Protocadherin-19 and N-cadherin act synergistically during brain morphogenesis in zebrafish.
Abstract
The protocadherins comprise the largest subgroup within the cadherin superfamily, yet their cellular and developmental functions are not well understood. In this study, we demonstrate that pcdh19 (protocadherin 19) acts synergistically with n-cadherin (ncad) during anterior neurulation in zebrafish. In addition, Pcdh19 and Ncad interact directly, forming a protein–protein complex both in vitro and in vivo. Although both molecules are required for calcium-dependent adhesion in a zebrafish cell line, the extracellular domain of Pcdh19 does not exhibit adhesive activity, suggesting that the involvement of Pcdh19 in cell adhesion is indirect. Quantitative analysis of in vivo two-photon time-lapse image sequences reveals that loss of either pcdh19 or ncad impairs cell movements during neurulation, disrupting both the directedness of cell movements and the coherence of movements among neighboring cells. Our results suggest that Pcdh19 and Ncad function together to regulate cell adhesion and to mediate morphogenetic movements during brain development.
Introduction
The development of the vertebrate nervous system is orchestrated through a complex interplay of cell–cell interactions that coordinate cell migration, morphology, and proliferation in a time- and place-appropriate manner. Although the molecular machinery underlying these cellular interactions and the mechanisms coordinating dynamic developmental events are not well understood, cadherins have been shown to play important roles in early embryonic development. The cadherin superfamily is a collection of calcium-dependent cell surface receptors that includes both the classical cadherins and the more recently discovered protocadherins (Takeichi, 1988; Nollet et al., 2000; Redies, 2000; Hulpiau and van Roy, 2009). Several studies have shown that N-cadherin (Ncad) plays an important role in neurulation. Loss of Ncad in zebrafish impairs convergence movements in the anterior neural plate (Lele et al., 2002; Hong and Brewster, 2006) and results in subsequent abnormalities in brain and retinal morphology (Lele et al., 2002). More recently, Ncad has been shown to mediate actin assembly in the neural plate of Xenopus laevis (Nandadasa et al., 2009) and, in concert with Nectin-2, to play a role in apical constriction (Morita et al., 2010).
Like the classical cadherins, several protocadherins have been implicated in early developmental events. Pcdh18a (Protocadherin 18a) contributes to cell movements during epiboly (Aamar and Dawid, 2008). Additionally, paraxial protocadherin (PAPC) has also been implicated in morphogenetic processes, including convergent extension (Chen and Gumbiner, 2006; Kim et al., 1998; Unterseher et al., 2004; Wang et al., 2008; Yamamoto et al., 1998), somitogenesis (Kim et al., 2000) and tissue separation (Medina et al., 2004). Intriguingly, PAPC has been shown to modulate cell adhesion by antagonizing the function of C-cadherin (Chen and Gumbiner, 2006). Although the mechanism by which PAPC regulates C-cadherin is not known, a study in cultured hippocampal neurons has shown that induction of the related molecule Pcdh8 by electroconvulsive shock treatment results in binding of Pcdh8 to Ncad and internalization of the complex (Yasuda et al., 2007). Together, the studies on PAPC/C-cadherin and Pcdh8/Ncad suggest an intimate functional relationship between these two branches of the cadherin superfamily.
Mutations in pcdh19 result in a female-limited form of epilepsy, which is characterized by seizures in infancy and early childhood and cognitive impairment in adults (Dibbens et al., 2008; Depienne et al., 2009; Marini et al., 2010). We have previously shown that pcdh19 plays an essential role during early stages of brain morphogenesis during zebrafish development (Emond et al., 2009). Embryos that are morphant for pcdh19 exhibit severely disrupted brain morphology, which is caused at least in part by a defect in cell movements in the anterior neural plate. The phenotype of pcdh19 morphants is very reminiscent of ncad mutants and morphants (Lele et al., 2002; Hong and Brewster, 2006), as each exhibit a failure in the convergence of the lateral neural plate and abnormal morphology in the developing midbrain and hindbrain regions. This similarity suggests that pcdh19 and ncad could be functioning in a cooperative manner during zebrafish brain development.
In this study, we show that Pcdh19 and Ncad act in concert to coordinate cell movements during neurulation of the anterior neural plate in zebrafish embryos. Morpholinos directed against pcdh19 and ncad act synergistically at subthreshold doses. Pcdh19 and Ncad also associate physically through their extracellular domains. The physical relationship between these two proteins may impact Ncad adhesion, as loss of either Pcdh19 or Ncad eliminates calcium-dependent cell aggregation in vitro, despite the fact that Pcdh19 does not exhibit intrinsic adhesive activity on its own. Finally, we demonstrate that knockdown of pcdh19 or ncad has nearly identical effects on cell movements in vivo. Our results demonstrate both a physical and functional interaction of Pcdh19 and Ncad that is essential for brain morphogenesis and underscore the idea that protocadherins may act as cofactors for cadherin function during vertebrate development.
