Abstract
Store-operated Ca2+ entry is controlled by the interaction of stromal interaction molecules (STIMs) acting as endoplasmic reticulum ER Ca2+ sensors with calcium release–activated calcium (CRAC) channels (CRACM1/2/3 or Orai1/2/3) in the plasma membrane. Here, we report structural requirements of STIM1-mediated activation of CRACM1 and CRACM3 using truncations, point mutations, and CRACM1/CRACM3 chimeras. In accordance with previous studies, truncating the N-terminal region of CRACM1 or CRACM3 revealed a 20–amino acid stretch close to the plasma membrane important for channel gating. Exchanging the N-terminal region of CRACM3 with that of CRACM1 (CRACM3-N(M1)) results in accelerated kinetics and enhanced current amplitudes. Conversely, transplanting the N-terminal region of CRACM3 into CRACM1 (CRACM1-N(M3)) leads to severely reduced store-operated currents. Highly conserved amino acids (K85 in CRACM1 and K60 in CRACM3) in the N-terminal region close to the first transmembrane domain are crucial for STIM1-dependent gating of CRAC channels. Single-point mutations of this residue (K85E and K60E) eliminate store-operated currents induced by inositol 1,4,5-trisphosphate and reduce store-independent gating by 2-aminoethoxydiphenyl borate. However, short fragments of these mutant channels are still able to communicate with the CRAC-activating domain of STIM1. Collectively, these findings identify a single amino acid in the N terminus of CRAC channels as a critical element for store-operated gating of CRAC channels.
INTRODUCTION
Stromal interaction molecules (STIMs) STIM1 and STIM2 in the ER function as luminal Ca2+ sensors that activate the calcium release–activated calcium (CRAC) channels CRACM1/2/3 (Orai1/2/3) in the plasma membrane after store depletion and give rise to the store-operated CRAC current (ICRAC) (Liou et al., 2005; Roos et al., 2005; Zhang et al., 2005; Feske et al., 2006; Spassova et al., 2006; Vig et al., 2006b; Lis et al., 2007; Parvez et al., 2008). Only the combined overexpression of sensor and channel proteins reconstitutes amplified CRAC currents (Mercer et al., 2006; Peinelt et al., 2006; Soboloff et al., 2006; Zhang et al., 2006; Parvez et al., 2008). All three mammalian CRAC channel homologues (CRACM1, CRACM2, and CRACM3) represent functional store-operated channels, but with distinct characteristics (Lis et al., 2007). STIM1 functions as a Ca2+ sensor in the ER (Liou et al., 2005; Roos et al., 2005; Zhang et al., 2005), and store depletion causes it to redistribute from a diffuse localization throughout the ER into puncta near the plasma membrane, followed by CRAC channel migration to these areas (Luik et al., 2006). The distance between ER-resident STIM1 puncta and the plasma membrane is in the range of 10–25 nm (Wu et al., 2006), enabling a direct interaction as evident from fluorescence energy transfer signals from fused fluorophores (Barr et al., 2008; Muik et al., 2008; Navarro-Borelly et al., 2008) or coimmunoprecipitation (Co-IP) (Vig et al., 2006a; Yeromin et al., 2006; Yuan et al., 2009). It has also been suggested that CRACM1 is part of a larger molecular complex, as CRACM1 requires larger space between the two membranes (11–14 nm) compared with STIM1 (4–6 nm) (Várnai et al., 2007). A recent study shows that STIM1–CRACM1 interaction is sufficient to gate the CRAC channel into the open state in the absence of other proteins of the CRACM1 channel complex (Zhou et al., 2010). For the cytosolic part of STIM1, a direct interaction has been suggested by at least three independent studies that identified a minimal region in STIM1 that is needed for coupling to and activation of CRACM1. When overexpressed, these soluble fragments of STIM1, termed CRAC-activating domain (CAD) (Park et al., 2009), STIM1 Orai-activating region (Yuan et al., 2009), or Orai-activating small fragment (Muik et al., 2009), induce constitutive CRAC currents.
Within CRACM proteins, the C-terminal cytoplasmic coiled-coil region has been reported to play a critical role in the binding of STIM1, and single-point mutations within this region disrupt STIM1 coupling (Muik et al., 2008; Navarro-Borelly et al., 2008; Frischauf et al., 2009). However, the role of the N terminus of CRACM1 in the STIM1/CRACM1 signaling machinery is less clear. Deletion of the entire N-terminal portion of CRACM1 inhibits activation of CRAC, but not co-clustering of the mutant with STIM1 (Li et al., 2007). Furthermore, STIM1 inefficiently activates a CRACM1 variant in which the majority of the N-terminal residues are deleted (Δ1–73) (Li et al., 2007; Yuan et al., 2009), whereas the STIM1 Orai-activating region and CAD domains are both capable of coupling to and activating this deletion variant (Park et al., 2009; Yuan et al., 2009). In addition, the highly conserved region proximately to the first transmembrane domain (74–91) not only plays an important role in the binding of CAD and activating ICRAC (Park et al., 2009) but also in calcium-dependent inactivation via calmodulin (CaM) binding (Mullins et al., 2009). Recently, this region has been implicated to interact with another protein that may be part of the CRACM-STIM coupling machinery, called CRAC regulator 2A (CRACR2A), as this Ca2+-binding protein has been shown to regulate store-operated Ca2+ entry (SOCE) by interacting with both CRACM1 and STIM1, and possibly participating in the assembly and stabilization of the CRACM1–STIM1 interaction (Srikanth et al., 2010).
A significant problem in assessing CRAC channel mutations is that channel opening is the final step of a complex series of events, and any defect in the cascade results in lack of channel opening. In particular, it remains unknown whether any molecular manipulation that disrupts STIM1-mediated CRAC channel activity does so because of the inability of STIM1 to interact with the channel or because the mutation prevents channel gating. The highly conserved region between amino acids 74 and 91 in the N-terminal region of the CRACM proteins is very important for CRAC channel activation and STIM1 binding. Because this region contains positively charged amino acids, we wondered if they might play a role either in the communication between STIM1 and CRACM proteins or in gating of the channel. We assessed this question using site-directed mutagenesis and chimeric constructs of CRACM1 and its homologue CRACM3, and by taking advantage of recent observations that CRACM3 channel function can be probed independently of store depletion and STIM1 signaling (DeHaven et al., 2008; Peinelt et al., 2008; Schindl et al., 2008; Zhang et al., 2008). Our results suggest that a single residue near the first transmembrane segment of CRAC channels might be a crucial element for CRAC channel function.
MATERIALS AND METHODS
Molecular cloning, mutagenesis, and overexpression
The N-terminal deletion mutants of CRACM1, ΔN35, ΔN49, ΔN66, and ΔN87, and CRACM3, ΔN41 and ΔN62, were prepared by PCR method using appropriate primers. The chimeras CRACM1-N(M3) (M1N3) and CRACM3-N(M1) (M3N1) were obtained by replacing the N-terminal region of CRACM1 (1–87) with the N-terminal region of CRACM3 (1–64) and vice versa by using overlapping primers with introduced restriction sites and PCR method. Junction sites of all chimeras were chosen based on conserved amino acid residues to avoid missing, changed, or additional residues at the junction sites. Full-length human CRACM1 and CRACM3 were subcloned as described previously (Lis et al., 2007). All PCR products were subcloned into HA-pCAGGSM2-IRES-GFP vector for the expression in mammalian cells. CRACM1 (amino acids 48–91) and CRACM3 (amino acids 23–66) constructs were generated by PCR with appropriate primers and cloned into pECFP-C1 vector using XhoI and BamHI restriction sites. All point mutations were introduced using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies), and PCR products and constructs were verified by sequencing. The FLAG-Myc-CAD construct was provided by R. Lewis (Stanford University, Stanford, CA).
For electrophysiological analysis, CRACM proteins were overexpressed in HEK293 cells stably expressing STIM1 (Soboloff et al., 2006) or transiently expressed FLAG-Myc-CAD using lipofectamine 2000 (Invitrogen), and the GFP-expressing cells were selected by fluorescence. Experiments were performed 24–72 h after transfection. Cells cotransfected with FLAG-Myc-CAD (Park et al., 2009) and CRACM constructs were cultured in low calcium media (20 µM) to minimize the toxicity of constitutively active ICRAC.
