermal fluctuations maximize their entropy by forming intramolecular territories and limiting intermolecular intermingling (9–16). While general physical principles leading to formation of chromosomal territories seem to be understood, it is still unclear why the territories of chromosomes that are more transcriptionally active are usually located closer to the centre of the nucleus than the territories of chromosomes that are less transcriptionally active (6,8,15). There is also a need to understand the underlying principles causing preferential localization of transcribed genes at the periphery of their chromosomal territories (3,6,17–19). Although the peripheral location of actively transcribed genes, with respect to their chromosomal territories, explains why interchromosomal contacts involve most frequently actively transcribed genes, it is more difficult to understand why transcriptionally non-active regions more frequently enter into contact with each other than with transcriptionally active regions (20). Here we show, using numerical simulations, that the mere capability of transcription factories to simultaneously engage active genes belonging to different chromosomal territories (3,21,22) is sufficient to generate the known dependence between the transcriptional activity of a given chromosomal territory and its radial position within cells’ nuclei. The same property of transcription factories also permits us to explain why the actively transcribed genes are located at the periphery of their chromosomal territories and why interchromosomal contacts between transcriptionally inactive regions are more frequent than those between transcriptionally active and inactive regions (20).
MATERIALS AND METHODS
The details of the simulation procedure are described in Supplementary Data and schematically presented in Supplementary Figures. In essence, we measure equilibrium properties of thermally fluctuating polymeric chains (chromatin fibres of individual chromosomes) confined within a small sphere (nucleus). This is achieved by the Metropolis Monte Carlo simulations based on crankshaft moves (14,23,24). We test how inter-chain attachments mimicking transcription factories-mediated interchromosomal contacts affect such characteristics of the simulated system as: (i) the average radial positions of simulated chains within the sphere. (ii) The average position of points of attachment with respect to the territories formed by polygonal chains. (iii) The probability of contacts between non-attached chains or between attached and non-attached chains.
In the G1 phase, decondensed chromatin fibres serve as templates for RNA synthesis by RNA polymerases. Interestingly, RNA polymerases are team workers and associate into groups containing about eight active polymerases, where each of them is firmly bound to a different transcription unit (25,26). By simultaneous binding to active genes belonging to different chromosomes, transcription factories determine what portions of different chromosomes contact each other in interphase nuclei (3,21,22). We decided to model the effect of transcription factories-mediated interchromosomal contacts on the overall organization of chromosomal territories within interphase nuclei. The transcription factories were not modelled as such but were replaced by a set of links between chains representing transcriptionally active chromosomes (see Supplementary Figure S1). To operate with computationally manageable and relatively easily interpretable setup, we decided to study thermally equilibrated systems consisting of 16 freely jointed equilateral polygonal chains placed together within a small sphere (for more details, see Supplementary Data).
Traditionally, long polymeric chains under conditions where the intersegmental repulsion is greatly screened (like it is believed to be the case of chromatin) are modelled using equilateral ideal chains, where each segment is infinitely thin (27,28). On the other hand, we know that chromatin fibres are not infinitely thin and the ratio between the statistical segment length and its thickness can vary according to particular conditions like counterions concentration, state of chromatin fibres, etc. Therefore, self-avoiding models are also frequently used to model chromatin behaviour (12,15).
To show that our results are general, i.e. they do not depend on the particular model used, we performed the simulations for two diametrically different models. In the first case, we studied properties of simple polygonal chains with segments that were practically infinitely thin. In the second case, we operated with a beaded-chain model where each vertex of equilateral polygon was placed in the centre of a hard-core sphere that had the same diameter as the segment length. In the case of polygonal chains composed of infinitely thin segments, each polygon was composed of 40 segments and the radius of the sphere enclosing all 16 polygons was set to about 3.6 segment lengths, ensuring a sufficient density of packing to model interactions between decondensed but highly crowded chromatin fibres in cell nuclei. The diameter of the enclosing sphere was adjusted to give the same density of segments as in the case of polygons with excluded volume where the radius of each segment was set to 0.1 of the segment length and where such polygonal chains constitute 10% of the sphere's volume. In the case of beaded chains, each chain was composed of 100 beads and the radius of the sphere enclosing all 16 beaded chains was set to 12.6 diameters of individual beads as this gave 10% volume occupation within the sphere. In interphase nuclei, chromatin occupies 10–20% of nuclear volume (15); therefore, our model is subjected to similar crowding effects as these believed to act on interphase chromosomes in living cells. We modelled the transcription factory-mediated interchromosomal attachment by imposing an elastic potential that effectively brought eight pre-selected vertices belonging to eight different polygons to a distance <1/10 of the segment length from each other for simple polygonal chains (Supplementary Figures S1 and S2) and to a distance <2 segment lengths for beaded chains. The remaining eight polygons were free. To characterize the equilibrium properties of simulated systems, we used Metropolis Monte Carlo procedure based on crankshaft moves (14,23,24). It was shown earlier that when modelled chromatin fibres cannot pass through each other, they spontaneously form chromosomal territories (13,14). To further investigate these topological effects on nuclear architecture, we compared systems where intersegmental passages between the segments belonging to the same or to different polygons were allowed or not allowed. Although, chromosomes are linear in a formal sense, in a topological sense they behave as circular molecules as they are organized into loops of various sizes (29–36). In addition, diverse regions of chromosome territories are frequently attached to the nuclear matrix or to the nuclear membrane (37). All these features ensure that chromatin fibres forming one chromosome territory do not have free ends that could pierce and thread through another chromosome territory. To reflect the absence of free ends of chromosome fibres, we modelled them as closed chains (14). When we do not permit intersegmental passages, we restrict the topology of modelled chains to unknotted and uncatenated configurations. When passages are permitted, the topology of modelled chains is unrestricted and we can observe formation of knots and catenanes.
