Abstract
Amplification of the 5′ ends of cDNA, although simple in theory, can often be difficult to achieve. We describe a novel method for the specific amplification of cDNA ends. An oligo-dT adapter incorporating a dUTP-containing PCR primer primes first-strand cDNA synthesis incorporating dUTP. Using the Cap finder approach, another distinct dUTP containing adapter is added to the 3′ end of the newly synthesized cDNA. Second-strand synthesis incorporating dUTP is achieved by PCR, using dUTP-containing primers complimentary to the adapter sequences incorporated in the cDNA ends. The double-stranded cDNA-containing dUTP serves as a universal template for the specific amplification of the 3′ or 5′ end of any gene. To amplify the ends of cDNA, asymmetric PCR is performed using a single gene-specific primer and standard dNTPs. The asymmetric PCR product is purified and non-target transcripts containing dUTP degraded by Uracil DNA glycosylase, leaving only those transcripts produced during the asymmetric PCR. Subsequent PCR using a nested gene-specific primer and the 3′ or 5′ T-RACE primer results in specific amplification of cDNA ends. This method can be used to specifically amplify the 3′ and 5′ ends of numerous cDNAs from a single cDNA synthesis reaction.
INTRODUCTION
The genomes of an increasing number of organisms have been sequenced; however, the exact structure of many transcripts remain unresolved, particularly their 5′ untranslated regions (UTRs). Many open-reading frames are predicted and need to be supported by experimental data to obtain an accurate annotation of the genome. For example, Salehi-Ashtiani et al. (1) performed large-scale rapid amplification of cDNA ends (RACE) on Caenorhabditis elegans, identified UTRs, new exons and redefined previously annotated exons. The lack of experimental data supporting gene structure is in part due to the methodologies employed in cloning gene sequences. Cloning of novel cDNAs typically results in sequences that only partially represent the mRNA species, with the 3′ ends usually well represented and the 5′ end of the sequence often incomplete. This tendency can be due to RNA degradation or is a result of the cDNA synthesis reaction not reaching completion. When working with novel organisms, where the genome has not been sequenced, obtaining the 5′ end of the cDNA, although in theory quite simple, can often prove to be technically challenging.
The unknown 5′ sequence from the ends of cDNA molecules can be cloned using a PCR technique know as rapid amplification of cDNA ends (RACE) (2). In Classic RACE, homopolymer tailing of the cDNA is used to append a linker sequence to the cDNA terminus. Extension of unknown regions of the cDNA is then achieved through PCR using a gene-specific primer and a primer that can bind and prime DNA synthesis from the linker sequence. Different versions of RACE have been developed, making significant improvements to the original methodology, including New RACE (3) and Cap finder RACE (4,5). In New RACE, an anchor primer is ligated to the 5′ end of the mRNA molecule prior to reverse transcription, which subsequently synthesized cDNA molecules incorporate into their sequence (3). Therefore, only full-length cDNA molecules are amplified by PCR using the anchor primer and gene-specific primer. The Cap-switch RACE (Cap finder) protocol exploits the Murine moloney leukaemia virus (MMLV) reverse transcriptase ability to add extra 2–4 cytosine residues to the 3′ ends of cDNA molecules after reaching the cap structure at the 5′ end of the mRNA template. When a primer with multiple guanine residues at its 3′ end is present in the reaction mix, annealing of the poly G to poly C sequences occurs, allowing for copying of the annealed primer sequence to the cDNA, adding an adapter sequence to the end of the cDNA (4,5). These methods have been developed into commercially available RACE kits including Clontech (Cap switching Smart RACE), Invitrogen (5′ RACE system) and Ambion (First Choice RLM-RACE kit).
Many of the commercially available kits offer protocols that allow the amplification of 5′ cDNA ends of many genes from a single cDNA synthesis reaction. Despite claims of specific amplification of the desired 5′ region of a target gene, a common problem that occurs in RACE reactions is obtaining non-specific amplification products (5,6,7). The use of a second nested PCR reaction has been shown to increase the specificity of the reaction; however, non-specific amplification and false positive products are still often obtained. When cDNA synthesis is primed using an oligo-dT primer, all mRNA species are reverse transcribed, and the 3′ primer site will subsequently be incorporated into the 3′ end of all newly transcribed cDNAs. When cDNA templates such as these are used for PCR, non-specific amplification is likely due to the incorporation of the primer sequence in every cDNA molecule in the universal pool of full-length cDNAs.