Electrophysiology
Patch clamp experiments were performed in the tight-seal whole cell configuration at 21–25°C. High-resolution current recordings were acquired using an EPC-9 amplifier (HEKA). Voltage ramps of 50-ms duration spanning a range of −150 to +150 mV were delivered from a holding potential of 0 mV at a rate of 0.5 Hz over a period of 100–300 s. All voltages were corrected for a liquid junction potential of 10 mV. Currents were filtered at 2.9 kHz and digitized at 100-µs intervals. Capacitive currents were determined and corrected before each voltage ramp. We assessed the low-resolution temporal development of currents by extracting the current amplitude at −80, +80, and +130 mV from individual ramp current records. Where applicable, statistical errors of averaged data are given as means ± SEM, with n determinations. Standard external solutions were as follows (in mM): 120 NaCl, 2 MgCl2, 10 CaCl2, 10 TEA-Cl, 10 HEPES, 10 glucose, pH 7.2 with NaOH, and 300 mOsm. In the CAD experiments, the cells were kept in nominal calcium-free external solution, and the standard external solution containing 10 mM Ca2+ was applied. In some experiments, 2-aminoethoxydiphenyl borate (2-APB) was added to the standard external solution at a final concentration of 50 µM. Standard internal solutions were as follows (in mM): 120 Cs-glutamate, 20 Cs · BAPTA, 3 MgCl2, 10 HEPES, 0.02 inositol 1,4,5-trisphosphate (IP3), pH 7.2 with CsOH, and 300 mOsm. All chemicals were purchased from Sigma-Aldrich.
Immunofluorescence staining and confocal microscopy
HEK293 cells stably expressing STIM1 were transfected with various CRACM1 and CRACM3 constructs (see above). For staining, cells were fixed with ice-cold methanol for 20 min at −20°C. Unspecific binding sites were blocked with 2% BSA in 1× PBS for 1 h at room temperature. Thereafter, cells were stained with an anti-HA antibody (1:600 in blocking solution; rat α-HA; Roche) for 1.5 h at 37°C and a detecting antibody conjugated with Alexa 658 (1:1,200 in PBS; goat α-rat; Invitrogen) for 40 min at 37°C and visualized via confocal microscopy (LSM510; Carl Zeiss, Inc.) using z-stack imaging and a Plan-Apochromat 63×/1.4 oil DIC objective with optical zoom of three. Stacks ranged between 1.5 and 2 µm, and the pinhole was kept constant at 274 µm. For excitation, a HeNe laser (70%) was used at 543-nm wavelength. Emission was collected with a 560-nm long-pass filter, and images were analyzed with LSM Image Browser Software (Carl Zeiss, Inc.). Quantifications of plasma membrane and cytosolic fluorescence levels were performed in individual cells by analyzing fluorescence integrals of a narrow band of the cells’ contour (cortical plasma membrane region, 4–8-pixels wide) and central cell area enclosed by the band. Fluorescence densities per unit area were obtained by dividing the cumulative fluorescence intensities by the respective areas.
Western blotting and Co-IP
HEK293 cells were transiently cotransfected with FLAG-Myc-CAD and CFP-CRACM constructs (as described above). 24 h after transfection, cells were harvested in PBS and lysed in 0.5 ml of lysis buffer (in mM): 150 NaCl, 20 Tris-HCl, 10% glycerol, 1% Triton, and protease inhibitor cocktail (Sigma-Aldrich). The lysates were spun down at 8,000 g for 10 min, and the supernatant was precipitated with anti-FLAG M2 agarose beads (Sigma-Aldrich) for 4 h at 4°C. The total protein amount was determined by Bradford assay, and 50 µg of the lysates was loaded as a control. Beads were washed four times with lysis buffer, and complexes were released with SDS-loading buffer. Lysates and immunoprecipitations were resolved by SDS-PAGE and analyzed using anti-GFP goat polyclonal antibody (Santa Cruz Biotechnology, Inc.) at a dilution of 1:1,000 or an anti-FLAG mouse monoclonal antibody at a dilution of 1:7,500 (Sigma-Aldrich). Anti–mouse IgG (1:12,500; GE Healthcare) and anti–goat IgG (1:80,000) peroxidase conjugate (Sigma-Aldrich) were used as secondary antibodies according to the manufacturer’s instructions. Proteins were detected by using the ECL Plus Western Blotting Detection System (GE Healthcare).
Online supplemental material
Fig. S1 shows small but enhanced 2-APB currents in the overexpression of CRACM1-N(M3) and CRACM1. Fig. S2 illustrates the function of double mutants in CRACM1, CRACM3, and CRACM3-N(M1). Fig. S3 summarizes the analysis of positively charged amino acid residues in the N-terminal region of CRACM1. Fig. S4 indicates that CRACM1-K85E and CRACM3-K60E reduce the store-operated current in co-overexpression with the CRACM-wt proteins but not as efficiently as the SCID mutation CRACM1-R91W. Figs. S1–S4 are available at http://www.jgp.org/cgi/content/full/jgp.201010484/DC1.
RESULTS
N-terminal region truncations
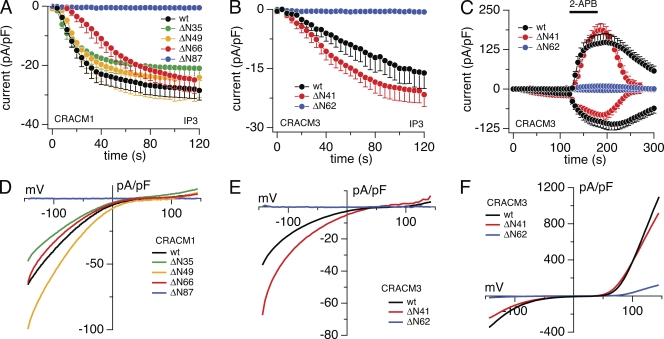
To investigate the role of the N-terminal region of CRACM1 and CRACM3 in STIM1-mediated channel activity, we constructed mutants in which the proximal end of the N-terminal region was sequentially deleted (refer to Materials and methods). Fig. 1 shows average current amplitudes of inward currents in stable STIM1-expressing HEK293 cells transiently overexpressing the various deletion mutants. With increasing N-terminal region truncations up to amino acid residue 50 in CRACM1, the kinetics and current densities of inward currents were essentially identical to those obtained from wild-type (wt) CRACM1 (Fig. 1 A), and the I-V relationships were typical of CRAC currents (Fig. 1 D; currents were extracted at the end of the experiment at 120 s). Even the CRACM1-ΔN66 mutant reached similar maximal currents as wt CRACM1, although the kinetics of activation was slower. However, the complete removal of the cytoplasmic N-terminal region (CRACM1-ΔN87) resulted in the complete loss of CRAC currents, demonstrating that CRACM1 N-terminal region deletions remain functional up to amino acid residue 66 and suggesting that ∼20 amino acids close to the plasma membrane are important for CRAC channel activity. These observations are in line with previous studies (Liao et al., 2007; Takahashi et al., 2007; Muik et al., 2008).
Figure 1.
Effects of N-terminal region truncations in CRACM1 and CRACM3. Average CRAC current densities at −80 mV induced by 20 µM IP3 in stable STIM1-expressing HEK293 cells transiently overexpressing truncation mutants. (A) CRACM1-wt (black; n = 45), CRACM1-ΔN35 (green; n = 6), CRACM1-ΔN49 (orange; n = 5), CRACM1-ΔN66 (red; n = 5), and CRACM1-ΔN87 (blue; n = 6). (B) Magnified plot of the initial 120-s data from C. (C) CRACM3-wt (black; n = 9), CRACM3-ΔN41 (red; n = 7), and CRACM3-ΔN62 (blue; n = 5). Bars indicate external application of 50 µM 2-APB. (D–F) Average I-V relationships of CRAC currents extracted from representative HEK293 cells shown in A–C obtained at 120 s (D and E) and 180 s (F). Data represent leak-subtracted current densities (pA/pF) evoked by 50-ms voltage ramps from −150 to +150 mV. Error bars indicate SEM.