Chains composed of 40 or even 100 segments may seem to be much too short as models of chromosomes. However, it needs to be understood that, in our simulations, one segment corresponds to one Kuhn's statistical segment of decondensed chromosomes. Studies of interphase nuclei revealed that decondensed chromosomes behave like 30-nm chromatin fibres with Kuhn's statistical segment length of ∼400 nm and linear density of ∼150 bp/nm (38). Therefore, in our simulations using infinitely thin polygonal models, each segment represents a chromatin fragment with ∼60 000 bp and each chain composed of 40 segments represents ∼2 400 000 bp. In total, 16 chains each composed of 40 statistical segments can model a genome composed of ∼40 Mb which is nearly half of the genome size of Caenorhabditis elegans.
RESULTS
Radial position of chromosomal territories
Infinitely thin polygonal model
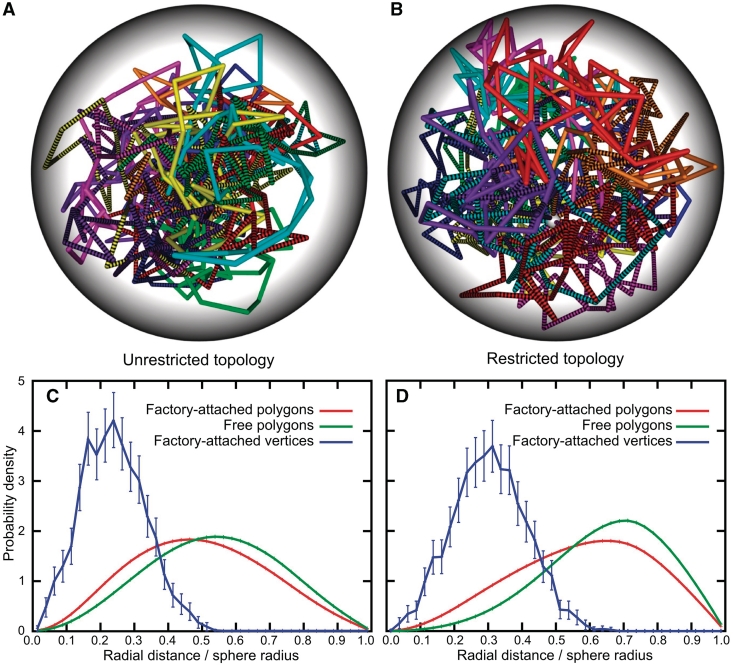
Figure 1A and B show typical snapshots obtained in simulations using the infinitely thin polygonal model. We can see that when the topology was unrestricted (Figure 1A), the polygons were concentrated in the centre of the sphere and formed an interlinked network, in which the individual polygons were extensively catenated with other polygons. In the topologically restricted case (Figure 1B), in which the polygons always maintain their starting topology of unknotted and non-catenated circular chains, the polygons fill the sphere much more uniformly since they maximize their entropy by limiting interpolygonal intermingling (13,14,39). To visually distinguish free polygons from those that are attached to each other, the latter ones are coloured using a striped pattern. A closer look at the snapshots of topologically unrestricted and restricted systems gives an impression that, on average, the attached polygons occupy a more central position in the sphere than free polygons. To verify this impression, we measured the probability of finding points belonging to inter-attached or non-attached polygons at a given distance from the centre of the confining sphere. Figure 1C and D present the probability density distributions of distances from the centre of the sphere for points belonging to attached polygons (red profiles), free polygons (green profiles) and for the vertices that, in our simulations, correspond to points of attachment to the transcription factory (blue profiles). It is clearly seen that, in topologically unrestricted (Figure 1C) and restricted (Figure 1D) systems, the mere attaching of several polygons to each other causes that these polygons tend to occupy a more central position in the sphere than free polygons present in the same sphere. Looking now at the radial positions occupied by vertices connected to the modelled transcription factory, we can clearly see that these vertices are very strongly shifted towards the centre of the sphere when compared with random points on free polygons or on polygons that were attached to each other.
Figure 1.
Radial positions of infinitely thin polygons confined within a small sphere depend on whether these polygons are attached to each other or not. (A and B) Representative snapshots from simulations involving topologically unrestricted and restricted polygons, respectively. Individual polygons are coloured differently and in addition these that were attached to each other to simulate binding to the same transcription factory are coloured using a stripped pattern. To help visualize individual polygons that were modelled as infinitely thin, they are presented as having a significant thickness. (C and D) Radial distributions of points on free polygons (green), points on the polygons attached together (red) and the vertices that were representing points of attachment to a transcription factory (blue). Panels C and D correspond to simulations without and with topological restriction, respectively. For red and green profiles the error bars are smaller than the width of the lines.