Prevention of PCR carryover contamination by incorporation of dUTP during PCR and uracil-DNA glycosylase (UDG) treatment is a widely used technique (8). dUTP is incorporated into all amplicons by substituting dUTP for dTTP in PCR reactions. Treatment of PCR products with UDG prior to thermal cycling cleaves DNA at uracil bases thereby inhibiting amplification of carryover contamination from previous PCR reactions (9). This function can be exploited to remove unwanted DNA sequences from reaction mixtures. For example, the use of dUTP and UDG for selective RNA amplification has been reported (10); however, no methods using dUTP/UDG for RACE, and in particular, the technically more challenging 5′ RACE are currently available. We describe a modified version of the RACE protocol that enables the PCR reaction to be greatly simplified by degrading non-target cDNA sequences using UDG. Double-stranded cDNA is synthesized to incorporate dUTP, and this then serves as a template for subsequent amplification of cDNA ends of any target gene. During synthesis of the double-stranded cDNA, PCR primer sites (where dUTP replaces dTTP) are incorporated at the 3′ and 5′ ends of each cDNA molecule. These primer sites are incorporated at the 3′ end as an oligo-dT adapter sequence and at the 5′ end through the Cap select/Cap finder approach of the newly synthesized first-strand cDNA. Second-strand synthesis using a dUTP containing nucleotide mix is performed through PCR using primers (which also contain dUTP) complementary to the 5′ and 3′ priming sites. This preamplification step increases the abundance of rare transcripts and importantly allows small amounts of input RNA to be used. The double-stranded cDNA, incorporating dUTP, now serves as a universal pool for PCR amplification of 5′ and/or 3′ cDNA ends for any gene of interest. To specifically amplify the ends of target genes, an asymmetric PCR using a single gene-specific primer (GSP) and standard dNTPs (without dUTP) is performed. The reaction mixture is then treated with UDG to simplify the PCR reaction mixture by degrading non-target cDNAs. A subsequent PCR using a nested gene-specific primer in combination with a primer (T-RACE primer) incorporated into the 5′ or 3′ end of the cDNA during cDNA synthesis results in specific amplification of cDNA ends for target sequences. This technique provides a novel and reliable method to specifically amplify the 5′ and 3′ ends of cDNAs from a single pool of cDNA.
MATERIALS AND METHODS
RNA extraction
Total RNA was extracted from 100 mg of Atlantic salmon (Salmo salar L.) or zebrafish (Danio rerio) fast skeletal muscle tissue using Lysing matrix D (Qbiogene, Irvine, CA) with 1 ml Tri Reagent (Sigma, Gillingham, Dorset, UK) and homogenized using a Fast Prep instrument (Qbiogene, Irvine, CA). The total RNA was quantified based on absorbance at 260 nm. Genomic DNA contamination was removed by treatment with Turbo DNA-free (Ambion, Austin, TX, USA), and the integrity of purified RNA confirmed by agarose gel electrophoresis.
T-RACE ready cDNA synthesis
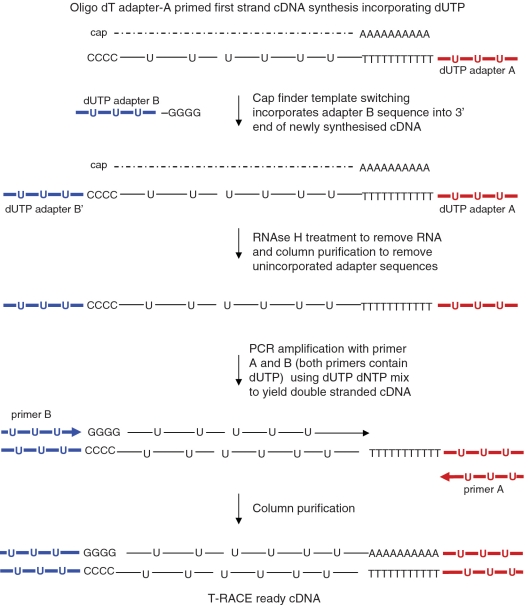
The T-RACE ready protocol is depicted in Figure 1 and the primers used are listed in Table 1.