Results
Pcdh19 and Ncad exhibit similar phenotypes
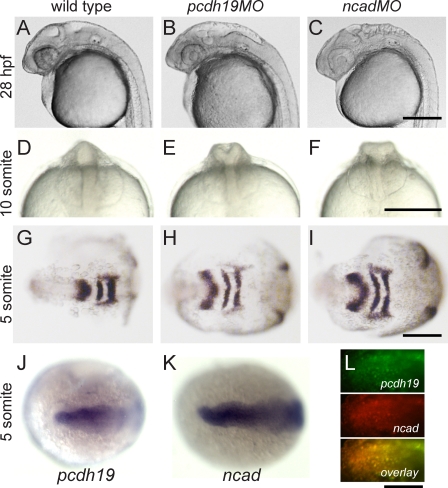
It has previously been shown that disruption of either Pcdh19 or Ncad impairs brain morphogenesis in zebrafish (Lele et al., 2002; Emond et al., 2009). The midbrain–hindbrain regions generally have a bumpy and/or misfolded appearance at 28 h postfertilization (hpf; Fig. 1, A–C), although the trunk is largely unaffected (not depicted). At earlier stages, both pcdh19 and ncad morphants exhibit characteristic malformations of the anterior neural rod (Fig. 1, D–F). The brains of pcdh19 morphants appear Y shaped in anterior views (Fig. 1 E), whereas ncad mutants and morphants (Lele et al., 2002) exhibit a characteristic T- or mushroom-shaped brain (Fig. 1 F). In both cases, convergence cell movements in the lateral neural plate are impaired. In contrast to wild-type embryos, embryos injected with antisense morpholino oligonucleotides against either pcdh19 or ncad (pcdh19MO or ncadMO, respectively) exhibit a substantially wider neural plate, as shown by labeling of five-somite stage embryos with riboprobes against pax2.1 and krox20 (Fig. 1, G–I). The expression of pcdh19 and ncad overlaps substantially in the anterior neural plate (Fig. 1, J–L). Thus, knockdown of pcdh19 and ncad exhibit very comparable phenotypes throughout early stages of neural development: late convergence of the anterior neural plate is compromised, leading to aberrant brain morphology, whereas trunk neurulation is unaffected.
Figure 1.
Pcdh19 and Ncad morphants exhibit similar phenotypes. (A–C) Lateral views of the head of 28-hpf embryos. Wild-type embryos (A) exhibit a smooth hindbrain and well-formed midbrain/hindbrain and optic tectum. The midbrain and hindbrain regions are severely perturbed in pcdh19 (B) and ncad (C) morphants. Bar, 250 µm. (D–F) Anterior views of wild-type embryos (D) show a smooth neural rod along the dorsal midline. The brain of pcdh19 morphants (E) has a Y shape. Embryos morphant for ncad (F) have an appearance similar to that of pcdh19 morphants, exhibiting a T or mushroom shape. Bar, 150 µm. (G–I) Five-somite embryos labeled for pax2.1 and krox20. The neural plate of wild-type embryos (G) has converged substantially and is much narrower than those of pcdh19 (H) or ncad (I) morphants. Bar, 250 µm. (J–L) Expression of pcdh19 and ncad overlaps in the anterior neural plate. Embryos at the five-somite stage were labeled with riboprobes against pcdh19 (J) or ncad (K). Although ncad is expressed throughout the neural plate, the expression of pcdh19 is restricted to the anterior region of the neural plate. This overlapping expression is also observed by two-color in situ hybridization (L). Bar, 50 µm. Anterior is to the left.
Knockdown of Pcdh19 and Ncad is synergistic
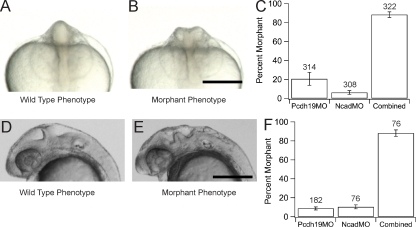
As depletion of either Pcdh19 or Ncad results in similar defects in brain morphogenesis and because they have overlapping patterns of expression, we hypothesized that these molecules may be acting in concert during morphogenesis of the anterior neural plate. To address this, we used a subthreshold morpholino approach to probe for a functional interaction between these two molecules (Fig. 2). Although typical doses of morpholino used to knock down ncad and pcdh19 are 1–2 ng and 2–3 ng, respectively, embryos were injected with subthreshold doses of ncadMO (0.2 ng), pcdh19MO (0.5 ng), or both together. At the 10-somite stage, the brain of wild-type or unaffected embryos appears as a smooth, solid rod in anterior views (Fig. 2 A). In contrast, the brains of both ncad and pcdh19 morphants appear T or Y shaped, as mentioned in the previous paragraph, allowing live embryos to be scored (Fig. 2 B). Injection of subthreshold doses of ncadMO or pcdh19MO on their own resulted in low proportions of morphant phenotype (Fig. 2 C). However, coinjection of both pcdh19MO and ncadMO resulted in a dramatic increase in embryos with aberrant brain morphology (Fig. 2 C). Similar results were obtained when scoring 28-hpf embryos for wavy or bumpy brains (Fig. 2, D–F). Subthreshold doses of either pcdh19MO or ncadMO alone yielded a low proportion of embryos with morphant phenotype compared with coinjection of morpholinos (Fig. 2 F). These results indicate that knockdown of Pcdh19 and Ncad is synergistic during anterior neurulation.
Figure 2.
Pcdh19 and Ncad act synergistically. (A and B) Embryos were scored at the 10-somite stage for wild-type (A) or morphant (B) phenotypes. (C) Quantification of the interaction between pcdh19 (pcdh19MO) and ncad (ncadMO) morpholinos. Injection of 0.5 ng pcdh19MO results in 20.6 ± 6.8% (314 embryos from four experiments) morphant embryos, whereas injection of 0.2 ng ncadMO yields 6.2 ± 1.9% (308 embryos from four experiments) morphants. However, 88.4 ± 3.0% of embryos (322 embryos from four experiments) were morphant when coinjected with 0.5 ng pcdh19MO and 0.2 ng ncadMO. (D and E) Lateral view of embryos exhibiting wild-type (D) or morphant (E) phenotype at 28 hpf. (F) Injection of 1 ng pcdh19MO results in 8.8 ± 1.6% morphant embryos (182 embryos from five experiments). Similarly, injection of 0.25 ng ncadMO yields 10.5 ± 2.1% (76 embryos from three experiments) morphant phenotype. In contrast, coinjection of 1 ng pcdh19MO and 0.25 ng ncadMO yields 88.2 ± 3.4% of morphant embryos (76 embryos from three experiments). Error bars represent SEM. Bars, 250 µm.