Beaded chain model
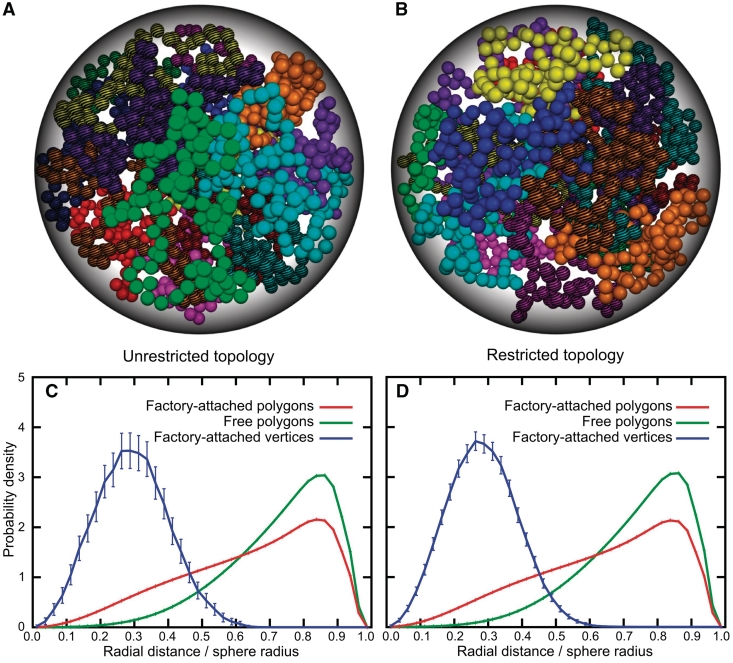
Figure 2 shows typical snapshots of equilibrated beaded chains obtained in simulations that permitted or not intersegmental passages, A and B, respectively. We can see that in both cases the beaded chains fill the available space quite uniformly. This contrasts with the results obtained with infinitely thin polygonal models, where a relatively uniform distribution was only observed when the topology of polygons was restricted to unknotted and uncatenated polygons (Figure 1). The observation that beaded chains show very similar overall space occupation both in the absence and presence of topological restriction suggests that beaded chains with this relatively large thickness (determined as the ratio between the beads’ diameter and the length of the chain) are, to a great extent, confined to the topology of unknotted and uncatenated closed chains even when simulations permit intersegmental passages. Indeed, the analysis of topological states of equilibrated beaded chains with unrestricted topology revealed that only 28% of individual chains were catenated with some other chains, while in the case of infinitely thin polygons this value exceeded 99%. In addition, the formed catenanes were almost exclusively composed of the simplest two component catenanes in case of beaded chains, while infinitely thin polygonal chains were typically forming multicomponent catenanes where each chain was, on average, catenated with ∼10 other chains. Therefore, our results indicate that for relatively thick self-avoiding chains, the presence or absence of the topological restriction has only a minor effect on their statistical characteristics. This result agrees with recent theoretical studies of topological and entropic repulsion between self-avoiding chains under semi-dilute conditions (40). The topological confinement of beaded chains by the excluded volume effect could be expected since several earlier studies have demonstrated that the probability of knotting and catenation decreases rapidly with increasing thickness of the modelled chains (41–43); however, these earlier studies did not investigate probabilities of catenation in a situation where many modelled thick chains are highly crowded in a limited volume.
Figure 2.
Radial positions of circular beaded chains confined within a small sphere depend on whether these chains are attached to each other or not. (A and B) Representative snapshots from simulations involving topologically unrestricted and restricted beaded chains, respectively. Individual beaded chains are coloured differently and in addition these that were attached to each other to simulate binding to the same transcription factory are coloured using a stripped pattern. (C and D) Radial distributions of points on the underlying polygons of beaded chains that were free (green), or were attached with other chains (red). Blue profiles show radial distribution of centres of beads representing points of attachment to the transcription factory. C and D correspond to simulations without and with topological restriction, respectively. For red and green profiles the error bars are smaller than the width of the lines.
In Figure 2A and B, to visually distinguish free chains from inter-attached ones the latter were coloured using striped patterns. Upon visual inspection of these figures it is hard to say whether inter-attached chains locate more close to the centre of the modelled nuclei than free chains. However, upon plotting the distributions of distances from the centre of the sphere for points belonging to beaded chains that were attached to each other (red profiles) or not (green profiles), we can clearly see that the former occupy a more central position in modelled nuclei, (see Figure 2C and D). Analysing now the radial positions occupied by points connected to the modelled transcription factory (blue profiles), it is obvious that these points are very strongly shifted towards the centre of the sphere when compared with average positions of random points on all beaded chains confined within the sphere. Nearly identical profiles of distances shown in Figure 2C and D support our earlier conclusion that at the thermodynamic equilibrium beaded chains of relatively short lengths tend to remain unknotted and uncatenated even if intra- and inter-chain passages are possible.
Position of active genes within their chromosome territories
We tested whether transcription factories-mediated interchromosomal attachments could also explain observations that transcription factories are preferentially located on the peripheries of chromosomal territories (3,17) rather than being engulfed within the territories.