Figure 1.
Diagrammatic representation of double-stranded template (T-RACE cDNA) synthesis protocol. First-strand cDNA synthesis using a dNTP mix that includes dUTP is primed using an oligo-dT adapter-A primer. After reaching the 5′ end of the mRNA (dashed line), oligo(dC) is added to the end of the cDNA by the MMLV reverse transcriptase. Base pairing between the oligo(dC) and the oligo(dG) of adapter-B occurs, and through the process of template switching, the reverse compliment of adapter-B (adapter B′) is incorporated into the 3′ end of the newly synthesized cDNA as previously described (4,5). Adapters A and B both contain dUTP instead of dTTP. Adapters A and B serve as priming sites for PCR amplification using primers A and B, which also have dTTP replaced by dUTP. PCR is performed using a dNTP mix that contains dUTP, so the resulting double-stranded cDNA (T-RACE ready cDNA) contains dUTP residues throughout and dUTP-containing adapters at the 3′ and 5′ ends.
Table 1.
Primer and adapter sequences used for first- and second-strand cDNA synthesis and specific amplification of cDNA ends
| Primer | Sequence (5′ – 3′) | Application |
|---|---|---|
| Adapter A | AUCUCGAGUUCGCGCCGGAUCC(T)25VN | cDNA synthesis and adapter-A incorporation |
| Adapter B | AUAUGCACUGCCGCGUCUGAGGGGGGGG | Cap finder adapter-B incorporation |
| Primer A | CUCGAGUUCGCGCCGGAUC | Second-strand cDNA synthesis |
| Primer B | AUAUGCACUGCCGCGUCUGA | Second-strand cDNA synthesis |
| 5′ T-RACE primer | ATATGCACTGCCGCGTCTGA | 5′ specific amplification of cDNA ends |
| 3′ T-RACE primer | CTCGAGTTCGCGCCGGATC | 3′ specific amplification of cDNA ends |
First-strand cDNA synthesis incorporating dUTP and 5′ and 3′ adapter sequences
First-strand cDNA synthesis was performed using a modified dNTP mixture that contained dUTP and a MMLV reverse transcriptase. cDNA synthesis was primed with Adapter A (Table 1) using 1 μg of total RNA. The RNA, Adapter A and dNTPs were incubated at 65°C for 5 min and then chilled on ice before addition of the remaining components in the following reaction mixture (20 μl): 2.5 μM Adapter A, 50 mM Tris–HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 1.0 mM dUTP, 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.125 mM dTTP, 40 U RNAse inhibitor (RNase OUT, Invitrogen, Carlsbad, CA, USA), 200 U SuperscriptII (Invitrogen, Carlsbad, CA, USA) and incubated for 1 h at 42°C. After incubation, 0.4 μl of fresh 100 mM MnCl2 and 1 μl of Adapter B (10 μM) were added to the reaction that was further incubated for 15 min at 42°C. The reaction was then heat inactivated at 70°C for 10 min. Degradation of the RNA in the RNA/cDNA hybrid was achieved by the addition of 1 μl RNAse H (Invitrogen, Carlsbad, CA, USA) and incubation at 37°C for 15 min. The reaction was purified through a QIAquick PCR purification column and eluted in 30 μl elution buffer as per manufacturer’s guidelines (Qiagen Inc., Chatsworth, CA, USA).
Second-strand synthesis incorporating dUTP by PCR
Second-strand synthesis was achieved by PCR using the priming sites incorporated at the 3′ (Adapter B) and 5′ (Adapter A) ends of the first-strand cDNA. Of the purified first-strand cDNA 2 μl was used as template and amplified in the following reaction (50 μl): 2.5 mM MgCl2, 1.0 mM dUTP, 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.125 mM dTTP, 0.5 μM primer A and 0.5 μM primer B (Table 1). The reaction mixture was mixed and heated to 95°C before addition of five units of Taq DNA polymerase (Bioline, London, UK). The reaction mixture was then incubated at 95°C for a further 2 min, followed by 21 cycles of 95°C denaturing for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 6 min. The reaction was then chilled on ice and purified through a QIAquick PCR purification column (Qiagen Inc., Chatsworth, CA, USA) and eluted in a 100 µl elution buffer as per manufacturer’s guidelines. The purified double-stranded cDNA, incorporating dUTP and 5′ and 3′ adapter sequences (T-RACE ready cDNA) serves as template for the following PCR reactions.
