Abstract
Summary: GView is a Java application for viewing and examining prokaryotic genomes in a circular or linear context. It accepts standard sequence file formats and an optional style specification file to generate customizable, publication quality genome maps in bitmap and scalable vector graphics formats. GView features an interactive pan-and-zoom interface, a command-line interface for incorporation in genome analysis pipelines, and a public Application Programming Interface for incorporation in other Java applications.
Availability: GView is a freely available application licensed under the GNU Public License. The application, source code, documentation, file specifications, tutorials and image galleries are available at http://gview.ca
Contact: gary.van.domselaar@phac-aspc.gc.ca
1 INTRODUCTION
The commercial introduction of next-generation sequencing technology has reduced the time and expense of genome sequencing to such an extent that it is now possible for virtually any microbiology lab to generate genome sequences for the microorganisms that they study. Visualizing the genomic features contained in these sequences is important to aid biologists in understanding the genetic basis of microbial traits, and to convey this information to others. A number of software packages already exist for this purpose (Supplementary Table 1). Early software packages like GenomePlot (Gibson and Smith, 2003) and GenoMap (Sato and Ehira, 2003) generate circular genome maps in bitmap (PNG, JPG) formats, but do not support standard sequence file formats and have limited customizability. CGView (Stothard and Wishart, 2005) and Circos (Krzywinski et al., 2009) can generate very high quality, highly customizable circular maps in bitmap and scalable vector graphics (SVG) formats, and can be incorporated in pipelines, but have limited interactivity, and like their predecessors, require custom input formats. DNAPlotter (Carver et al., 2009) generates interactive, high quality, customizable circular and linear maps, but does not support SVG image output, and cannot save the customizations applied to a genome map for later use. The motivation behind GView is to provide in a single application the best features of all these microbial genome viewing packages: ease-of-use; customizability; interactivity; ability to generate circular and linear maps; ability to read standard sequence file formats; ready incorporation in pipelines and third-party applications; and the ability to generate aesthetically pleasing, publication quality image output.
2 IMPLEMENTATION
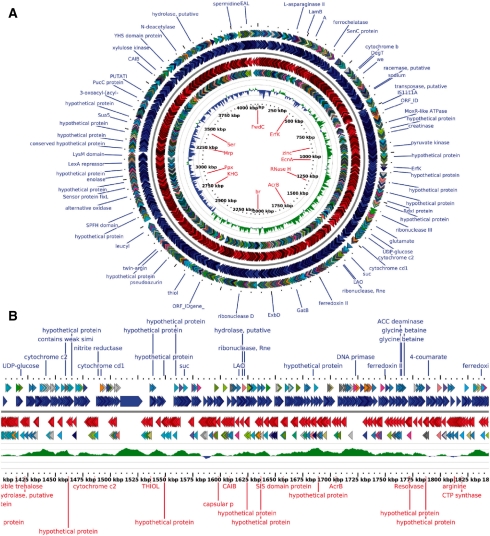
GView is a complete rewrite of the CGView Java package that preserves the best features of the original application. Genome features and feature labels are displayed in tracks above and below the genome backbone. Graphs can be generated to display feature-level properties (e.g. BLAST atlases) or sequence-level properties (e.g. GC content, GC skew). Optional feature legends can be displayed inline with the map image or as a separate image. Maps can be output in bitmap (PNG, JPG) or vector (SVG) formats for use in web pages and publications, or for additional customization in image editing programs (Fig. 1).
Fig. 1.
(A) A circular genome plot for Roseobacter dentrificans with tracks showing genes on forward and reverse strands, COG categories, GC skew and automatically generated labels. (B) A linear view of a subregion from (A).
GView maintains support for pipelining applications via a command-line interface with options to define image dimensions, zoom level and a subregion of the map on which to centre the display. The Application Programming Interface (API) has been substantially reworked to make incorporation in third party Java programs simpler and more straightforward. The new API includes additional support for making selections to the features, tracks and regions of the map. GView also contains substantial new functionality relative to the original CGView. Genome maps can now be generated with circular or linear layouts. A ‘wrapped’ linear layout, similar to that available in DNAPlotter, is planned for a future release. Dynamic panning and zooming allows direct interaction with the map via familiar keyboard and mouse controls, and is useful to quickly navigate to biologically interesting subregions of the map for detailed exploration. To aid in navigating maps at high zoom levels, an optional ‘bird's eye view’ is available that displays and adjusts the current view in the context of the entire genome map. A new feature label placing algorithm keeps label positions stable while zooming in and out of the map (Been, 2006).
File compatibility has been improved considerably compared to CGView. GView incorporates the BioJava libraries (Holland et al., 2008) to read common genome sequence file formats (EMBL, GFF, Genbank). Backwards compatibility is maintained with the original CGView XML to customize the appearance of the genome map. In addition, GView introduces a new style specification file format—termed ‘Genome Style Sheet’ (GSS)—designed to make genome map customization easier for biologists without computer programming backgrounds and less cumbersome than most GUI-based style customization interfaces. Based on the Cascading Style Sheet specification already in widespread use for applying style information to HTML documents, the GSS specification provides a clean separation of content and style, thus a single GSS file can be used to apply a common style to a set of genome sequences and associated graphs. The GSS format supports the arbitrary grouping of the various map elements into sets, and styles can be quickly applied to that set. By combining this grouping ability with the ability to operate on the feature keys, specifiers, and genome coordinates contained in the genome sequence file (and graph data files), the GSS format provides a highly compact, intuitive and powerful style application ability while retaining the high customizability of the original CGView XML.
Funding: Canada's National Microbiology Laboratory.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Been K. Dynamic map labeling. IEEE Trans. Visual. Comp. Graph. 2006;12:773–780. doi: 10.1109/TVCG.2006.136. [DOI] [PubMed] [Google Scholar]
- Carver T, et al. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R, Smith DR. Genome visualization made fast and simple. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- Holland RC, et al. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24:2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–1584. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.