Infinitely thin polygonal model
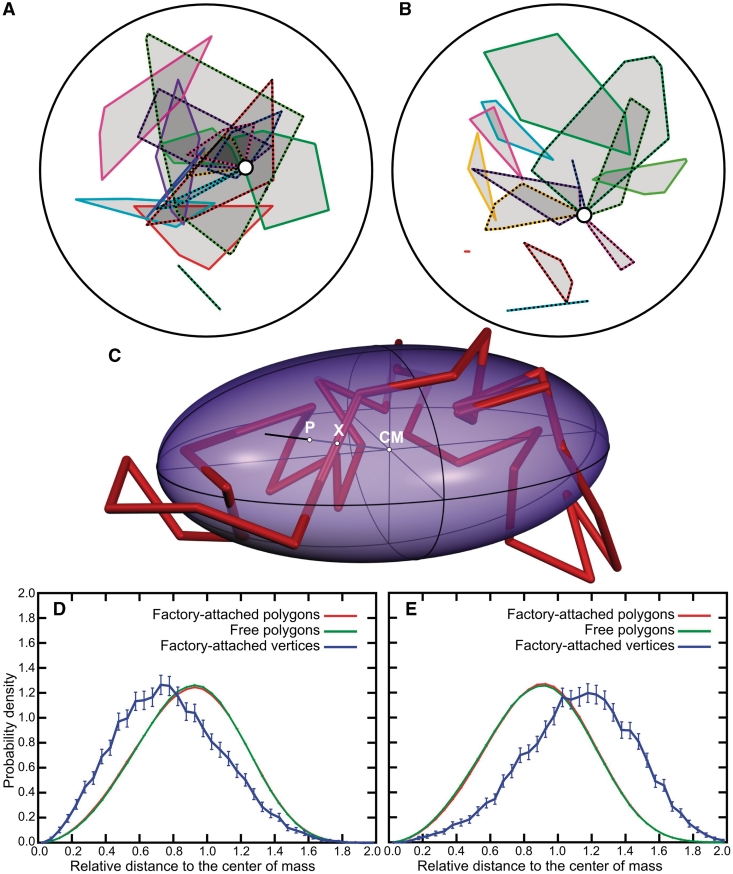
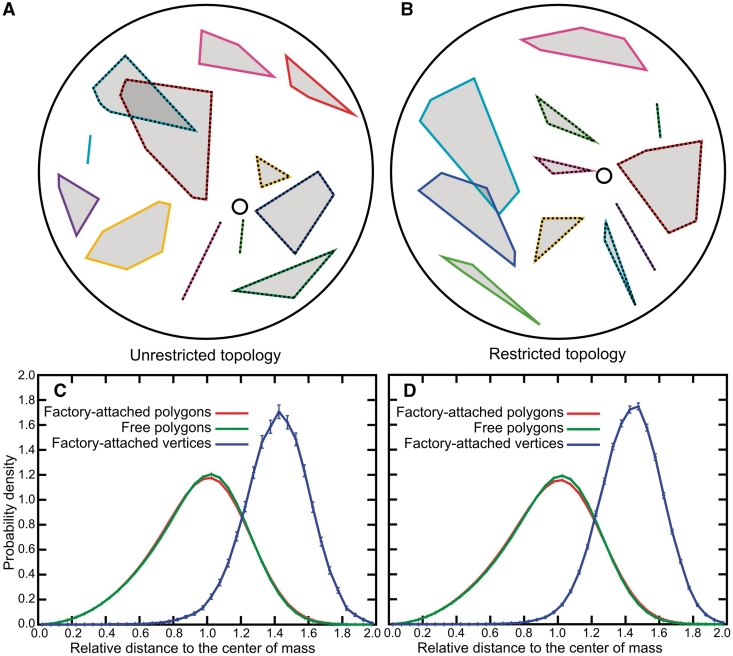
Figure 3A and B show cross sections of equilibrated configurations of infinitely thin polygons with unrestricted or restricted topology, respectively (the cross sections are of the configurations shown earlier in Figure 1A and B, respectively). Each cross-sectional plane passes through the centre of the enclosing spheres and through the position of transcription factory. To help visualize the territories occupied by different polygons, we enclosed the intersection points of individual polygons with the plane of the cross section within differently coloured convex envelopes. Similarly to the colouring applied in Figure 1A and B, the convex envelopes enclosing factory-attached polygons are drawn with striped lines. Some of the convex envelopes drawn with striped lines seem to be disconnected from the central circle that indicates the position of transcription factory. However, this is only due to the fact that the region of attachment of those territories to the transcription factory is in a different plane than the one of the shown cross section. The convex envelopes presented as straight segments indicate that a given polygon intersected the plane of cross section at just two points. Visual inspection of the cross sections gives an impression that points of mutual attachment are located at the peripheries of territories formed by individual polygons with restricted topology (Figure 3B). However, this does not seem to be the case of polygons with unrestricted topology (Figure 3A). To collect quantitative data on localization of inter-attachment points within their respective chromosomal territories, we characterized the internal spatial distribution of points in mutually attached and free polygons and compared them with the corresponding spatial distribution of vertices that served as the points of attachments (see Figure 3C, D and E). Since random configurations of polygons do not have the spherical symmetry but are better approximated by ellipsoids (44–46), we renormalized the actual radial distances so that points located on the surface of characteristic ellipsoids of inertia constructed for each analysed configuration of the polygons have their normalized radial distance values equal to 1 (Figure 3C; for more detailed description see Supplementary Data and Supplementary Figure S6). The characteristic ellipsoid of inertia is an imaginary ellipsoidal surface that would have the same inertial property as the polygon it represents, if the polygon's mass were equally redistributed along that surface (47). Figure 3D shows that, in the case of polygons with unrestricted topology, the vertices that were attached to each other are on average at a smaller relative distance from the centre of mass of their polygons than random points of these polygons. Such behaviour is at odds with cytological observations and indicates that models based on topologically unrestricted infinitely thin polygons are unlikely to reflect the behaviour of chromatin fibres (3,17). However, simulations using polygons with restricted topology clearly show properties consistent with the cytological observations. As shown in Figure 3E, the vertices that are attached to each other have a very strong tendency to locate further from the centre of mass of their polygons than random points of these polygons. The observation that, only in polygons with restricted topology, the modelled points of inter-territorial attachment preferentially occupied peripheral positions within their chromosomal territories supports our earlier proposal that decondensed chromatin fibres in interphase nuclei behave as uncatenated and unknotted polymeric rings (14).
Figure 3.
Localization of vertices representing points of attachment to the transcription factory with respect to the territories formed by topologically unrestricted (A and D) and topologically restricted infinitely thin polygons (B and E). (A and B) Cross sections of the configurations are shown in Figure 1A and B, respectively. Points of intersection of individual polygons with the plane of the cross section define convex envelopes that are analogous to cross sections through chromosomal territories (7,14). Territories formed by polygons modelled as attached to a transcription factory (indicated with a circle) are demarcated with striped lines. (C) The principle of determining whether a given point on a polygon has a peripheral or central position within a territory formed by this polygon. For a given point X, the relative distance to the centre of mass CM of the polygon was defined as the ratio between the distance X-CM and P-CM, where P is the intersection between the ray X-CM and the characteristic ellipsoid of inertia defined by a given polygon (47). (D, E) Relative radial distributions with respect to centres of mass of polygons for: points on free polygons (green), points on the polygons attached together (red) and for the vertices that were representing points of attachment to a transcription factory (blue) are shown. For red and green profiles the error bars are smaller than the width of the lines.