Overexpression of Pcdh19 fails to rescue Ncad morphants and vice versa
As both pcdh19 and ncad morphants exhibited an arrest of neural plate convergence, measurements of neural plate width provide a quantitative method to assess pcdh19 and ncad function (Fig. 1, G–I). To further investigate the functional interaction of Pcdh19 and Ncad, we began by characterizing the effects of Pcdh19 or Ncad overexpression. Heat shock induction of either Pcdh19-GFP or Ncad-GFP expression at tailbud stage had no effect on neural plate widths at the five-somite stage (Fig. 3, A and B). To determine whether the Pcdh19-GFP and Ncad-GFP were functional in these assays, we coinjected them with pcdh19MO or ncadMO, respectively. Expression of each GFP fusion was induced at tailbud stage, and embryos were allowed to develop until the five-somite stage. Expression of Pcdh19-GFP was able to compensate for the depletion of endogenous Pcdh19, resulting from morpholino knockdown (Fig. 3 C). Similarly, expression of Ncad-GFP was able to rescue ncad morphants (Fig. 3 D).
Figure 3.
Overexpression of Pcdh19 or Ncad fails to reciprocally rescue. Cumulative probability distributions of neural plate widths in five-somite stage embryos. (A and B) Expression of Pcdh19-GFP (A) or Ncad-GFP (B) does not affect neural plate width. (C and D) Expression of Pcdh19-GFP (C) or Ncad-GFP (D) can compensate for the loss of endogenous Pcdh19 or Ncad, respectively. (E and F) Expression of Pcdh19-GFP (E) or Ncad-GFP (F) cannot reciprocally rescue ncad morphants or pcdh19 morphants, respectively. HS, heat shocked; Un-HS, un-heat shocked.
To probe the functional relationship of pcdh19 and ncad in more detail, we performed reciprocal rescue experiments to determine whether overexpression of Pcdh19-GFP could compensate for Ncad loss and vice versa. In neither case did overexpression of Pcdh19 or Ncad compensate for the loss of the other molecule (Fig. 3, E and F). One interpretation of these results is that Pcdh19 and Ncad act in parallel molecular pathways to mediate distinct cellular processes that each contribute to cell convergence in the neural plate. Alternatively, if Pcdh19 and Ncad form a functional protein complex, overexpression of one should not compensate for the loss of the other.
Pcdh19 and Ncad interact physically
Several recent studies have indicated that protocadherins are capable of interacting with classical cadherins (Chen and Gumbiner, 2006; Yasuda et al., 2007; Chen et al., 2009). Given the possibility that Pcdh19 and Ncad might interact physically, we performed coimmunoprecipitation (coIP) experiments. GFP- or myc-tagged Pcdh19 and Ncad (Fig. 4 A) were cotransfected into COS cells, and either Pcdh19 or Ncad was immunoprecipitated. In each case, Pcdh19 or Ncad was able to coimmunoprecipitate with the other protein (Fig. 4, B–D). The cytoplasmic domains of these proteins were not required for this interaction, as deletion mutants lacking their respective cytoplasmic domains (Pcdh19ΔCP and NcadΔCP) coimmunoprecipitated (Fig. 4 E). In contrast, fusions of just the cytoplasmic domains to human CD4 did not coimmunoprecipitate (Fig. 4, F and G). To determine whether Pcdh19 interacts with classical cadherins in vivo, we used antibodies against Pcdh19 to perform immunoprecipitations from extracts prepared from five-somite stage embryos and Zem2S cells (Fig. 4 H). The Zem2S cell line was derived from blastula-stage embryos (Collodi et al., 1992; Ghosh and Collodi, 1994) and expresses both Ncad and Pcdh19, as determined by RT-PCR (Fig. S1). Immunoprecipitation of endogenous Pcdh19 was able to coimmunoprecipitate classical cadherins from both embryo and Zem2S cell extracts.
Figure 4.
Pcdh19 physically interacts with Ncad. (A) Diagram of the constructs used for coIPs and BiFC. We constructed plasmids containing full-length Pcdh19, Pcdh19 lacking the cytoplasmic domain (Pcdh19ΔCP) or the cytoplasmic domain of Pcdh19 fused to human CD4 (CD4-Pcdh19CP), and the corresponding Ncad plasmids (Ncad, NcadΔCP, and CD4-NcadCP). (B–D) CoIPs show that Pcdh19 interacts with Ncad when cotransfected into COS cells. (B) Immunoprecipitation of Pcdh19-myc pulls down Ncad-GFP. (C) Immunoprecipitation of Ncad-GFP brings down Pcdh19-myc. (D) Immunoprecipitation of Ncad-myc also pulls down Pcdh19-GFP. (E) The cytoplasmic domains of Ncad and Pcdh19 are not required for their interaction. Immunoprecipitation of Pcdh19ΔCP-myc pulled down NcadΔCP-GFP. (F and G) The extracellular domains of Pcdh19 and Ncad are required for their interaction, as immunoprecipitation of CD4-NcadCP-myc fails to pull down CD4-Pcdh19CP-GFP (F), and pulling down CD4-Pcdh19CP-GFP does not coimmunoprecipitate CD4-NcadCP-myc (G). (H) Immunoprecipitation of endogenous Pcdh19 pulls down classical cadherins in vivo. Zem2S extracts and embryo extracts were immunoprecipitated with (+) or without (−) anti-Pcdh19 primary antibody, and proteins were blotted with anti–pan-cadherin antibody. (I–L) Bimolecular fluorescence complementation performed in HEK293 cells. Green, Venus fluorescence; blue, DAPI. (I and J) Pcdh19 and Ncad directly associate in cells transfected with the full-length constructs Pcdh19-VenusC and Ncad-VenusN (I) or with NcadΔCP-VenusN and Pcdh19ΔCP-VenusC (J). (K and L) HEK293 cells transfected with Ncad-VenusN and CD4-Pcdh19-VenusC (K) or CD4-NcadCP-VenusN and CD4-Pcdh19CP-VenusC (L) do not exhibit any fluorescence. (M and N) A maximum intensity projection of two optical sections of live cells expressing Pcdh19-GFP (M) or Ncad-GFP (N) in a five-somite stage embryo. (O) Low power image of two five-somite stage embryos coexpressing Ncad-VenusN and Pcdh19-VenusC. Fluorescence is evidence of efficient and widespread interaction of Pcdh19 and Ncad. (P) A maximum intensity projection of two optical sections of cells coexpressing Ncad-VenusN and Pcdh19-VenusC. Arrows point to accumulations at cell–cell contacts. Bars: (I and J) 25 µm; (K and L) 100 µm; (O) 150 µm; (M, N, and P) 10 µm.