We next analyzed the CRACM3 protein, which is also activated in a store-operated and STIM1-dependent manner but can be activated and probed for channel function by 2-APB, which can gate the channel independently of store depletion, even in the absence of STIM1 overexpression (DeHaven et al., 2008; Peinelt et al., 2008; Schindl et al., 2008; Zhang et al., 2008). We generated two truncation mutants, CRACM3-ΔN41 and CRACM3-ΔN62, which are equivalent mutants to CRACM1-ΔN66 and CRACM1-ΔN87, respectively. We assessed the ability of both mutants to be activated via STIM1 by IP3-induced store depletion (Fig. 1 B) as well as pharmacological STIM1-independent gating by 2-APB (Fig. 1 C). Similar to CRACM1, in CRACM3 the partial truncation of the N-terminal region up to amino acid residue 41 did not change IP3-mediated currents compared with wt CRACM3, whereas the full truncation mutant CRACM3-ΔN62 resulted in complete loss of IP3-dependent activation (Fig. 1, B and E). The current arising from the CRACM3-ΔN41 truncation was even slightly larger (−20 vs. −15 pA/pF; Fig. 1 B) compared with wt CRACM3 currents but showed the characteristic ICRAC I-V relationship (Fig. 1 E). Because CRACM3 can be activated independently of STIM1, we tested the truncated mutations for 2-APB–activated CRAC currents (Fig. 1, C and F). The application of 2-APB induced similar current amplitudes in the CRACM3-ΔN41 construct as in the wt CRACM3 (Fig. 1 C) with similar I-V relationships (Fig. 1 F). The CRACM3-ΔN62 construct, however, showed an inward current of about −4 pA/pF at −80 mV and an outward current of about +11 pA/pF at +80 mV (Fig. 1, C and F). The reduced current amplitudes compared with wt channels were likely a result of lower expression levels and fewer channels being incorporated into the plasma membrane, as evident from immunofluorescence staining and confocal analysis (Fig. 2, B and E). Nevertheless, this demonstrates that this mutant can in principle form functional channels that can be activated by 2-APB but remain refractory to store depletion and STIM1-mediated gating.
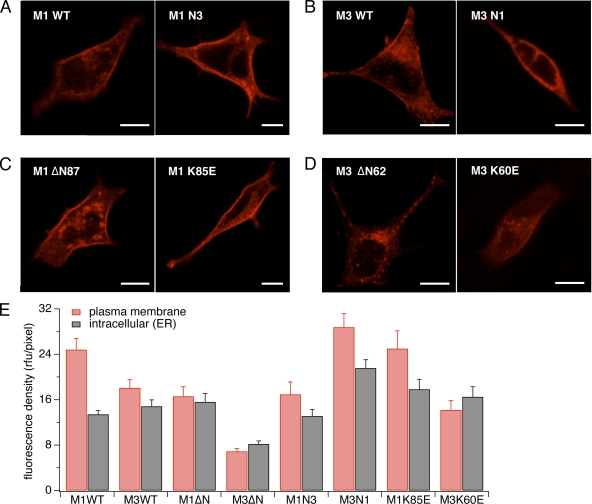
Figure 2.
Subcellular localization of CRAC channel variants. Stable STIM1-expressing HEK293 cells were transfected with pCAGGSM2 vectors of various constructs. Cells were stained with an anti-HA antibody and a detecting antibody conjugated with Alexa 658 and visualized via confocal microscopy (LSM510; Carl Zeiss, Inc.). Bars, 10 µm. Images show representative examples of at least 10 individual cells for each construct. (A) CRACM1-wt construct (left) and chimera of CRACM1 with the CRACM3 N-terminal region (right). Both proteins, including the chimera, are expressed in the plasma membrane. (B) CRACM3-wt construct (left) and chimera of CRACM3 with the CRACM1 N-terminal region (right). Both proteins are expressed in the plasma membrane, although the expression of the latter appears to be less. (C) CRACM1-ΔN87 (left) and CRACM1-K85E (right). Both constructs are inserted into the plasma membrane. (D) CRACM3-ΔN62 (left) has reduced plasma membrane expression, and the CRACM3-K60E construct (right) shows expression in the plasma membrane. (E) Fluorescence densities obtained from confocal images, as shown in A–C. Values represent the average fluorescent intensities normalized to the unit area of the cortical and intracellular regions of 8–15 cells each.
The above results could be interpreted to mean that the N-terminal 20 amino acids proximal to the plasma membrane are involved in STIM1-mediated gating of CRACM1 and CRACM3, or that they are required for adequate transport, localization, and/or formation of functional channels. To resolve this question, we assessed the subcellular distribution of the full CRACM1 N-terminal region truncation (CRACM1-ΔN87) and found that the protein was at least targeted properly and localized within the plasma membrane (Fig. 2, B and E). Immunofluorescence also showed that although the CRACM3-ΔN41 construct efficiently translocated to the plasma membrane (not depicted), the full truncation mutant (CRACM3-ΔN62) largely remained intracellularly (Fig. 2, B and E). Thus, it appears that STIM1-dependent gating of CRACM1 and CRACM3 requires some 20 amino acids of their N-terminal regions right next to the plasma membrane. In addition, it appears that these 20 residues may be important for proper targeting of CRACM3 to the plasma membrane.
Chimeras
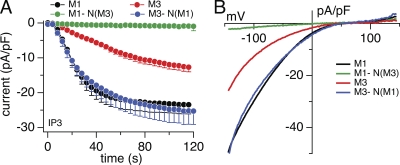
We have previously reported differences in kinetics and amplitude of STIM1-dependent activation of CRACM1 and CRACM3 (Peinelt et al., 2008). CRACM1 currents typically activate more rapidly and to higher amplitudes compared with CRACM3, and we hypothesized that this might reflect differences in CRACM/STIM1 coupling. We addressed this question through chimeric constructs in which we mutually exchanged N- and C-terminal regions of both proteins and where the N-terminal region of CRACM1 (amino acids 1–87) was substituted with that of CRACM3 (amino acids 1–62) and vice versa. Data obtained from these chimeras are shown in Fig. 3. The N-terminal region of CRACM1 conferred a significant increase in current amplitude and acceleration of activation kinetics on CRACM3 channels (Fig. 3 A), whereas the typical I-V relationships of currents remained unchanged. The CRACM3-N(M1) chimera (M3N1) localized in the plasma membrane (Fig. 2, B and E) but was expressed at higher levels than the parent CRACM3 channel, which likely accounts for this chimera’s larger current amplitudes. The accelerated kinetics of activation of this chimera (τ = 26.9 ± 4.3 s) was similar to that of CRACM1 (τ = 24.7 ± 4.4 s) rather than CRACM3 (τ = 66.2 ± 8.3 s), suggesting that the N-terminal region harbors crucial structural elements mediating STIM1-dependent activation of CRACM1 and CRACM3. Conversely, exchanging the N-terminal region of CRACM1 with that of CRACM3 (CRACM1-N(M3), short M1N3) did not reproduce the CRACM3 current behavior, but instead resulted in severely reduced store-operated currents of just −0.8 pA/pF (Fig. 3 A), despite normal expression levels and localization to the plasma membrane (Fig. 2, A and E). This current is ∼20-fold reduced compared with CRACM3-wt and just slightly larger than endogenous CRAC currents observed in control cells stably overexpressing STIM1 (−0.5 pA/pF; see Fig. S1). These currents are unlikely to represent endogenous channels alone because the application of 2-APB to STIM1 cells overexpressing CRACM1-N(M3) produced significantly larger currents than in STIM1 control cells (Fig. S1). Collectively, these data suggest that the N-terminal region of CRACM channels determines the STIM1-mediated kinetics of activation.
Figure 3.
Function of N terminus chimeras of CRACM1 and CRACM3. (A) Average CRAC current densities at −80 mV induced by 20 µM IP3 in stable STIM1-expressing HEK293 cells transiently overexpressing CRACM1-wt (black; n = 43), CRACM3-wt (red; n = 70), CRACM1-N(M3) (green; n = 5), and CRACM3-N(M1) (blue; n = 5). Error bars indicate SEM. (B) Average I-V relationships of CRAC currents extracted from representative HEK293 cells shown in A and obtained at 120 s. Data represent leak-subtracted current densities (pA/pF) evoked by 50-ms voltage ramps from −150 to +150 mV.
CRACM1-K85E and CRACM3-K60E mutants
The 20-residue stretch proximal to the plasma membrane in the cytosolic portion of the N-terminal region of CRACM proteins is crucial for STIM1-dependent gating of CRACM1 (amino acids 67–87) and CRACM3 (amino acids 42–62) channels (Fig. 1). Sequence analysis of that part of the protein identifies it as a conserved region of high homology among CRACM proteins that contains a putative CaM-binding motif. This stretch of amino acids is able to bind CaM in a Ca2+-dependent manner and plays a role in Ca2+-dependent inactivation (Mullins et al., 2009). Therefore, we first created a double-point mutation construct that would disrupt the CaM-binding motif in CRACM1 and the CRACM3-N(M1) chimera to investigate the potential effects on STIM1-mediated channel activation (Fig. S2). This double mutation in CRACM1 (K78E-K85E) failed to activate large IP3- (Fig. S2, A and C) or 2-APB–induced currents (Fig. S2, B and D). This indicates a loss of function effect that not only prevents STIM1-mediated activation but also abolishes STIM1-independent gating of the channel because HEK293 cells transfected with wt CRACM1 still yield small IP3-induced currents that can be enhanced by 2-APB (Fig. S1).