Beaded chain model
Figure 4A and B shows cross sections of equilibrated configurations of beaded-chains models that were equilibrated under conditions that did (Figure 4A) or did not permit inter- and intra-chain passages (Figure 4B) (the cross sections are of the configurations shown in Figure 2A and B, respectively). Similarly to Figure 3A and B, we indicated positions of modelled chromosomal territories by convex envelopes enclosing all intersection points of lines connecting the centres of beads constituting a given modelled territory with the plane of the cross section. Different territories are marked with different colours and striped lines demarcate positions of the territories that are attached to each other via transcription factory (position of which is indicated by a circle). In both cases (topologically unrestricted or restricted), we can clearly see that the position of transcription factories is located outside of chromosomal territories. In contrast to the situation presented in Figure 3A and B, we can see relatively large distances between positions of transcription factories (open circles) and chromosomal territories that are attached to them. This is the consequence of a large excluded volume of beaded chains. In addition, as discussed for the infinitely thin polygonal model, the region of attachment of territories to the transcription factory can be in a different plane than those shown in the figure. Figure 4A and B do not reveal clear differences between positions of points of attachment within modelled chromosomal territories when these are topologically restricted or not. In both cases the points of inter-territorial attachment locate at the periphery of implicated modelled chromosome territories. To collect more quantitative data, we characterized the average spatial distribution of attached and non-attached beads within equilibrated beaded chains. The method of this characterization was the same as that used in the case of infinitely thin polygons (Figure 3C). As can be clearly seen from Figure 4C and D, the vertices that were attached to each other locate preferentially at the peripheries of chromosomal territories to which they belong. This result agrees with the literature data (3,6,17–19). Almost identical profiles of internal spatial distribution of points belonging to chains that may or may not be permitted to undergo intra- and inter-chain passages (compare Figure 4C and D) indicate again that beaded circular chains with a relatively large diameter/length ratio preferentially remain unknotted and uncatenated even if changes of the topology are permitted.
Figure 4.
Localization of beads representing points of attachment to the transcription factory with respect to the territories formed by topologically unrestricted (A and C) and topologically restricted beaded chains (B and D). A and B, cross sections of the configurations shown in Figure 2A and B, respectively. Points of intersection of lines connecting centres of beads of individual chains with the cross-sectional plane define convex envelopes that approximate the extent of territories formed by these chains in the cross section. Territories formed by beaded chains attached to a transcription factory (indicated with a circle) are demarcated with striped lines. (C, D) Relative radial distributions with respect to centres of mass of beaded chains for points on the underlying polygons of beaded chains that were unattached to other chains (green) or were attached to other chains (red) and for centres of beads that were representing points of attachment to the transcription factory (blue). For red and green profiles the error bars are smaller than the width of the lines.
Interchromosomal contact probabilities
Recent study of long-range interactions in human genome provided high-quality data showing that chromosomes with elevated transcriptional activity preferentially contact other chromosomes with elevated transcriptional activity while chromosomes with low transcriptional activity prefer to contact other chromosomes with low transcriptional activity (20). It is somewhat trivial that transcription factories are ideally suited to mediate contacts between chromosomal territories with many active genes. However, it is less clear what brings together chromosomal territories with low transcriptional activity. We, therefore, decided to check whether transcription factories-mediated interchromosomal contacts could also explain why transcriptionally non-active regions are more likely to contact other non-active regions than transcriptionally active ones (20).
Infinitely thin polygonal model
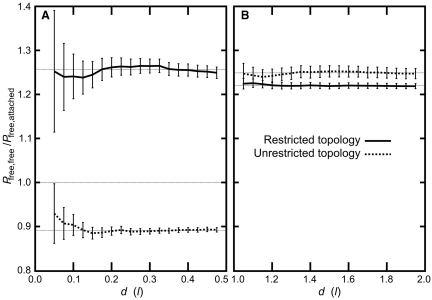
We first tried to visually assess whether, in the case of infinitely thin polygons, the free polygons are grouped together in the presence of polygons that are attached to each other. In the case of topologically unrestricted infinitely thin polygons, these effectively behave like phantom polygons and therefore their spatial redistribution within the sphere are of course unaffected by the presence of phantom polygons that were attached to each other (Figure 3A). However, in the case of infinitely thin polygons that are not permitted to undergo inter-segmental passages, these effectively exclude each other and we indeed observed frequent grouping of the free-polygons due to their exclusion from the space occupied by the polygons attached to each other (Figures 3B and 6). To obtain quantitative data, we measured the average probability of contacts involving all possible pairs of vertices belonging to two different infinitely thin polygons that were not attached to each other as well as the average probability of contacts involving all possible pairs of vertices when one belonged to free polygons and another to polygons attached to each other. Two vertices were considered in contact when the distance between them was smaller than a given distance d. We observed that the ratio between the probability of contact between two vertices belonging to two different free polygon and the probability of contact between two vertices where one belongs to a free and another to an attached polygon was practically independent of the distance d when d ranged from 0.05 to 0.5 of the segment length (Figure 5A). In the case of phantom systems, this ratio had a value slightly <0.9 while for non-phantom polygons this ratio was of ∼1.25. Only the latter result is consistent with the recent demonstration that chromosomal domains with low transcription activity preferably contact other chromosomal domains with low transcription activity (20).
Figure 6.