Although coIPs demonstrated that Pcdh19 and Ncad interact physically, they do not answer the question of whether this interaction is direct. Additionally, we wanted to determine whether the Pcdh19–Ncad interaction occurred within the same cell (cis). To address these questions, we used bimolecular fluorescence complementation (BiFC; Kerppola, 2006, 2008). BiFC relies on the ability of YFP fragments (VenusN, amino acids 1–172; and VenusC, 155–end) to associate noncovalently when brought into close proximity and to form a functional, fluorescent YFP molecule (in this case, the Venus variant of YFP). When cotransfected into COS cells, full-length Pcdh19-VenusC and Ncad-VenusN fusions yielded fluorescence (Fig. 4 I). An identical result was obtained when the tags were reversed (unpublished data). Similarly, expression of Pcdh19ΔCP-VenusC with NcadΔCP-VenusN also resulted in fluorescence complementation (Fig. 4 J). To confirm that the Ncad and Pcdh19 cytoplasmic domains were not able to interact, we cotransfected cells with Ncad-VenusN and CD4-Pcdh19CP-VenusC or CD4-NcadCP-VenusN and CD4-Pcdh19CP-VenusC (Fig. 4, K and L), observing fluorescence in neither case. Our results demonstrate that Pcdh19 binds directly to Ncad and that this cis-interaction does not require the cytoplasmic domains of these two proteins.
To verify that Pcdh19 can interact with Ncad in vivo, we performed BiFC in live embryos. First, we characterized the expression of Pcdh19-GFP (Fig. 4 M) and Ncad-GFP (Fig. 4 N) by expressing them under the control of the hsp70 promoter and imaging them between the two- and five-somite stages. Pcdh19-GFP is present on the plasma membrane, labeling fine processes (Fig. 4 M). Although expression was weaker than for Pcdh19-GFP, Ncad-GFP also labeled the cell surface and accumulated at cell–cell junctions (Fig. 4 N). To perform BiFC, Pcdh19-VenusC was coexpressed with Ncad-VenusN, and embryos exhibited widespread fluorescence (Fig. 4 O). When imaged with a two-photon microscope, BiFC fluorescence was found at the surface of labeled cells and was concentrated at cell–cell junctions (Fig. 4 P), a distribution similar to that exhibited by Ncad-GFP.
Knockdown of either Pcdh19 or Ncad disrupts cell aggregation
Because Pcdh19 is a member of the cadherin superfamily, we hypothesized that it could participate in cell adhesion. To investigate this possibility, we performed cell aggregation assays using Zem2S cells (Collodi et al., 1992; Ghosh and Collodi, 1994). Zem2S cells were scrape loaded with morpholinos against either ncad or pcdh19 and used in a standard cell aggregation assay (Takeichi, 1977). As a control, Zem2S cells were scrape loaded in the absence of morpholinos. In all conditions, cells failed to aggregate in the absence of calcium (Fig. 5, A–C), remaining as a monodisperse suspension. Mock-loaded cells showed pronounced cell aggregation in the presence of calcium (Fig. 5 D), which was manifested as a clear shift in the distribution of aggregate sizes (Fig. 5 G). In contrast, cells loaded with either ncad or pcdh19 morpholinos did not aggregate in the presence of calcium (Fig. 5, E and F). The ratio of mean aggregate size in the presence or absence of calcium was 7.6 for mock-loaded cells, whereas cells loaded with ncadMO or pcdh19MO showed ratios of 1.1 and 1.4, respectively. Thus, knockdown of either Ncad or Pcdh19 was sufficient to block the vast majority of calcium-dependent cell adhesion.
Figure 5.
Knockdown of either Ncad or Pcdh19 abolishes calcium-dependent cell adhesion in Zem2S cells. (A–C) Zem2S cells in the absence of calcium. Cells were mock scrape loaded (A) or loaded with ncad morpholino (B) or pcdh19 morpholino (C). (D–F) Zem2S cells in the presence of calcium. Cells were mock scrape loaded (D) or loaded with ncad morpholino (E) or pcdh19 morpholino (F). Bar, 100 µm. (G) The distribution of aggregate sizes for each experimental condition. (H) The ratio of mean aggregate size in the presence and absence of calcium provides a measure of calcium-dependent cell adhesion for mock (7.6 ± 0.6; n = 3), ncadMO (1.1 ± 0.3; n = 3), and pcdh19MO (1.4 ± 0.2; n = 3). Error bars represent SEM.
