Abstract
Summary: New software tools for graphical genotyping are required that can routinely handle the large data volumes generated by the high-throughput single-nucleotide polymorphism (SNP) platforms, genotyping-by-sequencing and other comparable genotyping technologies. Flapjack has been developed to facilitate analysis of these data, providing real time rendering with rapid navigation and comparisons between lines, markers and chromosomes, with visualization, sorting and querying based on associated data, such as phenotypes, quantitative trait loci or other mappable features.
Availability: Flapjack is freely available for Microsoft Windows, Mac OS X, Linux and Solaris, and can be downloaded from http://bioinf.scri.ac.uk/flapjack
Contact: flapjack@scri.ac.uk
1 INTRODUCTION
The concept of a graphical genotype to visualize haplotype diversity between chromosomes has been widely adopted since Young and Tanksley (1989) used it in the context of restriction fragment length polymorphism (RFLP) mapping populations. Existing software tools to display graphical genotypes include GGT (van Berloo, 2008) and Geneflow (geneflowinc.com). The advent of new high-throughput genotyping technologies have given a renewed stimulus to the concept of graphical genotyping, through a combination of dramatic reduction in cost per data point and vastly increased marker density and throughput. The resultant high-density data underpin new genetic approaches such as genome-wide association analysis (Rostoks et al., 2006). It also leads to the possibility of visually comparing many lines (e.g. samples or individuals) or sorting and selecting based on phenotype, identified groupings and genome features, such as quantitative trait loci (QTL) or gene models mapped to the genetic or physical genome. However, the ability to generate datasets with many thousands of markers (McMullen et al., 2009) on many thousands of lines imposes a significant demand on both software tools and the underlying computer hardware. Flapjack provides a high performance visual interface into graphical genotyping applications in genetics and plant breeding.
2 FEATURES
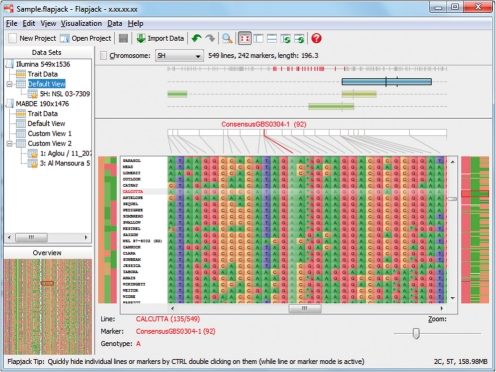
Flapjack's main display (Fig. 1) consists of a genotype rendering canvas that shows the data for a given chromosome. The alleles are plotted as a grid, with lines/germplasm running horizontally across the screen and markers/loci running vertically. The line names are shown in a list to the left, and across the top we provide a graphical view of the positions of the markers on the currently selected chromosome (from either physical or genetic maps). Several alternative map displays are provided, including a global view that shows where the currently visible markers are located on the chromosome, and a local view, that scales and optimizes the map to concentrate only on the region containing the currently visible on-screen markers. Hovering the mouse over an allele highlights not only the data under the point but also the name of the line in the lines list, the position on the chromosome display, and graphically displays the entire dataset for the line and marker at that position.
Fig. 1.
Flapjack's main interface, showing SNP genotypes, QTL and a trait-data heat map (additional screenshots can be seen online at http://bioinf.scri.ac.uk/flapjack/screenshots.shtml).
Flapjack provides several customizable colour schemes for data display, and will attempt to auto-select a suitable scheme based on the type of data loaded. The schemes include a four-colour nucleotide model (homozygous genotypes get a single colour; heterozygous genotypes are split diagonally); similarity models, that use one colour for every allele of a reference line or marker, and a second colour for any data that differs from the reference; and a model that performs frequency-based colouring that can be used to highlight rare alleles and haplotypes on a per-marker basis. Random colour schemes also exist that are applicable to datasets with a large number of possible values per allele position, such as SSR data. Flapjack's subtle use of colours and gradients allows for pattern recognition and structure to still be seen, even at the highest levels of overview.
Once a project is in use, additional data types can be imported and visualized alongside the main display. Information on phenotypic traits—both numerical and categorical (per line)—is displayed as a heat map running alongside the lines. This can also be used to reorder the lines, for example, by yield or flowering date.