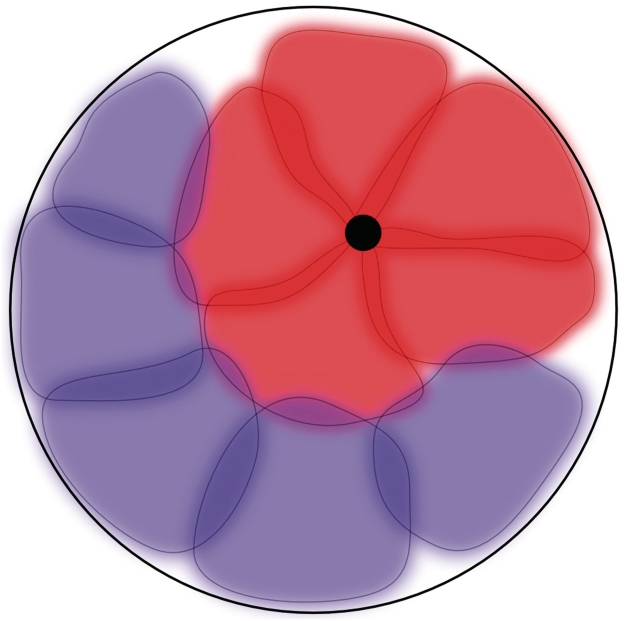
Graphic summary. Transcription factories-mediated attachments between transcriptionally active chromosomal territories are sufficient to explain experimentally observed features of nuclear architecture: (i) Territories formed by transcriptionally active chromosomes (marked in red) are on average located closer to the centre of the nucleus than the territories formed by transcriptionally non-active chromosomes (marked in blue). (ii) Actively transcribed genes interacting with a transcription factory (black circle) are located at the periphery of their chromosomal territories. (iii) Transcriptionally active and also transcriptionally non-active chromatin domains have a tendency for self-association.
Figure 5.
Contact probabilities ratios between two random points located on two free polygons Pfree,free and between two random points where one is located on a free and another on an attached polygon Pfree,attached. The Pfree,free/Pfree,attached ratio is plotted as a function of the distance d, where d is the threshold distance below which a pair of points is considered in contact. In case of infinitely thin polygons (A) the topological restriction was needed to favour contacts between random points located on different free polygons. In the case of beaded chains (B) the contacts between random points located on different free polygons was favoured irrespectively of whether the topological restriction was imposed or not.
Beaded chain model
Figure 5B shows that, in the case of beaded-chain models, a pair of two randomly chosen beads located on two different unattached chains is more likely to be in a contact than a pair of randomly chosen beads where one belongs to a free chain and another to a chain that is attached to other chains. Similarly to other results obtained with the beaded-chain model, the interchromosomal contact probabilities are largely independent of whether the topological restriction was imposed or not.
DISCUSSION
Figure 6 helps us to explain how known characteristics of nuclear architecture stem from transcription factories-mediated interchromosomal attachments. It is well established that individual chromosomes form separated chromosomal territories that hardly interpenetrate each other (7,32). We, therefore, schematically presented them as relatively well demarked regions that show some overlaps in their peripheral regions. Chromosomal territories that show high transcriptional activity (red colour) are attached to each other via a transcription factory (black small circle). The chromosomal territories that are less transcriptionally active (blue colour) are less likely to become attached to other territories and are, therefore, shown as free. Simple geometrical reasons cause that some of the territories that are attached together have to be placed far from the nuclear envelope even if some others can still stay close to the nuclear envelope. Placement of inactive territories does not have this constraint and each of them can be placed close to the nuclear membrane. The competition between the territories that are either attached together or not ensure that, on average, the territories that are transcriptionally active find themselves closer to the centre of the nucleus than the territories of transcriptionally inactive chromosomes.
We propose that attachments between transcriptionally active chromosomal territories make them constitute bigger objects, which win the competition with smaller objects for a more central location within the nucleus. However, even without inter-attachments, transcriptionally active territories swell due to changes of chromatin structure and effectively occupy a larger fraction of the nuclear volume as compared to non-active chromosomal territories with a similar DNA content (48). Recent modelling studies showed that this swelling effect alone is sufficient to push the bigger territories towards the centre of nuclei (15). We believe that the inter-attachment effect is stronger than the swelling effect but as they both act in the same direction, it is likely that in living cells the effective swelling of transcriptionally active territories and their interconnection via transcription factories both contribute to their more central location within cell nuclei. Figure 6 also helps to explain why the actively transcribed genes find themselves preferentially on the surface of their respective territories. Several recent papers demonstrated that intermingling of chromosome fibres belonging to different chromosomes is entropically costly (14–16). Therefore, if at least two of the genes transcribed in a given factory belong to two different chromosomes or even to two distant portions of the same chromosome, the easiest way to bring them together, without requiring entropically costly intermingling, is when these genes became retained at the periphery of their chromosomal territories. Looking at Figure 6 we can also understand why the pair-wise interchromosome contacts involve most frequently two transcriptionally active regions (regions around transcription factories) as well as why the interchromosome contacts between two inactive regions are more frequent than between active an non-active regions (20). Since the space in the nucleus is very limited, bringing together transcriptionally active regions not only sequesters them from contacting other regions but also groups together transcriptionally inactive regions in the remaining space, as depicted in Figure 6.
Figure 6 is only a cartoon and should not be taken as a quantitative representation of our results. We indicated there a limited intermingling between individual territories and indicated that transcription factory is in the centre of limited intermingling between contacting chromosome territories. The question arises as to how realistic this presentation is with the respect to our results? Do inter-attached polygons still form separate territories or the fact that they are forced to contact each other increases their intermingling? To estimate the extent of intermingling between chromosome territories that were attached to each other and between those that were free, we estimated the extent of overlap between convex envelopes enclosing all the intersection points of individual modelled territories with an equatorial cross section through the enclosing sphere (these convex envelopes can be seen on Figures 3A and B and 4A and B). Using the definition of intermingling that we introduced in a recent work (14) and briefly described in Supplementary Data, we could conclude that intermingling between territories that were attached together was stronger than between chromosome territories that were free. However, the extent of intermingling was still small in the case of beaded chains with and without topological restriction and infinitely thin polygonal model with topological restriction (see Supplementary Tables S1 and S2). These modelling results agree with cytological data showing clear chromosomal territories despite interterritorial attachments and limited local intermingling (3).