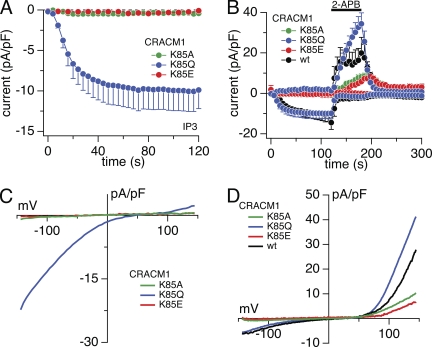
We then proceeded with single-point mutations of these residues and analyzed CRACM1 mutants in which we changed alkaline residue K78 to acidic glutamate residue (E) (Fig. S3) and K85 to glutamine (Q) or alanine (A) (Fig. 4). Mutation K78E in CRACM1 was able to produce sizeable IP3-induced inward currents of −17 pA/pF, similar in magnitude to those of wt channels (Fig. S3, B and C). However, the K85E mutant of CRACM1 essentially abolished IP3-induced currents, producing inward currents of just −0.2 pA/pF (Fig. 4, A and C), although this mutation did localize to the plasma membrane (Fig. 2, C and E). Nevertheless, 2-APB still evoked significant channel activation as evident from the outward currents in Fig. 4 B and the I-V curve in Fig. 4 D. The substitution of the alkaline amino residue (K) by alanine, a not particularly hydrophobic and nonpolar amino acid, yielded similarly small IP3-induced inward currents of around −0.5 pA/pF (Fig. 4, A and C). However, 2-APB enhanced the outward currents to 8 pA/pF (at +130 mV), indicating that this mutation was also present and properly assembled in the plasma membrane and could be gated STIM1-independently. A more conservative mutation of this residue is K85 to glutamine (Q), an amino acid with a polar side chain that differs from glutamate (E) only in that it contains an oxygen in place of the amino group. This point mutation, K85Q, was less efficient in suppressing STIM1-induced CRAC currents, yielding −10 pA/pF or ∼50% of the inward current typically seen with wt CRACM1. This indicates that the K85 residue in CRACM1 is indeed involved in tuning the STIM1-mediated gating process. Interestingly, the K85Q mutant, while yielding reduced IP3-induced inward current, actually produced larger 2-APB–induced currents compared with the CRACM1 wt channel (Fig. 4, B and D).
Figure 4.
Functional role of CRACM1-K85 mutation. Average CRAC current densities at −80 and + 130 mV induced by 20 µM IP3 in stable STIM1-expressing HEK293 cells transiently overexpressing point mutations. Black bars indicate the application of an external solution containing 50 µM 2-APB. Error bars indicate SEM. (A) Magnified plot of the initial 120-s data from B. (B) CRACM1-K85E (red; n = 6), CRACM1-K85Q (blue; n = 11), CRACM1-K85A (green; n = 10), and CRACM1-wt (black; n = 4: from a total of n = 12, four cells with the smallest current densities [induced by IP3] were averaged to approximate the lower current density of CRACM1-K85Q). (C and D) Average I-V relationships of CRAC currents extracted from representative HEK293 cells shown in A and B and obtained at 120 s (C) and 180 s (D), when current was fully activated by 2-APB. Data represent leak-subtracted current densities (pA/pF) evoked by 50-ms voltage ramps from −150 to +150 mV.
Therefore, we next asked whether this residue modified channel function above and beyond STIM1-mediated gating and also affected store-independent gating. To assess this, we used the CRACM3-N(M1) chimera, which confers the CRACM1 gating phenotype on CRACM3 (Figs. 3 A and 5 A) and can also be activated STIM1-independently by 2-APB (Fig. 5 B). The double mutation K78E-K85E in the CRACM3-N(M1) chimera produced reduced IP3-induced currents of −0.65 pA/pF (Fig. S2, A and C), which were not significantly different from those recorded in wt HEK293 cells overexpressing STIM1 alone (Fig. S1) (Peinelt et al., 2006). Although 2-APB was able to activate these channels (Fig. S2, B and D), indicating store-independent and direct gating of the channel, the current amplitudes did not reach the levels typically seen with the CRACM3 wt channel (DeHaven et al., 2007; Peinelt et al., 2008; Schindl et al., 2008; Zhang et al., 2008). The singular K78E mutation introduced into the CRACM1-based N-terminal region of the chimera behaved normally and produced large IP3-induced inward currents of −32 pA/pF at −80 mV, and these were further activated by 2-APB (Fig. 5, A and B). However, the K85E mutation in this chimera strongly suppressed STIM1-mediated activation of CRAC currents, producing just −1.7 pA/pF of inward current at −80 mV. Although strongly reduced compared with the unmutated chimera, this amount of current was still above the levels of endogenous CRAC currents, suggesting that channels were expressed in the plasma membrane but functionally impaired with respect to STIM1-dependent activation. Store-independent gating by 2-APB was clearly present, although smaller than with the K78E mutant, suggesting that this mutant may additionally have a STIM1-independent functional defect (Fig. 5, B and F).
Figure 5.
Functional role of N-terminal amino acid mutation in CRACM3-N(M1) chimera and CRACM3. Average CRAC current densities at −80 and +130 mV induced by 20 µM IP3 in stable STIM1-expressing HEK293 cells transiently overexpressing point mutation or chimera. Black bars indicate the application of an external solution containing 50 µM 2-APB. Error bars indicate SEM. (A and C) Magnified plot of the initial 120-s data from B and D. (B) CRACM3-N(M1) (black; n = 6), CRACM3-N(M1)-K78E (blue; n = 6), and CRACM3-N(M1)-K85E (red; n = 6). (D) CRACM3-R52E (green; n = 8), CRACM3-R53E (blue; n = 8), CRACM3-K60E (red; n = 7), and CRACM3-K60Q (orange; n = 9). (E–H) Average I-V relationships of CRAC currents extracted from representative HEK293 cells shown in A–D and obtained at 120 s (E and G) and at 180 s (F and H), when current was activated by 2-APB. Data represent leak-subtracted current densities (pA/pF) evoked by 50-ms voltage ramps from −150 to +150 mV.
Finally, we asked whether STIM1-mediated gating of CRACM3 involved a similar structural requirement as that observed for K85 in CRACM1. At first we tested whether the equivalent double-point mutation in CRACM3, CRACM3-R53E-K60E, was able to prevent IP3- and 2-APB–mediated currents like the CRACM1-K78E-K85E did. Indeed, the STIM1-dependent activation was reduced to −0.4 pA/pF (Fig. S2, A and C), and the store-independent gating by 2-APB was also dramatically suppressed (Fig. S2, B and D). The equivalent single-point mutation to K78E (CRACM1), R53E in CRACM3, produced 50% smaller IP3-induced currents (−6 pA/pF; Fig. 5, C and G) than the CRACM3 wt, and these were further activated by 2-APB application (Fig. 5, D and H). Next, we mutated the residue K60 in CRACM3 to glutamic acid (E) and tested for IP3- and 2-APB–induced activation. Just like the equivalent K85E mutant in CRACM1, the CRACM3-K60E mutant failed to produce store-operated currents (current amplitude at −80 mV was −0.4 pA/pF), suggesting that CRACM3 also relies on this residue for STIM1-mediated gating. Interestingly, the 2-APB–induced currents were also strongly reduced compared with the large currents evoked in wt CRACM3 (Fig. 5, D and H). This reduced amplitude, however, may not necessarily have been caused by impaired channel function but likely resulted from less efficient trafficking, as immunofluorescence staining indicated that this mutant is largely retained intracellularly with less prominent plasma membrane staining (Fig. 2, D and E). The substitution with a more conservative mutation of this residue, K60 to glutamine (Q), was less efficient in suppressing CRAC currents, reducing the inward current typically seen with wt CRACM3 by ∼75% to −3.5 pA/pF (Fig. 5, C and G), which indicates that this residue is also involved in tuning the STIM1-mediated gating process. Store-independent activation by 2-APB was also clearly present, although smaller than with the CRACM3 wt, suggesting that this mutant additionally affects the STIM1-independent activation (Fig. 5, D and H).
Because the N-terminal region close to the first transmembrane domain in CRACM1 and CRACM3 contains several basic amino acid residues (Fig. S3 A), we assessed their relevance for STIM1-mediated channel activation. The substitutions of arginine (R) at position 77 and lysine (K) at position 87 to glutamic acid reduce the IP3-induced current to −4 and −8 pA/pF (Fig. S3, B and C) compared with CRACM1. However, the overexpression of other point mutations at positions 69 and 87 in HEK293 cells stably expressing STIM1 resulted in normal current sizes comparable to the wt channel (Fig. S3, B and C). Collectively, these results suggest that the K85 residue in CRACM1 and K60 in CRACM3 are critical amino acids required for functional store-operated and STIM1-dependent activation of CRAC channels.