T-RACE PCR
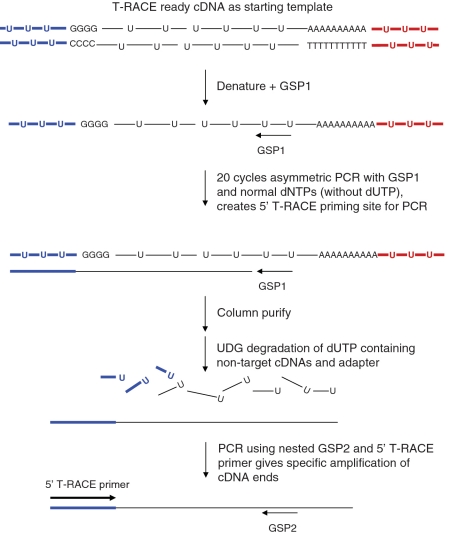
The PCR reactions for the targeted amplification of cDNA ends are shown in Figure 2, and the primers used are listed in Table 1.
Figure 2.
Diagrammatic representation of the 5′ specific amplification of cDNA ends using T-RACE. T-RACE ready cDNA serves as template for an asymmetric PCR reaction using a single gene-specific primer (GSP1) and standard dNTPs (without dUTP). This reaction produces single-stranded target cDNAs that include the adapter sequence comprising standard dNTPs. UDG treatment degrades all non-target cDNAs and adapters containing dUTP. Subsequent PCR using a nested GSP2 with the 5′ T-RACE primer results in the specific amplification of target cDNA ends.
Asymmetric PCR using standard dNTPs
Asymmetric PCR was performed using a gene-specific primer (GSP1) and standard dNTPs (dATP, dTTP, dGTP, dCTP) in the following hot-start 20 μl reaction: 0.5 μM GSP1, 2μl T-RACE ready cDNA, 16 mM (NH4)2SO4, 67 mM Tris–HCl, 3 mM MgCl2, 0.25 mM dNTPs. The reaction mixture was heated to 95°C for 2.5 min before the addition of one unit of Taq DNA polymerase (Bioline, London, UK), and then cycled 20 times with the following conditions: 95°C for 30 s, 60°C for 30 s, 72°C for 2 min. The reaction was chilled on ice, purified through a MinElute PCR purification column (Qiagen Inc., Chatsworth, CA, USA) and eluted in 11 μl water. The length of time for the 72°C extension step of the PCR should be varied according to the expected amplicon length.
Uracil DNA glycosylase degradation of non-target dUTP containing cDNAs
The entire asymmetric reaction is used in the following steps using a nested gene-specific primer (GSP2), and the primers used for PCR do not contain dUTP. cDNAs incorporating dUTP were removed by UDG treatment to leave only those cDNAs synthesized with standard dNTPs during the asymmetric PCR. The following 17.8 μl reaction was performed: 10 μl asymmetric PCR product, 2 μl (two units) Uracil DNA glycosylase (New England Biolabs, Hitchen, UK), 2 μl 10 × PCR buffer (160 mM (NH4)2SO4, 670 mM Tris–HCl), 1.2 μl 50 mM MgCl2, 1 μl nested GSP2, 1 μl of 10 μM T-RACE primer (Table 1), 0.6 μl water and incubated at 37°C for 30 min. Depending on whether 3′ or 5′ cDNA ends are targeted, the 3′ or 5′ T-RACE primer is used for amplification.
Targeted amplification of cDNA ends (T-RACE PCR)
The UDG-treated cDNA mixture was heated to 95°C before the addition of 2 μl 2.5 mM standard dNTPs and 0.2 μl (one unit) of Taq DNA polymerase (Bioline, London, UK). The reaction was denatured at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 60°C for 30s, 72°C for 2 min, followed by a final extension of 7 min at 72°C. The length of time for the 72°C extension step of the PCR should be varied according to the expected amplicon length.