Our proposal that transcription factories-mediated interchromosomal contacts lead to the equilibrium state of the nucleus in which transcriptionally active chromosomes are located closer to the centre of nucleus than less active chromosomes only applies to a default state of the nucleus which is attained without additional energy expenditure. However, cells can use the energy to change this default state and to remodel the usual nuclear architecture if that is needed for a particular purpose. For example, nuclei of photoreceptor cells of nocturnal vertebrates show the ‘reversed gradient’ of chromosomal position with inactive chromosomes preferentially occupying the centre of the nuclei (49). Interestingly, nuclei in those cells in addition to their standard function serve as specialized optical devices that optimize their optical properties when heterochromatin is located in their centres (50). Therefore, there was a strong evolutionary pressure to achieve this atypical nuclear organization of nuclei in photoreceptor cells of nocturnal vertebrates. This resulted in an evolutionary selection of additional mechanisms that at some energetic cost were able to override the default or economy state of nuclear organization. Nuclear actin and myosin are able to reposition chromosome territories and their action is needed when cells become quiescent and start to remodel their default nuclear organization (51). However, when the activity of nuclear actin is blocked, the quiescent cells maintain the default nuclear organization (51) that we were able to reproduce in our simulations.
It is generally accepted that transcription factories mediate contacts between transcriptionally active genes belonging to different chromosomal territories (6,8,15). However, it needs to be realized that transcription factories are composed not only of RNA polymerases but also of transcription factors such as EKFL, GATA-1 or FOG-1 and these factors also contribute to bridging of active genes belonging to different chromosomal territories (52). Therefore, our model of transcription factories-mediated attachment between active genes belonging to different chromosomes should not be understood that these contacts are mediated exclusively via RNA polymerases. In fact, studies using RNA polymerase inhibitors showed that the established contacts persist over several hours when most of RNA polymerases are not bound any more to their target genes (53).
In our simulations we considered cases of transcription factories that simultaneously transcribed genes belonging to different chromosome territories and for such transcription factories we observed their preferential localization on the peripheries of their chromosomal territories. However, some transcription factories may bind exclusively genes belonging to the same chromosomal territory. Such factories would not need to be preferentially located on the periphery of their territories and this may explain why some transcription factories can be found immersed within their territories (54,55).
In our simulations, transcription factories were permanently joining individual transcription units. A more realistic approach would be to model an equilibrium state where interacting DNA sites would be associated together only for a fraction of time (56). However, it is to be expected that when transcriptionally active chromosomes with hundreds of active genes have a substantial portion of them periodically involved in interchromosomal interactions then this will lead to a quasi-permanent interchromosomal interactions just like those that we modelled here.
CONCLUSIONS
Using a simple model of thermally fluctuating polymeric chains crowded within a sphere, we have shown that the mere fact that transcription factories mediate mutual attachment of independent transcriptionally active chromatin domains is sufficient to explain the following three principal features of nuclear architecture: (i) Transcriptionally active chromosomes are preferentially located closer to the centre of the nucleus. (ii) Actively transcribed genes are located at the periphery of their chromosomal territories. (iii) Transcriptionally non-active chromatin domains are grouped together similarly to the case of transcriptionally active chromatin domains. However, to observe all these features, it is critical that chromatin fibres belonging to different chromosome territories are prevented from extensive intermingling with each other. This can be spontaneously achieved when chromatin fibres were organized into loops of a relatively small chain length. Under such conditions, knotting and catenation probabilities are very low (41–43). Therefore, topoisomerase passages between chromatin fibres will be spontaneously directed towards simplification of loops topology, resulting in individual loops that are mostly unknotted and uncatenated with each other. The proposal that chromatin fibres are organized into relatively small loops is supported by a growing number of experimental data (35,36). An additional element that may be important for limiting intermingling of chromosomal territories could be provided by an ‘immunity’ of regular chromatin fibres to topo-II-mediated inter-fibre passages. Of course, type II DNA topoisomerases mediate very efficiently passages between protein-free DNA molecules. However, regular chromatin fibres may not be compatible with the steric requirements enabling one chromatin fibre to pass through a topo-II-spanned gate in the other chromatin fibre. If regular chromatin fibres were indeed poor substrates for topoII-mediated inter-fibre passages, then chromosome territories established after decondensation of mitotic chromosomes would be additionally prevented from intermingling (14).
We showed here that transcription factories-mediated association between transcriptionally active chromosomal territories is sufficient to explain that transcriptionally inactive territories are preferentially located near the nuclear membrane. However, other mechanisms may enhance this effect. For example an association of inactive chromatin with inner nuclear membrane proteins have been established in several studies (57,58). In fact, it seems reasonable that a mechanism that sequesters inactive chromatin does not need to act against the natural positional tendency of inactive chromosomal regions but just stabilizes this situation. Acting against the natural gradient would require energy input and would be difficult to maintain on a long term.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (31003A-116275 to A.S.). Funding for open access charge: The Open Access publication charge for this article has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. A. Bates, Prof. K. Millet and G. Witz for their comments on the manuscript. The computations were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the Swiss Institute of Bioinformatics.