Pcdh19 does not mediate calcium-dependent adhesion
The cell aggregation data suggest a role for Pcdh19 in cell adhesion. To directly assess the adhesive activity of Pcdh19, we fused the extracellular domain of Pcdh19 to the Fc region of human IgG (Pcdh19EC-Fc) for use in bead aggregation assays (Fig. S2). As a positive control, we also generated a similar NcadEC-Fc fusion protein. In the absence of calcium, NcadEC-Fc beads remained as single particles (Fig. 6 A). Although NcadEC-Fc–coated beads exhibited robust calcium-dependent bead aggregation (Fig. 6, B and C), we observed little evidence for calcium-dependent adhesion of Pcdh19EC-Fc–coated beads (Fig. 6, D–F). This result differs from a recent finding that chicken Pcdh19 can mediate weak adhesion (Tai et al., 2010). Although our data could be consistent with weak calcium-independent adhesion, the discrepancy could also be because of differences in methodology. Thus, despite evidence of a role for Pcdh19 in the aggregation of Zem2S cells, we find that the extracellular domain of Pcdh19 does not confer significant calcium-dependent, homophilic adhesion.
Figure 6.
Pcdh19 does not mediate significant calcium-dependent bead aggregation. (A and B) Beads coated with NcadEC-Fc in the absence (A) or presence (B) of calcium. (C) Distribution of NcadEC-Fc–coated bead aggregate sizes in the absence and presence of calcium. (D and E) Beads coated with Pcdh19EC-Fc in the absence (D) or presence (E) of calcium. Bar, 100 µm. (F) Distribution of Pcdh19EC-Fc–coated bead aggregate sizes in the absence and presence of calcium.
Defects in cell movements in pcdh19 and ncad morphants
To characterize cell movements in vivo, we used transgenic embryos expressing a Histone H2A–GFP fusion protein, Tg(h2afv:gfpkca66), which labels cell nuclei and allows precise cell tracking (Pauls et al., 2001; Geldmacher-Voss et al., 2003; England et al., 2006; McMahon et al., 2008). Using in vivo two-photon microscopy and automated time-lapse analysis, we characterized cell trajectories during morphogenesis of the anterior neural plate. In wild-type embryos, cells converge medially, moving ventrally at the midline to form a dorsal seam (Fig. 7 A and Video 1). In addition, there is a net anterior component to the trajectories. In contrast to wild-type embryos, pcdh19 morphants exhibited compromised cell convergence movements (Fig. 7 B and Video 2). As the initial formation of the dorsal seam and ventral movement still occurred, the arrest of cell convergence resulted in the formation of a central lumen (Video 2). A similar arrest of cell convergence was obtained in ncad morphants (Fig. 7 C and Video 3). However, in contrast to pcdh19 morphants, knockdown of ncad also blocked events at the midline and the formation of the dorsal seam, resulting in a uniform accumulation of cells across the midline. This difference in behavior likely accounts for the distinct phenotypes of pcdh19 and ncad morphants, i.e., the respective Y- and T-shaped brain morphologies apparent at the 10-somite stage (Fig. 1, E and F). As we have shown that coinjection of subthreshold doses of pcdh19MO and ncadMO acts synergistically, we also wanted to determine the effects of morpholino coinjection on cell trajectories. The total concentration of morpholino in coinjected embryos (0.7 ng) is substantially less than that in pcdh19 or ncad morphants (2.5 ng and 1.5 ng, respectively). As was observed in pcdh19 and ncad morphants, the coinjected embryos exhibited compromised convergence movements (Fig. 7 D and Video 4) and, similar to ncad morphants, converging cells did not respect the midline boundary. From these data, we measured the mean velocity of cell movements in each experimental condition (Fig. 7 E) and decomposed these measurements into anteroposterior (Fig. 7 F) and mediolateral components (Fig. 7 G). Although full knockdown of either pcdh19 or ncad or partial knockdown of both resulted in a decrease in overall cell velocities, this decrease was not significant.
Figure 7.
In vivo two-photon time-lapse microscopy and automated cell tracking in wild-type and morphant embryos. (A–D) Cell trajectories determined from time-lapse sequences from Tg(h2afv:GFP) transgenic embryos. Time lapses were collected from wild-type embryos (A), pcdh19 morphants (B), ncad morphants (C), and embryos coinjected with subthreshold doses of pcdh19 and ncad morpholinos (D). Bar, 50 µm. Anterior is to the left. (E–G) Graphs show the mean instantaneous cell velocities (E) and the mean cell velocities along the anteroposterior (F) and mediolateral axis (G). Error bars represent SEM.
One key feature of cell movement in wild-type embryos is the apparent directedness of the movements, with little unproductive motion (Fig. 8 A). To provide a measure of this directedness, we first compared the net distance traveled by a tracked cell (between the first and last time points of a cell trajectory) with the total distance (the sum of the individual distances moved at each time interval). A plot of the net distance versus the total distance for cells should result in a line with a slope of 1 if the cells moved in a straight line throughout their trajectory and a slope <1 if they exhibited any meandering. For cells in wild-type embryos, a scatter plot of individual cell distances results in points distributed around a line with a 0.75 slope (Fig. 8 B, n = 3,109 cells from three embryos). The ratio of net distance to total distance defines an index of directedness for each tracked cell. A histogram of the directedness indices for all measured cells showed that the movement was strongly directional (Fig. 8 C). In pcdh19 morphants, cells exhibited more complex trajectories that had an increased Brownian character (Fig. 8 D), yielding an increase in unproductive cell movements (Fig. 8, E and F; n = 3,669 cells from three embryos; P < 0.001 by Kolmogorov-Smirnov test). A fit to the scatter plot in Fig. 8 E had a 0.32 slope. A similar loss of directed movement was found in ncad morphants (Fig. 8, G–I; n = 7,056 cells from three embryos; P < 0.001). The scatter plot of distances in Fig. 8 H was best fit by a line with a 0.28 slope. Embryos coinjected with subthreshold doses of pcdh19MO and ncadMO also exhibit compromised directed cell movements (Fig. 8, J–L; n = 4,736 cells from four embryos; P < 0.001). Individual cell trajectories were less linear (Fig. 8 J) than wild-type embryos (Fig. 8 A). A plot of net distance against total distance for cell trajectories was best fit by a line with a 0.42 slope (Fig. 8 K), and the frequency distribution of directedness also revealed an increased propensity for cells to meander (Fig. 8 L).