To assess whether the K85E and K60E mutations can assemble into heteromeric channel complexes and to test for dominant-negative phenotype, we co-overexpressed K85E and K60E with respective wt constructs. Fig. S4 (A–C) illustrates that the co-overexpression of K85E and K60E in equal amounts with the wt homologues reduced IP3-mediated currents by ∼50% to −10.97 pA/pF of the wt CRACM1 current and ∼90% to −0.79 pA/pF of CRACM3. These results suggest that K85E and K60E mutants indeed form heterometric complexes with wt homologues. We also investigated another crucial amino acid within the N terminus of CRACM1, the previously identified R91W mutant that results in a human severe combined immunodeficiency (SCID) phenotype (Feske et al., 2006), and cotransfected it with CRACM1 wt protein into HEK-293 cells stably expressing STIM1. As illustrated in Fig. S4 D, CRACM1-R91W prevented the activation of large CRAC currents and produced current amplitudes of just 1–3 pA/pF (Fig. S4 E). This amounts to ∼10% of the normally observed current amplitudes observed with the wt channels in the absence of this mutant and results in a stronger suppression of channel activity than the K85E mutant, which results in ∼50% reduction. These data indicate that the R91W mutant can coassemble with the wt CRACM1 subunit (Fig. S4, D and E), exerting a dominant-negative effect on CRAC current in such heteromeric channels.
CRACM1-K85E and CRACM3-K60E mutants disrupt CRAC current activation by CAD but not by its binding
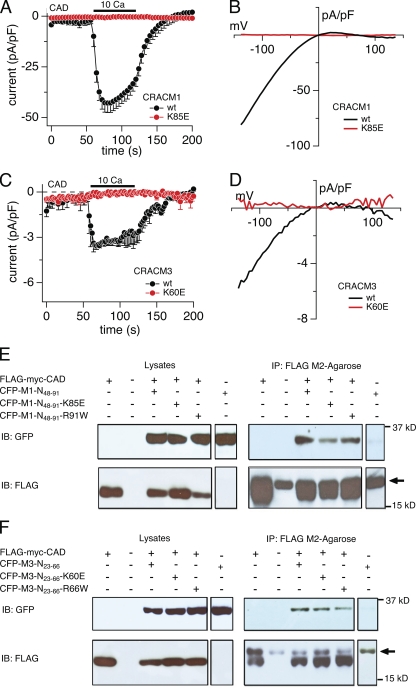
A minimal cytosolic region of STIM1 has been shown to functionally activate CRAC currents (Muik et al., 2009; Park et al., 2009; Yuan et al., 2009). This highly conserved CAD encompasses a putative coiled-coiled region and part of the ERM domain of STIM1. Park et al. (2009) also demonstrated that CAD binds directly to N and C termini of CRACM1/Orai1 and that the membrane-proximal region of the CRACM1 N terminus (amino acid residues 68–91) is required to activate the CRAC channel. Because the lysine residue we identified in the N terminus of CRACM1 lies exactly in this region (K85E), we asked whether it is required for CAD-mediated gating of CRAC currents. We obtained the FLAG-myc-CAD construct (Park et al., 2009) and confirmed that its coexpression with wt CRACM1 resulted in constitutive activation of CRAC currents (Muik et al., 2009; Park et al., 2009; Yuan et al., 2009) that were revealed when the cells were exposed to 10 mM of extracellular Ca2+, producing a CRAC-like inward current of −43 pA/pF (Fig. 6 A) and an I-V relationship characteristic of ICRAC (Fig. 6 B). Similar to CRACM1, the CRACM3-wt construct was able to produce a small inward current in response to 10 mM Ca2+ (−3.6 pA/pF; Fig. 6 C) with an ICRAC-like I-V relationship (Fig. 6 D). The introduction of point mutant CRACM1-K85E completely abolished CAD-mediated gating of this construct (Fig. 6, A and B), and so did the CRACM3-K60E mutant (Fig. 6, C and D). Thus, altering this particular lysine residue in the N-terminal region of both CRACM1 and CRACM3 channels creates a mutant that can no longer be gated by CAD.
Figure 6.
Lysine residue point mutations of short CRACM1 and CRACM3 N-terminal constructs bind to the CAD of STIM1 but suppress current activation. (A) HEK293 cells were transfected with the FLAG-myc-CAD and CRACM1 or CRACM1-K85E construct, respectively (refer to Materials and methods). Normalized current densities of CAD-mediated CRAC currents at −80 mV are plotted against time of the experiment. Cells were kept in nominal Ca2+-free solution, and the bar indicates the application of external solution containing 10 mM Ca2+. In control experiments, the application of Ca2+ produced an inward current, characteristic of ICRAC (black; n = 7). The K85E mutation completely abolished current activation upon Ca2+ application (red; n = 9). Data represent the average leak-subtracted current densities (pA/pF) evoked by 50-ms voltage ramps from −150 to +150 mV. Error bars indicate SEM. (B and D) Average I-V relationships of CRAC currents extracted at 120 s from representative cells shown in A and C. (C) Cells were transfected with FLAG-myc-CAD and CRACM3 or CRACM3-K60E constructs, respectively, and the average normalized current densities at −80 mV are plotted against time of the experiment. Similar to CRACM1, CRACM3 gave rise to ICRAC when 10 mM Ca2+ was applied (black; n = 5), indicated by the bar. Likewise, the K60E mutation in CRACM3 completely abolished current activation (red; n = 5). Error bars indicate SEM. (E and F) Representative Co-IP analysis of HEK-293 cells transfected with FLAG-myc-CAD construct and various short N-terminal constructs of CRACM1 (E) or CRACM3 (F), respectively. Immunoblotting of the corresponding lysates is shown in the left row. Lines represent control experiments of cells transfected with the CAD construct only (∼14 kD), no transfected HEK293 cells, and the short N-terminal construct of CRACM1 or CRACM3 (∼32 kD) alone or cotransfected with the CAD construct. Blots in the upper lane were immunoblotted with anti-GFP antibody, and those in the lower lane were immunoblotted with anti-FLAG antibody. The upper right lane shows the actual Co-IP, using agarose beads coated with FLAG antibody. For both CRACM1 and CRACM3, all three N-terminal constructs bound to CAD. The lower right lane represents the immunoprecipitated fraction immunoblotted with anti-FLAG. The arrow indicates the band representing the light chain of the FLAG antibody.
Because the above mutants abolished the CAD-mediated gating of CRAC currents, we asked whether this residue is part of a CAD-binding site at the N terminus of CRAC channels. To address this question, we designed a short CRACM1 wt (CFP-M1-N48-91) construct based on the previously identified membrane-proximal region of the CRACM1 N-terminal domain (amino acids 48–91) (Park et al., 2009). In addition, we designed a corresponding 44–amino acid construct (amino acids 23–66) of the wt CRACM3 protein (CFP-M3-N23-66). To test if the lysine point mutations interfere with CAD binding to CRACM1 and CRACM3, we performed site-directed mutagenesis on these constructs and assessed Co-IP of FLAG-myc-CAD and the various short CRACM1 and CRACM3 constructs, respectively. As demonstrated previously (Park et al., 2009), the short N-terminal CFP-M1-N48-91 construct (amino acid residues 48–91) can directly bind to the CAD construct (Fig. 6 E). We used this short construct to avoid binding to the C-terminal region of the full-length CRACM1 protein. The K85 mutant (CFP-M1-N48-91-K85E) retained the ability to bind to CAD, although possibly a little weaker than the wt construct, as seen in Fig. 6 E. Although we cannot rule out that a slightly weaker binding could be sufficient to disrupt STIM1’s ability to gate CRACM1, the binding activity that remains with K85E would indicate that the lysine residue does not fully account for the interaction of CAD to the N terminus of CRACM1. Because the mutation of the arginine amino acid residue 91, which gives rise to a SCID phenotype in humans (Feske et al., 2006), lies in the same membrane-proximal region that has been shown to be relevant for CRAC channel gating, we also tested whether a CFP-M1-N48-91-R91W construct could bind to CAD. Our Co-IP, using the CAD of STIM1 (Fig. 6 E), confirmed earlier results by Derler et al. (2009) that the arginine residue is not relevant for the binding of STIM1. Finally, we tested for the binding of CAD to the corresponding short CRACM3 constructs (Fig. 6 F). The binding assay of CAD to CFP-M3-N23-66 yielded similar results, as the short CRACM1 wt construct showed a clear Co-IP band. In addition, the K60E mutant (CFP-M3-N23-66-K60E) as well as the CFP-M3-N23-66-R66W mutation (corresponding to the CRACM1-R91W SCID mutant) also exhibited CAD binding (Fig. 6 F). Thus, although these mutations in the short CRACM1 and CRACM3 constructs do not significantly interfere with the binding of CAD, they represent crucial residues for the STIM1-dependent gating of the CRAC channel downstream of STIM1 binding.