REFERENCES
- 1.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 2.Belmont AS, Bruce K, Bulger RE, Beeuwkes R, 3rd, Saubermann AJ. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 5.Gasser SM. Nuclear architecture − visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 6.Kosak ST, Groudine M. Gene order and dynamic domains. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- 7.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. Plos Biol. 2005;3:826–842. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marenduzzo D, Micheletti C, Cook PR. Entropy-driven genome organization. Biophys. J. 2006;90:3712–3721. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki J, Takano A, Matsushita Y. Topological effect in ring polymers investigated with Monte Carlo simulation. J. Chem. Phys. 2008;129:034903. doi: 10.1063/1.2954018. [DOI] [PubMed] [Google Scholar]
- 11.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Nooijer S, Wellink J, Mulder B, Bisseling T. Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucleic Acids Res. 2009;37:558–568. doi: 10.1093/nar/gkp219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vettorel T, Grosberg AY, Kremer K. Statistics of polymer rings in the melt: A numerical simulation study. Phys. Biol. 2009;6:025013. doi: 10.1088/1478-3975/6/2/025013. [DOI] [PubMed] [Google Scholar]
- 14.Dorier J, Stasiak A. Topological origins of chromosomal territories. Nucleic Acids Res. 2009;37:6316–6322. doi: 10.1093/nar/gkp702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook PR, Marenduzzo D. Entropic organization of interphase chromosomes. J. Cell Biol. 2009;186:825–834. doi: 10.1083/jcb.200903083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki J, Takano A, Deguchi T, Matsushita Y. Dimension of ring polymers in bulk studied by Monte-Carlo simulation and self-consistent theory. J. Chem. Phys. 2009;131:6. doi: 10.1063/1.3247190. [DOI] [PubMed] [Google Scholar]
- 17.Scheuermann MO, Tajbakhsh J, Kurz A, Saracoglu K, Eils R, Lichter P. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp. Cell Res. 2004;301:266–279. doi: 10.1016/j.yexcr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Cremer T, Kupper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol. Cell. 2004;96:555–567. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl Acad. Sci. USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang CH, Belmont AS. Close encounters between active genes in the nucleus. Genome Biol. 2005;6:237. doi: 10.1186/gb-2005-6-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 23.Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 24.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faro-Trindade I, Cook PR. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value. Mol. Biol. Cell. 2006;17:2910–2920. doi: 10.1091/mbc.E05-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell University Press; 1979. [Google Scholar]
- 28.Dobay A, Dubochet J, Millett K, Sottas PE, Stasiak A. Scaling behavior of random knots. Proc. Natl Acad. Sci. USA. 2003;100:5611–5615. doi: 10.1073/pnas.0330884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J. Cell Biol. 1995;130:1239–1249. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munkel C, Langowski J. Chromosome structure predicted by a polymer model. Phys. Rev. E. 1998;57:5888–5896. [Google Scholar]
- 31.Munkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 32.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories − a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Bohn M, Heermann DW, van Driel R. Random loop model for long polymers. Phys. Rev. E. 2007;76:8. doi: 10.1103/PhysRevE.76.051805. [DOI] [PubMed] [Google Scholar]
- 34.Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, Indemans MH, Gierman HJ, Heermann DW, van Driel R, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc. Natl Acad. Sci. USA. 2009;106:3812–3817. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zlatanova J, Caiafa P. CCCTC-binding factor: to loop or to bridge. Cell. Mol. Life Sci. 2009;66:1647–1660. doi: 10.1007/s00018-009-8647-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papantonis A, Cook PR. Genome architecture and the role of transcription. Curr. Opin. Cell Biol. 2010;22:1–6. doi: 10.1016/j.ceb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. Review: the dynamics of the nuclear lamins during the cell cycle − relationship between structure and function. J. Struct. Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 38.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. USA. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller M, Wittmer JP, Cates ME. Topological effects in ring polymers: A computer simulation study. Phys. Rev. E. 1996;53:5063–5074. doi: 10.1103/physreve.53.5063. [DOI] [PubMed] [Google Scholar]
- 40.Marenduzzo D, Orlandini E. Topological and entropic repulsion in biopolymers. J. Stat. Mech. Theory Exp. 2009:L09002. [Google Scholar]
- 41.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 43.Hirayama N, Tsurusaki K, Deguchi T. Linking probabilities of off-lattice self-avoiding polygons and the effects of excluded volume. J. Phys. A Math. Theoret. 2009;42:105001. [Google Scholar]
- 44.Aronovitz JA, Nelson DR. Universal features of polymer shapes. J. Phys. 1986;47:1445–1456. [Google Scholar]
- 45.Haber C, Ruiz SA, Wirtz D. Shape anisotropy of a single random-walk polymer. Proc. Natl Acad. Sci. USA. 2000;97:10792–10795. doi: 10.1073/pnas.190320097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millett KC, Plunkett P, Piatek M, Rawdon EJ, Stasiak A. Effect of knotting on polymer shapes and their enveloping ellipsoids. J. Chem. Phys. 2009;130:165104. doi: 10.1063/1.3117923. [DOI] [PubMed] [Google Scholar]
- 47.Rawdon EJ, Kern JC, Piatek M, Plunkett P, Stasiak A, Millett KC. Effect of knotting on the shape of polymers. Macromolecules. 2008;41:8281–8287. [Google Scholar]
- 48.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 50.Ragoczy T, Groudine M. The nucleus inside out − through a rod darkly. Cell. 2009;137:205–207. doi: 10.1016/j.cell.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook PR. A model for all genomes: the role of transcription factories. J. Mol. Biol. 2009;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 53.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrai C, Xie SQ, Luraghi P, Munari D, Ramirez F, Branco MR, Pombo A, Crippa MP. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 8:e1000270. doi: 10.1371/journal.pbio.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicodemi M, Prisco A. Thermodynamic Pathways to Genome Spatial Organization in the Cell Nucleus. Biophys. J. 2009;96:2168–2177. doi: 10.1016/j.bpj.2008.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, Singh PB, Georgatos SD. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- 58.Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp. Cell. Res. 2009;315:1895–1903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.