QTL aligned against chromosome map positions may be visualized at the top of the screen, using a novel method of packing and displaying the features across a custom number of tracks, with the number controlled by a slider.
A user interacts with Flapjack using one of three modes: navigation mode, marker mode or line mode; with the latter two options enabling support for object highlighting and selection. This provides a graphical means of filtering the data, for example, to reorder the lines based upon their similarity across a specific subset of markers or to export sections of data into their own custom view. Selections can be made either manually or by markers under a QTL. Flapjack allows the user to create any number of these custom views, each containing its own set of lines, markers, ordering, colour schemes, bookmarked locations and so on.
The data for a given view—either in graphical or in its underlying raw format—can be exported back to disk. Images can be produced and saved in PNG format for the current view, or the on-screen subsections of a view, and the user can select whether to include all components (allele data, chromosome maps, line names, traits, etc.) or pick and choose only the ones of interest. When exporting the underlying data, similar options are available to export the entire dataset or to only include data from specific chromosomes or the currently selected lines and markers. The data are saved in tab-delimited plain text files identical in format to the files original imported into Flapjack.
Although completely standalone, with data imported via simple plain-text formats, integration with external data sources such as Germinate (bioinf.scri.ac.uk/germinate) is also possible. This provides easy selection and export of data directly into Flapjack, along with web-links back to the line and marker data in the original database. This feature has been designed to work with any external data source, by means of supplying Flapjack with a custom URL that can be queried with key/value pairs.
Flapjack projects are persistent, with all data, views, user selections and so on being saved to either an XML-based file or an experimental binary format more suited to very large datasets. The XML and text formats are documented on our web site, and are also currently supported by iMAS (icrisat.org/bt-biomatrics-imas.htm), QU-GENE (Podlich and Cooper, 1998), Gramene (Liang et al., 2008), Genstat (vsni.co.uk/software/genstat) and The Hordeum Toolbox (hordeumtoolbox.org). Projects can also be created using a command-line utility, which provides a convenient integration with custom analysis pipelines and databases.
3 IMPLEMENTATION
Flapjack is written in Java and is compatible with any system running Java 1.6 or higher. For convenience, we provide installable versions with everything required to run the application, including a suitable Java run-time. These are available for Windows, Mac OS X, Linux and Solaris. Flapjack regularly monitors our server for new versions and will prompt, download and update quickly and easily when a new release is available. The code is internationalized and is distributed with translations in English (UK/US) and German.
The code can take advantage of multicore processors, a feature especially significant for the rendering code, which—among its other optimizations—is capable of simultaneous rendering across all cores, greatly improving the end-user experience when navigating around large or complex datasets. We have designed Flapjack to be very memory efficient, and are confident that it can comfortably handle datasets with hundreds of millions of alleles even on a machine with just 1 GB of main memory.
4 FUTURE WORK
Future development with Flapjack will entail enhancing its visualizations to provide better support for very small datasets, primarily by enabling the display of all markers across the genome in a single view. We want to extend support for rendering features beyond QTL to include more generic features, such as gene models for SNP data anchored to physical maps, and to provide a graph track to display summary information such as PIC values or test statistics. We are also working with academic and breeding company partners to explore supporting additional data formats such as HapMap and PLINK, and on closer integration with Germinate, by allowing its databases to be automatically populated by the data imported into Flapjack.
ACKNOWLEDGEMENTS
We would like to thank colleagues within the Genetics and BioSS Programmes at SCRI for their input to this project.
Funding: Scottish Government (RERAD, Programme 1); Scottish Funding Council; Scottish Enterprise through the Scottish Bioinformatics Research Network (SBRN) project.
Conflict of Interest: none declared.
REFERENCES
- Liang C, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- Podlich DW, Cooper M. QU-GENE: a platform for quantitative analysis of genetic models. Bioinformatics. 1998;14:632–653. doi: 10.1093/bioinformatics/14.7.632. [DOI] [PubMed] [Google Scholar]
- Rostoks N, et al. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc. Natl Acad. Sci. USA. 2006;103:18656–18661. doi: 10.1073/pnas.0606133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. J. Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- Young ND, Tanksley SD. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor. Appl. Genet. 1989;77:101. doi: 10.1007/BF00292322. [DOI] [PubMed] [Google Scholar]