Figure 8.
Analysis of directional cell movements in wild-type and morphant embryos. (A–C) Subset of cell trajectories from wild-type embryos (A). The colored dots indicate the track origins. (B) A plot of net distance traveled against the sum of individual steps for each cell trajectory. Black line has a slope of 1. Red line is a fit to the data points and has a slope of 0.75. (C) A frequency histogram of cell directedness is shown. (D–F) Subset of cell trajectories derived from pcdh19 morphant embryos (D). (E) A plot of net distance against summed distance for cell trajectories in pcdh19 morphants. A line of best fit through the data points has a slope of 0.32. (F) A frequency histogram of the directedness is shown. (G–I) Subset of cell trajectories derived from ncad morphant embryos (G). (H) A plot of net distance against summed distance for cell trajectories in ncad morphants. A line of best fit through the data points has a slope of 0.28. (I) A frequency histogram of the directedness is shown. (J–L) Subset of cell trajectories derived from morphant embryos injected with low doses of both pcdh19MO and ncadMO (J). (K) A plot of net distance against summed distance for cell trajectories in coinjected morphants. A line of best fit through the data points has a slope of 0.43. (L) A frequency histogram of the directedness is shown. Bar, 15 µm.
As both Pcdh19 and Ncad are cell surface proteins that could mediate interactions among groups of neighboring cells, we hypothesized that there would also be a loss of correlation between neighboring cells in the morphant embryos. To provide a measure of correlation among cell movements, we determined the relative orientation (cosine Θ [cosΘ]) of cell displacement vectors for each cell and all neighboring cells within an approximately two-cell radius. CosΘ, which we call directional correlation, is a convenient metric, which ranges from 1 for parallel vectors to −1 for antiparallel vectors. In wild-type embryos, the distribution of the directional correlation was weighted heavily toward values of 1 (52,343 steps; Fig. 9 A), indicating that most neighboring cells were moving in parallel or nearly parallel trajectories. In contrast, both pcdh19 (62,593 steps; P < 0.001; Fig. 9 B) and ncad morphants (51,078 steps; P < 0.001; Fig. 9 C) exhibited a dramatic increase in uncorrelated movements, with values for directional correlation distributed all along the range from 1 to −1. Similarly, coinjected embryos also exhibited a decrease in cohesive cell movements (43,971 steps; P < 0.001; Fig. 9 D). Thus, knockdown of either Pcdh19 or Ncad or partial knockdown of both resulted in specific defects in cell motility in the anterior neural plate, including a decrease in the directedness of cell movements and a reduction in the coupled migration of neighboring cells.
Figure 9.
Morphant embryos exhibit a loss of correlated movement among neighboring cells. (A–D) Frequency distribution of directional correlation values (cos(Θ)) in wild-type embryos (A), pcdh19 morphants (B), ncad morphants (C), and embryos coinjected with subthreshold doses of pcdh19 and ncad morphlinos (D).
Discussion
The protocadherins comprise the largest subgroup within the cadherin superfamily, yet their cellular and developmental roles are not well understood, relative to the extensively studied classical cadherins. In this study, we show that Pcdh19 and Ncad interact both functionally and physically to coordinate cell adhesion and cell movements during zebrafish neurulation.
Pcdh19 and Ncad physical and functional interactions
Our data demonstrate that Pcdh19 binds directly to Ncad and that this cis-interaction likely occurs through the extracellular domains of these proteins. Previously, Pcdh8 was shown to bind directly to Ncad in cultured hippocampal neurons (Yasuda et al., 2007) and to negatively regulate Ncad at the synapse by stimulating its endocytosis. Similarly, PAPC negatively regulates C-cadherin adhesive activity during gastrulation in Xenopus laevis (Chen and Gumbiner, 2006). Considered with these earlier studies of Pcdh8 and PAPC, our data support the idea that the ability to interact with classical cadherins may be a general feature of the protocadherin family. Thus, protocadherins could contribute to a combinatorial code that modulates cell–cell interactions through the regulation of classical cadherins. This idea is particularly intriguing with regard to the clustered protocadherins, which have been proposed to play a role in specifying synaptic connections (Wu and Maniatis, 1999; Yagi, 2003).
In contrast to Pcdh8/Ncad and PAPC/Ccad, our data indicate that Pcdh19 and Ncad are not antagonistic but act cooperatively. Abrogation of either Ncad or Pcdh19 function using antisense morpholinos yields very similar developmental defects, and our subthreshold morpholino experiments demonstrate that these molecules act synergistically. Collectively, these results indicate that individual protocadherin family members may either act as positive or negative regulators of their target classical cadherin.