Collectively, our results indicate that the opening of CRACM1 and CRACM3 channels is completely abolished when modifying the newly discovered lysine residue in the N terminus (K85E and K60E, respectively). Both short-construct mutants (CFP-M1-N48-91-K85E and CFP-M3-N23-66-K60E) are able to bind to the nominal CAD in STIM1, indicating that the binding of STIM1 might be functional and not impaired. However, the lack of constitutively active CRAC currents in CAD-overexpressing cells indicates a defect in channel gating and/or stable channel assembly despite CAD binding. Additionally, the K60E mutation in CRACM3 (but not the K85E mutation in CRACM1) may also experience impaired channel trafficking to the plasma membrane (Fig. 2, D and E) and may contribute to reduced 2-APB–induced currents.
DISCUSSION
This study provides an additional and complementary assessment of the structural requirements of store-dependent as well as store-independent activation of CRACM1 and extends it to its close homologue CRACM3. Here, we confirm that the N-terminal region between amino acid residues 67 and 87 is crucial for the activation of CRAC channels via STIM1. Our main novel observations are (a) the same N-terminal region (amino acids 42–62) is also crucial for store-operated activation of CRACM3 channels; (b) chimeric constructs demonstrate that the efficacy of STIM1-mediated gating in terms of kinetics and magnitude depends on the N terminus; (c) point mutations identify a single crucial lysine residue in the N-terminal region of CRAC channels (K85 and K60 in CRACM1 and CRACM3, respectively) that participates in the gating of CRAC channels; (d) the K60E mutation in CRACM3 also shows reduced 2-APB–induced currents; (e) additionally, the K60E mutation in CRACM3 also seems to impair channel trafficking to the plasma membrane and may contribute to the strong suppression of STIM1- and 2-APB–induced currents; and (f) a short stretch of the CRAC N terminus (amino acids 48–91 and 23–66) hosting the lysine residues K85E and K60E, respectively, can still bind to the CAD region of STIM1 in an artificial binding assay, and so does the N terminus of CRAC with SCID mutations R91W and R66W, respectively.
Although the available data indicate a critical role of the C-terminal region in interacting with STIM1, the functional role of the N-terminal region is less clear. Previous studies have demonstrated that the cytoplasmic C-terminal region is required for its interaction with STIM1, and the C-terminal region of STIM1 is able to bind to the C-terminal region of CRACM1 but not to the N terminus (Muik et al., 2008). Also, deletion of the C-terminal region or an L273S mutation in CRACM1 completely suppresses STIM1 binding and activation of ICRAC (Li et al., 2007; Muik et al., 2008). On the other hand, Takahashi et al. (2007) proposed that the N-terminal region plays an essential role in STIM1-mediated SOCE. The chimeric constructs in which we exchanged N termini of both channels with each other support the notion that the N-terminal region of CRACM1 indeed may also be involved in STIM1-mediated gating because it can confer the gating phenotype of CRACM1 onto CRACM3 in terms of current kinetics and magnitude (Fig. 3).
Complete truncation of the N-terminal region up to residue 87 of CRACM1 results in failure of channel opening (Fig. 1). These constructs localize to the plasma membrane (Fig. 2) and can interact with STIM1 (Liao et al., 2007; Muik et al., 2008). Our data further identify a single amino acid, K85 in CRACM1 and K60 in CRACM3, as crucial residues for store-operated current activation. Although these residues are located within a putative CaM-binding domain, it seems unlikely that they mediate their effects by disrupting CaM binding, because structural modeling would require at least a double mutation to do so; the single lysine mutations still retain the highest possible score for a functional CaM-binding motif. In addition, although CaM binds to CRACM1 in a Ca2+-dependent manner (68–91), it is not thought to mediate CRAC channel activation, but rather plays a role in Ca2+-dependent inactivation (Mullins et al., 2009) and may also inhibit STIM2-dependent activation of CRACM1 (Parvez et al., 2008). Finally, it would appear that CRAC channels and STIM proteins are entirely sufficient to produce channel openings in a reconstituted system where CaM is likely absent (Zhou et al., 2010).
The dramatic reduction in store-operated activation of CRAC channels with mutations in the critical N-terminal lysine residue suggests that this residue is involved in conformational changes that lead to channel opening, despite the ability of STIM1 to bind. An alternative or contributing factor might be that these residues are required for the assembly or stability of CRAC channel complexes. Recently, Srikanth et al. (2010) identified a CRAC regulator protein (CRACR2A) that is thought to be essential for the assembly and/or stability of the CRACM-STIM clusters and whose binding sites on CRACM1 and STIM1 partially overlap with those of CaM. They demonstrated that a double mutation on CRACM1, K85A/K87A, completely disrupted CRACR2A binding. Furthermore, a minimal region of the STIM1 protein has been identified that is needed for coupling to and inducing constitutive activity of CRAC channels (Muik et al., 2009; Park et al., 2009; Yuan et al., 2009). C and N termini of CRACM1 are both necessary to activate ICRAC, but the first 73 amino acid residues in CRACM1 protein are not absolutely required for activation (Li et al., 2007; Park et al., 2009). However, the additional deletion of the amino acid residues 73–84 from CRACM1 eliminated CAD binding to the N-terminal region and resulted in suppressed Ca2+ influx induced by CAD (Park et al., 2009). Nevertheless, the reported interaction of CAD with both the C and N termini of CRACM1 (Park et al., 2009) suggests an essential regulatory role in the CRACM1 channel gating. In the present study, the short N-terminal fragments of the K85E mutant in CRACM1 and K60E (CRACM3), respectively, are still able to bind to the minimal binding domain of STIM1 (CAD) (Fig. 6, E and F), but the gating or stable assembly of the channel is impaired. This position K85 has been suggested to be important for the CRACM1 binding to the CRAC regulator (CRACR2A) and thus might destabilize the interaction of CRACM and STIM1 (Srikanth et al., 2010). Therefore, it is possible that STIM1 is still able to bind to the N-terminal region of CRACM1 or CRACM3; however, CRACR2A is not, possibly affecting SOCE. It should be noted, however, that the role of CRACR2A in SOCE requires further investigation because this protein is not required for STIM1 and CRAC channel interaction in reconstituted systems where the protein is likely absent (Zhou et al., 2010). In any event, our observations suggest that a simple binding of STIM1 to the C- and/or N-terminal region of CRACM1 or CRACM3 may not by itself be sufficient to trigger channel gating into the open state. Instead, at least one further conformational step appears to be required to open the channel pore, and the lysine residues within the N terminus identified here might be critical for this step.
Exchanging the positively charged and polar lysine residue for an uncharged and nonpolar amino acid such as alanine (CRACM1-K85A) completely abolished IP3-induced currents and strongly decreased store-independent responses with 2-APB. When switching to the uncharged but polar amino acid residue glutamine (CRACM1-K85Q), the IP3-induced currents could be restored, indicating that glutamine may create a sufficiently polar environment to support STIM1-mediated activation. Interestingly, however, this mutant exhibited enhanced outwardly rectifying currents when exposed to 2-APB compared with CRACM1 wt channels (Peinelt et al., 2008). The reason for this might be a better accessibility of the 2-APB compound to the channel, resulting in altered ion selectivity (Peinelt et al., 2008; Schindl et al., 2008). Alternatively, during 2-APB application, the N terminus may contribute to inward rectification of CRAC currents by impeding outward movement of monovalent cations, with mutations in this region being less effective in doing so and enabling limited outward currents in conjunction with changes in pore selectivity induced by 2-APB. Introducing a negative charge by mutating the lysine residue into the polar but negatively charged amino acid residue glutamic acid (CRACM1-K85E or CRACM3-K60E) again resulted in complete disruption of the IP3-mediated CRAC currents. In this case, the presence of a negative charge strongly impaired the gating function of the CRAC channel and almost completely abolished the store-independent response to 2-APB (Fig. 4). At the same time, position 85 in the N-terminal region close to the first transmembrane domain appeared to be particularly important for channel gating because the equivalent mutations of other positively charged amino acid residues at positions 69, 77, 78, 83, and 87 (Fig. S3) had no such strong inhibitory effect and were still able to elicit IP3-mediated current activation.