Cloning
PCR products were separated on a 1.2% (m/v) agarose gel, and the bands excised and purified using a QIAquick gel extraction kit (Qiagen Inc., Chatsworth, CA, USA). The PCR products were cloned into a T/A pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA) and transformed into chemically competent TOP10 Escherichia coli cells (Invitrogen, Carlsbad, CA, USA). Individual colonies were grown and plasmids purified using a QIAprep Spin Miniprep Kit (Qiagen Inc., Chatsworth, CA, USA). Sequencing was performed using T3 and T7 sequencing primers with Big Dye terminator v3.1 sequencing chemistry (Applied Biosystems, Foster City, CA, USA) at the University of Dundee Sequencing Facility.
RESULTS AND DISCUSSION
We report the development of a novel method for acquiring the 5′ ends of cDNA molecules using a modification of previously published methods. Using this approach, we have identified the 5′ ends of cDNA molecules expressed in the muscle of zebrafish and Atlantic salmon (Figure 3A). The novelty of the method involves the production of double-stranded cDNA incorporating dUTP, and the addition of 5′ and 3′ adapters that also contain dUTP (Figure 1). By producing the second strand of cDNA, a priming site for a gene-specific primer for 5′ RACE is introduced, allowing for an asymmetric PCR reaction to be performed using a standard dNTP mix. This asymmetric PCR using only the gene-specific primer produces a cDNA fragment that includes the 5′ adapter sequence and comprises standard dNTPs (Figure 2). The degradation of dUTP-containing cDNA molecules by Uracil DNA glycosylase treatment leaves only the molecules comprising standard dNTPs primed by the gene-specific primer (Figure 2). This now serves as the template for PCR amplification using a nested gene-specific primer and the 5′ T-RACE primer, thus specifically amplifying the 5′ end of the desired cDNA sequence.
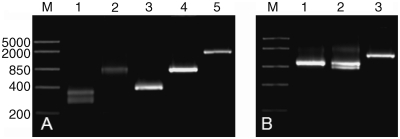
Figure 3.
(A) Specific amplification of 5′ cDNA ends from Atlantic salmon (Ss) and Zebrafish (Dr) fast muscle. Lane 1 STAC3 (Ss), Lane 2 IGF-I (Ss) Lane 3 CMYA5 (Dr), Lane 4 MAFbx (Ss), Lane 5 STAC3 (Ss). DNA size marker of 5000, 2000, 850, 400 and 200 bp is shown in Lane M. In all cases, a single amplicon was produced, except for STAC3 (Lane 1) where two alternative splice variants were obtained. 5′ T-RACE successfully amplified IGF-I (Lane 2) that is expressed at low levels in Atlantic salmon muscle, and a long transcript, CMYA5 (Lane 3), that is over 6 kb in length. To demonstrate that long amplicons can be amplified using the T-RACE method, we amplified ∼1.9 kb from STAC3 (Lane 5). (B) To demonstrate that the single pool of cDNA can be used for both 5′ and 3′ amplification of cDNA ends, we amplified the 3′ ends of Zebrafish CMYA5 (Lane 1) and Atlantic salmon MAFbx (Lane 2) and STAC3 (Lane 3). DNA size marker of 5000, 2000, 850, 400 and 200 bp is shown in Lane M.
To demonstrate the effectiveness of our approach, we amplified the 5′ ends of several genes of differing lengths whose mRNAs are present in varying abundance (Figure 3A). For example, we amplified the 5′ ends of zebrafish myospryn (CMYA5), a gene over 6 kb in length, Atlantic salmon MAFbx and insulin-like growth factor-I (IGF-I) that is expressed at low levels in muscle (Figure 3A). We also demonstrated that long amplicons can be generated using this method by amplifying the ∼1.9 kb 5′ end of STAC3 (Figure 3A). Using GSPs and the 5′ T-RACE primer, in all cases we were able to obtain a single amplicon (except for STAC3 where we identified two alternatively spliced transcripts) of the correct size (Figure 3A), the identity of which was confirmed by sequencing. We also used the same cDNA pool to amplify the 3′ ends for a number of genes (Figure 3B), demonstrating that a single cDNA pool can be used for both 5′ and 3′ amplification of cDNA ends.