Role of Pcdh19 and Ncad in cell adhesion
Knockdown of either Pcdh19 or Ncad abolishes calcium-dependent cell aggregation in the zebrafish cell line Zem2S, suggesting a role for Pcdh19 in cell adhesion, although Pcdh19 exhibits no intrinsic calcium-dependent adhesive activity in bead aggregation assays. These data are consistent with the idea that Pcdh19 and Ncad function in a protein–protein complex. If both Ncad and Pcdh19 independently conferred adhesive activity, knockdown of either protein alone should result in only a partial loss of cell aggregation in Zem2S cells. If a Pcdh19–Ncad complex is the functional unit, loss of either partner should be sufficient to block their adhesive activity. As Ncad does exhibit adhesive activity on its own, we hypothesize that Ncad may require cofactors, such as protocadherins, for optimal function in vivo. For instance, one possible way that Pcdh19 could contribute to Ncad-based adhesion is by competing with a negative regulator for binding to a common interaction site.
Involvement of Pcdh19 and Ncad in cell movements during neurulation
Previous studies revealed impairment in the convergence of the anterior neural plate in pcdh19 morphants (Emond et al., 2009) and ncad mutants and morphants (Lele et al., 2002; Hong and Brewster, 2006). The cellular mechanisms underlying these observations have not been reported for either molecule, although defects in radial intercalation and increased cell protrusive behavior have been observed in ncad morphants (Hong and Brewster, 2006). In this study, we provide the first detailed analysis of cell behaviors associated with the loss of either pcdh19 or ncad. In wild-type embryos, we find that cells exhibit smooth motility toward the midline. These movements are highly linear and directed, with little evidence for unproductive movement. Similarly, motion appears coordinated, as the direction of cell trajectories is highly coherent among neighboring cells. In contrast, both ncad and pcdh19 morphants exhibit highly compromised cell motility, with an increased propensity for cells to meander and a breakdown in the coordination of movement among neighboring cells. By our quantitative measurements, the loss of Pcdh19 or Ncad has a nearly indistinguishable impact on the movement of cells in the anterior neural plate of zebrafish embryos. Importantly, these quantitative effects are also observed in embryos with partial knockdown of both pcdh19 and ncad. This synergism supports the idea that these molecules cooperate to control coordinated cell movement in the anterior neural plate.
One significant difference between pcdh19 and ncad morphants occurs at the midline. In both wild-type and pcdh19 morphant embryos, cells form a dorsal seam at the midline as cells move ventrally. With the arrest of convergence in the lateral neural plate, pcdh19 morphants form a central lumen, giving the brain a Y-shaped appearance in anterior views. In contrast, ncad morphants appear defective in the ventral folding of the neural plate and formation of the dorsal seam. Therefore, cells accumulate uniformly across the width of the brain, resulting in a characteristic T- or mushroom-shaped brain. Ncad has recently been shown to play a role along with Nectin-2 in actin reorganization and apical constriction during neurulation in Xenopus (Nandadasa et al., 2009; Morita et al., 2010), suggesting that Ncad likely participates in multiple, distinct cellular processes during neurulation. It is possible that the participation of Ncad in these distinct processes requires distinct sets of molecular interactions. Although it is unlikely that all duties performed by Ncad require Pcdh19 and vice versa, our data provide evidence that Pcdh19 and Ncad interact both functionally and physically to control the movement of cells in the lateral neural plate.
Ncad is known to be an adhesion molecule but has also been shown to stimulate cell motility and is up-regulated in the epithelial–mesenchymal transition of several tumors and cell lines (Derycke and Bracke, 2004; Wheelock et al., 2008). Similarly, chicken Pcdh19 was found to interact with Nckap1, a component of the WAVE complex, an important regulator of actin dynamics (Tai et al., 2010). Thus, the in vivo phenotype of pcdh19 or ncad loss could be because of an impact on cell adhesion, cell motility, or both. A compromise in cell–cell interactions could disrupt the coherent movement of neighboring cells and the ability of cells to respond appropriately to spatial cues guiding their direction of movement. Alternatively, a failure to engage the actin cytoskeleton in a spatially appropriate manner would also disrupt the highly stereotyped and organized nature of cell movements in the neural plate. Most likely, some combination of the disruption of adhesion and actin reorganization contributes to the observed effects. Further studies will address the cellular roles of Pcdh19 and Ncad in more detail.
In this study, we established both a functional and physical link between Pcdh19 and Ncad, with both molecules playing essential roles in the morphogenesis of the anterior neural tube. In addition, quantitative in vivo observations provided the first detailed characterization of cell behaviors in response to Pcdh19 or Ncad loss and revealed strikingly similar effects on cell motility. Although classical cadherins and protocadherins would almost certainly have biological functions independent of each other, we propose that the ability to interact with and modulate the activity of classical cadherins is a general property of the protocadherin family, which would superimpose an additional level of combinatorial complexity onto that already suggested for the classical cadherins.
Materials and methods
Fish maintenance
Adult zebrafish (Danio rerio) and embryos of the Tübingen longfin and AB* strains were maintained at ∼28.5°C and staged according to Westerfield (1995). Embryos were raised in E3 embryo medium with 0.003% phenylthiourea (Sigma-Aldrich) to inhibit pigment formation.
In situ hybridization
In situ hybridization was performed using standard methods described previously (Westerfield, 1995). Antisense riboprobe, generated against the cytoplasmic domain of pcdh19 or ncad, was labeled with digoxigenin-dUTP (Roche) and added to a final concentration of 400 ng/ml. AP-conjugated anti-digoxigenin Fab fragments (Roche) were used at 1:5,000, and the in situ experiments were developed using Nitro blue tetrazolium chloride/BCIP (5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt; Roche). The probes pax2.1 (Krauss et al., 1991) and krox20 (Wilkinson et al., 1989) were also used. Two-color fluorescence in situ hybridization was performed according to the manufacturer’s instructions (Invitrogen) using fluorescein-labeled pcdh19 and digoxigenin-labeled ncad riboprobes.