N terminus truncations CRACM1-Δ87 and the corresponding CRACM3-Δ62 truncations (resulting in the deletion of crucial lysine residues K85 and K60, respectively) strongly suppress both channels but appear to affect CRACM1 and CRACM3 differentially in terms of trafficking, because CRACM1 localization appears normal, whereas CRACM3 is partly retained in intracellular compartments (Fig. 2). Although the trafficking defect reduces the number of CRACM3 channels in the plasma membrane, those channels remain functional, because 2-APB can gate the channels that do transfer to the plasma membrane. Previous publications dealing with CRACM1/STIM1 binding and regulation have not considered the two distinct gating modes of CRAC channels: a store-operated (STIM1-dependent) and non–store-operated gating by 2-APB (STIM1-independent) (DeHaven et al., 2007; Peinelt et al., 2008; Schindl et al., 2008; Zhang et al., 2008). The results in the present study now demonstrate that the CRACM3 N-terminal region is essential for store-operated STIM1-dependent gating but not essential for non–store-operated gating of CRACM3 channels by 2-APB. This is not entirely surprising, because the STIM1-dependent gating requires binding of STIM1 to the C-terminal region and subsequent conformational changes of the N-terminal region. In general, we found that CRACM3 trafficking may rely more heavily on the structural integrity of its N terminus because truncating or mutating this segment results in improper targeting to the plasma membrane (see Fig. 2).
Detailed analysis of the current densities supports the notion of a strong inhibitory effect of the CRACM3 point mutation, suggesting that a single K60E subunit is enough to strongly block the channel function. Assuming equal expression levels and unbiased assembly into tetramers, there are 16 possible assemblies. Under these conditions, homomeric wt CRACM3 assemblies remaining fully functional would then be able to generate 6.25% of wt current (or −0.79 pA/pF of −12.75 pA/pF in controls). Experimentally, the amount of current we observe is −1.37 pA/pF, suggesting that the insertion of a single mutant subunit into a tetrameric channel appears to be sufficient to reduce channel function almost completely. In the case of the CRACM1 mutant K85E, we see a 50% inhibition of current compared with wt (or −10.97 pA/pF of −23.4 pA/pF in controls), and this would indicate that at least two subunits are necessary to account for the inhibitory effect we observe.
Because a mutation in the K85/K60 residue disrupts store-operated calcium influx, we would assume that in humans the K85E mutation might yield a similar SCID phenotype as demonstrated for the R91W mutation. However, there are differences in the behavior of these residues because the mutation of a positive amino acid residue to a negatively charged residue K85E mutant completely disrupts STIM1-dependent current activation (−0.2 pA/pF), whereas the corresponding R91E mutation does not affect CRAC current activation (−6–8 pA/pF; Derler et al., 2009). The mutation to a neutral amino acid, K85A, also resulted in a nonfunctional channel (−0.37 pA/pF), whereas the corresponding R91G does not disrupt current activation (−6–8 pA/pF; Derler et al., 2009). Thus, on a molecular basis, the two residues are clearly distinct from each other.
Similar to the CRACM3-R66W (SCID), the CRACM3-K60E mutation disrupts STIM1-dependent current activation, but unlike R66W, it still can be activated in a store-independent manner by 2-APB to a certain amount. With respect to the CRACM1-R91W mutation, our results confirm that overexpression of this mutant alone results in almost complete suppression of CRAC currents below control levels and therefore may act as a dominant-negative subunit for endogenous channels (Fig. S4, A and B). The co-overexpression experiments further demonstrate that the CRACM1-R91W mutant can suppress CRACM1 channel subunits, presumably because they can form heteromultimers (Lis et al., 2007). These data (Fig. S4) are at odds with a recent report that showed relatively normal CRACM1 current amplitudes when coexpressing the mutant and wt channels, with just a slower kinetics after passive store depletion (Muik et al., 2008). A possible explanation for this discrepancy might be differences in the efficacy of mutant subunit expression in the two studies. Analysis of the amount of reduction suggests that a single R91W subunit is enough to essentially block channel function. Assuming equal expression levels and unbiased assembly into tetramers, there are 16 possible assemblies. Under these conditions, homomeric wt CRACM1 assemblies remaining fully functional would then be able to generate 6.25% of the wt current, namely 1.75 pA/pF of 28 pA/pF in controls, which is almost exactly the amount of current we observed: 1.76 pA/pF. Based on the above considerations and with the caveat of necessary assumptions in the calculations, we propose that incorporation of just one R91W subunit into the CRAC channel complex may be sufficient to essentially block its current flow. Hence, the effect of this mutant could result from a dominant-negative effect if it formed heteromeric channels with endogenous channel subunits, which has been demonstrated to be the case for wt CRACM proteins (Vig et al., 2006a; Lis et al., 2007).
So far, four single amino acids in CRACM1 have been identified as crucial for channel function, and point mutations in these residues result in complete loss of function. (1) E106Q is a pore mutant that disrupts ion conduction and acts as a dominant-negative subunit (Prakriya and Lewis, 2006; Vig et al., 2006a; Yeromin et al., 2006). (2) R91W is a mutation that disrupts SOCE in T cells and underlies the SCID pathology in human patients (Feske et al., 2006). The protein expresses in the plasma membrane, but it cannot support ion currents. The increased hydrophobicity at the interface of the N terminus–membrane has been claimed as the main cause for the nonfunctional channel (Derler et al., 2009). (3) L273S (Muik et al., 2008) and (4) L276D (Navarro-Borelly et al., 2008) are C-terminal–region mutants that have been proposed to be crucial for STIM1-dependent activation of CRACM1 because they localize to the plasma membrane but fail to interact with STIM1. The new K85 residue does not appear to be a part of the selectivity filter because the K85Q mutation develops IP3-induced currents with the distinctive I-V relationship of ICRAC (Fig. 4 C), whereas the K85E mutant appears to be a loss-of-function mutation. Because the same mutation in the chimera (CRACM3-N(M1)-K85E) remains unavailable for STIM1-dependent gating but can still be activated in a store-independent manner with 2-APB (Fig. 5, B and F), we propose that this residue is essential for the gating and/or stable assembly of CRAC channels.
CRAC channel opening is the final step of a complex series of events (Muik et al., 2009; Park et al., 2009; Yuan et al., 2009), and any defect in the cascade results in lack of channel opening. The initial step in STIM1/CRAC communication seems to be preserved, as judged from the Co-IP experiments that suggest intact binding properties of mutants that render CRAC channels silent. Nevertheless, these experiments might not represent the actual mechanism of interaction of full-length CRACM and STIM1 proteins because only the minimal fractions are used for our binding studies. However, the lack of channel opening supports the concept of the K85 and K60 residues in CRACM1 and CRACM3, respectively, as critical for the gating of the channel. Finally, our results support the hypothesis that the opening of CRAC channels may entail a sequential process that involves the binding of STIM1 to the coiled-coil region within the C-terminal region, which then causes a bridging toward the N-terminal region, resulting in a conformational change in CRAC channels to accomplish channel gating (Park et al., 2009). In light of recent work that proposes a tetrameric assembly of CRAC channel subunits interacting with dimeric STIM1 (Li et al., 2008; Mignen et al., 2008), and the direct binding of CAD to N and C termini of CRACM1 (Park et al., 2009), we further propose that the basic amino acid lysine (K) at the position 85 in CRACM1 (and K60 in CRACM3) is indispensable for the conformational changes to drive the opening of the CRAC channels and Ca2+ entry.
Acknowledgments
We thank L. Addington, L. Tsue, S. Johne, and M. K. Monteilh-Zoller for excellent technical support. The authors thank R. Lewis for the FLAG-Myc-CAD construct.
This work was supported by National Institutes of Health (NIH) grants R01-AI050200, R01-GM080555 (to R. Penner), and 5G12 RR003061-22 (UH imaging core); Queen Emma Research Fund (to A. Lis); the German Research Foundation (DFG) grants Li 1750/1-1 (to A. Lis) and PE 1478/5-1 CP; and Austrian Science Fund (FWF) grant J2784 (to S. Zierler).
Angus C. Nairn served as editor.