The reported T-RACE method has several advantages over current methodologies. Standard RACE methods can require large amounts of RNA for template that may not be possible to acquire from certain samples. Since the T-RACE protocol uses PCR during second-strand synthesis that amplifies the cDNA pool and increases the abundance of rare transcripts, it enables small amounts of starting material to be used. Additionally, standard RACE methods often result in non-specific amplification giving false positive results (5,6,7). When cDNA is synthesized using oligo-dT, the subsequent incorporation of the adapter sequence for 5′ RACE (through terminal transferase tailing or cap finder methods) occurs in every cDNA molecule. Due to this, mispriming events during PCR will often lead to exponential amplification, and so smears rather than distinct bands are often obtained. The use of a nested PCR reaction should increase the specificity of the RACE reaction; however, non-specific products are frequently obtained. The T-RACE method described herein degrades all of the non-target cDNA molecules, but leaves intact those that were primed by the gene-specific primer and synthesized using standard dNTPs, thereby significantly simplifying the templates present in the PCR reaction. As the gene-specific primer is used in an asymmetric PCR, where higher annealing temperatures are permitted, rather than during cDNA synthesis, the specificity of the reaction is increased. By using a nested primer in the second PCR reaction, specific amplification of the desired molecule is achieved. To demonstrate the increased specificity obtained by the T-RACE protocol, we amplified the same 5′ cDNA ends using the Classic RACE protocol (2,11). We were only able to obtain bands of equivalent size to the confirmed (sequenced) 5′ cDNA ends in two of the five reactions (for STAC3, Lane 1, and CMYA5, Lane 3, Figure 4B), and this was accompanied by high background in all reactions, a likely consequence of RNA self-priming as total RNA was used as the template (11).
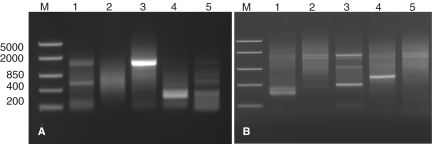
Figure 4.
(A) Non-specific amplification of products without UDG treatment. To demonstrate the increased specificity obtained by UDG treatment, we performed the same reactions to those in Figure 3A but without UDG treatment prior to thermal cycling. (B) Amplification of 5′ cDNA ends using the Classic race protocol. To demonstrate an improvement over existing techniques, PCRs were designed to amplify the 5′ cDNA ends of the same genes as in Figure 3A. Two of the five reactions (Stac3, Lane 1 and CMYA5, Lane 3) gave bands of the equivalent size to those obtained using the T-RACE protocol, with significantly higher background observed in all reactions.
As the various 5′ RACE methods such as Capfinder RACE and Classic RACE have their limitations, we tested the compatibility of our approach with the Classic RACE method. By modifying the Classic RACE protocol (2,11), to use dUTP-containing dNTP mix and uracil-containing adapters, we were able to produce double-stranded T-RACE ready cDNA containing uracil, which we then successfully performed T-RACE PCR from (data not shown). This alternative approach might be necessary when trying to perform T-RACE on certain transcripts. For example, the cap finder method can be problematic if the dG-containing switch primer binds to C-rich sequences in the mRNA yielding incomplete 5′ sequences (11). Similarly, the Classic RACE method can sometimes yield products that do not contain the complete 5′ sequence when the reverse transcriptase fails to reach the end of the mRNA (11). The T-RACE method should also be compatible with the New RACE method (3), if dUTP-containing dNTPs are used during the first-strand cDNA synthesis, and then double-stranded cDNA is produced as described by the protocol herein.
Recently, Olivarius et al. (12) demonstrated the use of high-throughput sequencing to identify transcriptional start sites via 5′ RACE. The T-RACE method can also be adapted for large-scale applications. To achieve this, the asymmetric PCR should be performed as described above, but several GSPs corresponding to genes of interest should be used in a single reaction. Following the UDG treatment, the T-RACE reaction should be performed using a mixture of nested GSPs that contain the illumina adapter attached at the 5′ end and the T-RACE primer that also has an illumina adapter attached at the 5′ end (for details of illumina adapters, see (12)). PCR using these primers will yield products that can be directly sequenced using high-throughput sequencing technologies.