Footnotes
Abbreviations used in this paper:
- 2-APB
- 2-aminoethoxydiphenyl borate
- CAD
- calcium release–activated calcium–activating domain
- CaM
- calmodulin
- Co-IP
- coimmunoprecipitation
- CRAC
- calcium release–activated calcium
- CRACR2A
- CRAC regulator 2A
- IP3
- inositol 1,4,5-trisphosphate
- SCID
- severe combined immunodeficiency
- SOCE
- store-operated Ca2+ entry
- STIM
- stromal interaction molecule
- wt
- wild type
References
- Barr V.A., Bernot K.M., Srikanth S., Gwack Y., Balagopalan L., Regan C.K., Helman D.J., Sommers C.L., Oh-Hora M., Rao A., Samelson L.E. 2008. Dynamic movement of the calcium sensor STIM1 and the calcium channel Orai1 in activated T-cells: puncta and distal caps. Mol. Biol. Cell. 19:2802–2817 10.1091/mbc.E08-02-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHaven W.I., Smyth J.T., Boyles R.R., Putney J.W., Jr 2007. Calcium inhibition and calcium potentiation of Orai1, Orai2, and Orai3 calcium release-activated calcium channels. J. Biol. Chem. 282:17548–17556 10.1074/jbc.M611374200 [DOI] [PubMed] [Google Scholar]
- DeHaven W.I., Smyth J.T., Boyles R.R., Bird G.S., Putney J.W., Jr 2008. Complex actions of 2-aminoethyldiphenyl borate on store-operated calcium entry. J. Biol. Chem. 283:19265–19273 10.1074/jbc.M801535200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derler I., Fahrner M., Carugo O., Muik M., Bergsmann J., Schindl R., Frischauf I., Eshaghi S., Romanin C. 2009. Increased hydrophobicity at the N terminus/membrane interface impairs gating of the severe combined immunodeficiency-related ORAI1 mutant. J. Biol. Chem. 284:15903–15915 10.1074/jbc.M808312200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., Rao A. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441:179–185 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Frischauf I., Muik M., Derler I., Bergsmann J., Fahrner M., Schindl R., Groschner K., Romanin C. 2009. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J. Biol. Chem. 284:21696–21706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sukumar P., Milligan C.J., Kumar B., Ma Z.Y., Munsch C.M., Jiang L.H., Porter K.E., Beech D.J. 2008. Interactions, functions, and independence of plasma membrane STIM1 and TRPC1 in vascular smooth muscle cells. Circ. Res. 103:e97–e104 10.1161/CIRCRESAHA.108.182931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu J., Xu P., Xie X., Chen L., Xu T. 2007. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J. Biol. Chem. 282:29448–29456 10.1074/jbc.M703573200 [DOI] [PubMed] [Google Scholar]
- Liao Y., Erxleben C., Yildirim E., Abramowitz J., Armstrong D.L., Birnbaumer L. 2007. Orai proteins interact with TRPC channels and confer responsiveness to store depletion. Proc. Natl. Acad. Sci. USA. 104:4682–4687 10.1073/pnas.0611692104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E., Jr, Meyer T. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis A., Peinelt C., Beck A., Parvez S., Monteilh-Zoller M., Fleig A., Penner R. 2007. CRACM1, CRACM2, and CRACM3 are store-operated Ca2+ channels with distinct functional properties. Curr. Biol. 17:794–800 10.1016/j.cub.2007.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik R.M., Wu M.M., Buchanan J., Lewis R.S. 2006. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J. Cell Biol. 174:815–825 10.1083/jcb.200604015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J.C., Dehaven W.I., Smyth J.T., Wedel B., Boyles R.R., Bird G.S., Putney J.W., Jr 2006. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J. Biol. Chem. 281:24979–24990 10.1074/jbc.M604589200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignen O., Thompson J.L., Shuttleworth T.J. 2008. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J. Physiol. 586:419–425 10.1113/jphysiol.2007.147249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., et al. 2008. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 283:8014–8022 10.1074/jbc.M708898200 [DOI] [PubMed] [Google Scholar]
- Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. 2009. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J. Biol. Chem. 284:8421–8426 10.1074/jbc.C800229200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins F.M., Park C.Y., Dolmetsch R.E., Lewis R.S. 2009. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. USA. 106:15495–15500 10.1073/pnas.0906781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Borelly L., Somasundaram A., Yamashita M., Ren D., Miller R.J., Prakriya M. 2008. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J. Physiol. 586:5383–5401 10.1113/jphysiol.2008.162503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., Lewis R.S. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S., Beck A., Peinelt C., Soboloff J., Lis A., Monteilh-Zoller M., Gill D.L., Fleig A., Penner R. 2008. STIM2 protein mediates distinct store-dependent and store-independent modes of CRAC channel activation. FASEB J. 22:752–761 10.1096/fj.07-9449com [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinelt C., Vig M., Koomoa D.L., Beck A., Nadler M.J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J.P. 2006. Amplification of CRAC current by STIM1 and CRACM1 (Orai1). Nat. Cell Biol. 8:771–773 10.1038/ncb1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinelt C., Lis A., Beck A., Fleig A., Penner R. 2008. 2-Aminoethoxydiphenyl borate directly facilitates and indirectly inhibits STIM1-dependent gating of CRAC channels. J. Physiol. 586:3061–3073 10.1113/jphysiol.2008.151365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M., Lewis R.S. 2006. Regulation of CRAC channel activity by recruitment of silent channels to a high open-probability gating mode. J. Gen. Physiol. 128:373–386 10.1085/jgp.200609588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J., DiGregorio P.J., Yeromin A.V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J.A., Wagner S.L., Cahalan M.D., et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169:435–445 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindl R., Bergsmann J., Frischauf I., Derler I., Fahrner M., Muik M., Fritsch R., Groschner K., Romanin C. 2008. 2-Aminoethoxydiphenyl borate alters selectivity of Orai3 channels by increasing their pore size. J. Biol. Chem. 283:20261–20267 10.1074/jbc.M803101200 [DOI] [PubMed] [Google Scholar]
- Soboloff J., Spassova M.A., Tang X.D., Hewavitharana T., Xu W., Gill D.L. 2006. Orai1 and STIM reconstitute store-operated calcium channel function. J. Biol. Chem. 281:20661–20665 10.1074/jbc.C600126200 [DOI] [PubMed] [Google Scholar]
- Spassova M.A., Soboloff J., He L.P., Xu W., Dziadek M.A., Gill D.L. 2006. STIM1 has a plasma membrane role in the activation of store-operated Ca(2+) channels. Proc. Natl. Acad. Sci. USA. 103:4040–4045 10.1073/pnas.0510050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Jung H.J., Kim K.D., Souda P., Whitelegge J., Gwack Y. 2010. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12:436–446 10.1038/ncb2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Murakami M., Watanabe H., Hasegawa H., Ohba T., Munehisa Y., Nobori K., Ono K., Iijima T., Ito H. 2007. Essential role of the N-terminus of murine Orai1 in store-operated Ca2+ entry. Biochem. Biophys. Res. Commun. 356:45–52 10.1016/j.bbrc.2007.02.107 [DOI] [PubMed] [Google Scholar]
- Várnai P., Tóth B., Tóth D.J., Hunyady L., Balla T. 2007. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J. Biol. Chem. 282:29678–29690 10.1074/jbc.M704339200 [DOI] [PubMed] [Google Scholar]
- Vig M., Beck A., Billingsley J.M., Lis A., Parvez S., Peinelt C., Koomoa D.L., Soboloff J., Gill D.L., Fleig A., et al. 2006a. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr. Biol. 16:2073–2079 10.1016/j.cub.2006.08.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M., Peinelt C., Beck A., Koomoa D.L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J.P. 2006b. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 312:1220–1223 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.M., Buchanan J., Luik R.M., Lewis R.S. 2006. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174:803–813 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeromin A.V., Zhang S.L., Jiang W., Yu Y., Safrina O., Cahalan M.D. 2006. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 443:226–229 10.1038/nature05108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.P., Zeng W., Dorwart M.R., Choi Y.J., Worley P.F., Muallem S. 2009. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11:337–343 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., Yu Y., Roos J., Kozak J.A., Deerinck T.J., Ellisman M.H., Stauderman K.A., Cahalan M.D. 2005. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 437:902–905 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., Yeromin A.V., Zhang X.H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K.A., Cahalan M.D. 2006. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc. Natl. Acad. Sci. USA. 103:9357–9362 10.1073/pnas.0603161103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., Kozak J.A., Jiang W., Yeromin A.V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M.D. 2008. Store-dependent and -independent modes regulating Ca2+ release-activated Ca2+ channel activity of human Orai1 and Orai3. J. Biol. Chem. 283:17662–17671 10.1074/jbc.M801536200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Meraner P., Kwon H.T., Machnes D., Oh-hora M., Zimmer J., Huang Y., Stura A., Rao A., Hogan P.G. 2010. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat. Struct. Mol. Biol. 17:112–116 10.1038/nsmb.1724 [DOI] [PMC free article] [PubMed] [Google Scholar]