During the development of this method, we made several changes to our original protocol that improved the efficiency of the reactions. Taq polymerase is less efficient at incorporating dUTP than dTTP (9) that could impact on the second-strand cDNA synthesis. A ratio of dUTP:dTTP of 8:1 improved the efficiency of the T-RACE reaction but still resulted in sufficient degradation of non-target cDNAs to give specific amplification. We also observed an improvement in the specificity of the PCR reactions by adding the MnCl2 and adapter-A after cDNA synthesis has completed. Presumably this modification decreases the chances of adapter-A priming cDNA synthesis during the first-strand cDNA reaction that could lead to spurious amplification products in subsequent reactions. Performing all reactions as a hot-start (adding Taq DNA polymerase after reactions have reached 95°C) improved specificity and this could also be achieved using a commercially available hot-start enzyme. Originally our method did not use a PCR (exponential) amplification step during double-stranded cDNA synthesis, and we were unable to amplify rare transcripts such as IGF-I from this template. This is likely to be due to the fact that the asymmetric PCR performed using standard dNTPs, which is critical to the success of this method, does not result in exponential amplification of products. However, performing 21 cycles of PCR for the second-strand synthesis improved the outcomes for amplifying rare transcripts, most likely by greatly increasing the number of targets available for the asymmetric PCR. The use of Uracil DNA glycosylase and dUTP has been widely used to control PCR product carryover contamination (8,9), and its effectiveness in simplifying 5′ RACE reactions is clearly demonstrated here (Figures 3A and 4A). Nested PCR reactions, which were not treated with UDG prior to thermal cycling resulted in smears and non-specific amplification products (Figure 4A), whereas treatment with UDG resulted in specific amplification (Figure 3A).
The method described herein offers several advantages over other protocols that are currently in use. Using UDG-treated PCR products as template results in specific amplification, which in most cases results in a single amplicon of the correct size. We have now used this technique to amplify the 5′ ends of several cDNAs, each one from the same T-RACE ready cDNA pool using total RNA as the starting material.
FUNDING
Funding for open access charge: Biotechnology and Biological Science Research Council (grant number BB/D015391/1).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
All S. salar samples were provided by Landcatch, Lochgilphead, UK. The authors thank Hung-Tai Li and Ian Amaral for zebrafish RNA.
REFERENCES
- 1.Salehi-Ashtiani K, Lin CW, Hao T, Shen Y, Szeto D, Yang XP, Ghamsari L, Lee H, Fan CY, Murray RR, et al. Large-scale RACE approach for proactive experimental definition of C. elegans ORFeome. Genome Res. 2009;19:2334–2342. doi: 10.1101/gr.098640.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts—amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromont-Racine M, Bertrand E, Pictet R, Grange T. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res. 1993;21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt WM, Mueller MW. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999;27:e31. doi: 10.1093/nar/27.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schramm G, Bruchhaus I, Roeder T. A simple and reliable 5′-RACE approach. Nucleic Acids Res. 2000;28:e96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaefer BC. Revolutions in rapid amplification of cDNA ends—new strategies for polymerase chain-reaction cloning of full-length cDNA ends. Anal. Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 7.Lipovich L, Lynch ED, Lee MK, King MC. A novel sodium bicarbonate cotransporter-like gene in an ancient duplicated region: SLC4A9 at 5q31. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-research0011. research0011.1–0011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aslanzadeh J. Preventing PCR amplification carryover contamination in a clinical laboratory. Ann. Clin. Lab. Sci. 2004;34:389–396. [PubMed] [Google Scholar]
- 9.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain-reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 10.Buchman GW, Schuster DM, Rashtchian A. Selective Rna amplification—a novel method using dump-containing primers and uracil DNA glycosylase. PCR Methods Appl. 1993;3:28–31. doi: 10.1101/gr.3.1.28. [DOI] [PubMed] [Google Scholar]
- 11.Scotto-Lavino E, Guangwei D, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- 12.Olivarius S, Plessy C, Carninci P. High-throughput verification of transcriptional starting sites by Deep-RACE. BioTechniques. 2009;46:130–132. doi: 10.2144/000113066. [DOI] [PubMed] [Google Scholar]