Morpholino and DNA injections
Antisense morpholino oligonucleotides were purchased from Gene Tools, LLC. The morpholinos were dissolved in dH2O at ∼8 ng/nl, aliquoted, and stored at −80°C. For use, morpholinos were diluted to a working concentration of 0.2–4 ng/nl in dH2O, and 1 nl was injected into one-cell stage embryos. The following doses were used in this study: full, effective dose of pcdh19MO, 2.5 ng; full, effective dose of ncadMO, 1.5 ng; subthreshold dose of pcdh19MO, 0.5 ng; subthreshold dose of ncadMO, 0.2 ng. Both functional morpholinos and control morpholinos directed against Ncad and Pcdh19 have been described previously (Lele et al., 2002; Emond et al., 2009). Both morpholinos (pcdh19MO and ncadMO) have substantial overlap with the 5′ untranslated regions of the respective genes, which is not present in the expression plasmids, and do not affect expression of Pcdh19-GFP or Ncad-GFP, respectively.
For rescue experiments, full doses of morpholino (2.5 ng pcd19MO or 1.5 ng ncadMO) were coinjected with 25 pg hsp70:pcdh19-gfp or 25 pg hsp70:ncad-gfp. Embryos were allowed to develop to tailbud stage and were then divided into two groups, one group was heat shocked at 37°C for 1 h, and the other was a control. Both groups were allowed to develop to the five-somite stage, fixed in 4% PFA in PBS, and processed for in situ hybridization.
For in vivo fluorescence complementation experiments, 10 pg each of two heat shock plasmids, one expressing a pcdh19 fusion to one-half of venus and the other expressing an ncad function to the other half of venus (hsp70:pcdh19-venusC and hsp70:ncad-venusN). Embryos were allowed to develop until the tailbud/one-somite stage and were heat shocked at 37°C for 1 h. Embryos were imaged on a two-photon microscope at the five-somite stage.
Zem2S cell culture, RT-PCR, and immunocytochemistry
The zebrafish embryo fibroblast cell line Zem2S (CRL-2147; American Type Culture Collection) was maintained at 28.5°C and grown in LDF medium (50% Leibovitz’s L-15 medium, 35% high-glucose DME, 15% F12 medium, 0.18 g/liter sodium bicarbonate, 15 mM Hepes, 100 U/ml penicillin, and 100 ug/ml streptomycin). Growth media were supplemented with 10% heat-inactivated fetal bovine serum 30 min after plating. To confirm expression of Ncad and Pcdh19, total RNA was isolated using Trizol (Invitrogen). First-strand synthesis was performed using reverse transcription (SuperScript III; Invitrogen) to generate cDNA, which was used as a template for PCR (Platinum Taq Polymerase High Fidelity; Invitrogen).
Two-photon time-lapse imaging and analysis
Transgenic embryos, Tg(h2afz:GFP)kca66, expressing Histone H2A–GFP (Pauls et al., 2001) were mounted in 0.5% low melting point agarose and placed on a custom-built laser-scanning two-photon microscope (Emond and Jontes, 2008; Emond et al., 2009). Excitation was provided by a Ti:Sapphire laser (Chameleon-XR; Coherent, Inc.) tuned to 920 nm. We also used a 20× NA 0.5 objective (Achroplan; Carl Zeiss, Inc.). The software for control of the microscope was provided by N. Ziv (Technion–Israel Institute of Technology, Haifa, Israel). Images were scanned with a pixel size of 0.4–0.6 µm. For long-term time-lapse imaging, 15–25 optical sections were collected (2.5–5-µm spacing) at 2-min intervals for 2–6 h. At least three time-lapse sequences were collected for each experimental condition. After the collection of time-lapse sequences, morphant embryos were confirmed by visual inspection.
Automated nucleus tracking was performed using custom-written MATLAB (The MathWorks) scripts. In brief, images were filtered using a Gaussian blur (radius = 1 pixel). A maximum intensity projection was made from two adjacent optical sections near the dorsal surface of the embryo. A template nucleus was cropped from an initial time point and used to identify the positions of nuclei by cross-correlation at each time point. To assemble trajectories from the ordered lists of nuclei positions, we modified the track MATLAB script of D. Blair and E. Dufresne (Georgetown University, Washington, D.C.), which is an implementation of the particle-tracking algorithm of Crocker and Grier (1996). Trajectories shorter than four time intervals were excluded from further analysis.
Quantitative analysis of cell trajectories was performed using custom MATLAB scripts. For each experimental condition, measurements were pooled from time-lapse sequences collected from at least three embryos. To measure instantaneous velocities, the distances between successive nuclei positions were divided by the time interval. The mean velocity was calculated by averaging all individual cell velocities over the first 10 time intervals.
To provide a measure of the directedness of cell trajectories, the net distance moved by each cell was determined, as was the sum of each individual step comprising the track. The ratio of the net distance to the summed distance (the directedness) can vary from 0 to 1 and provides a measure of the linearity of cell migration.
The coherence of neighboring cell movements was determined by defining displacement vectors for each tracked cell at each time point. Corresponding displacement vectors were calculated for all cells within a 50-µm radius of the anchor cell (2–3 cell radius). The inner product was used to calculate cos(Θ): cos(Θ) = (a1 × b1 + a2 × b2)/|a|×|b|. A value of 1 indicates that the vectors are parallel, and a value of −1 indicates that the vectors are antiparallel. Statistical significance was assessed using the Komolgorov-Smirnov goodness of fit test in Igor Pro (WaveMetrics).
Generation of Ncad and Pcdh19 constructs
The coding regions of Pcdh19 and Pcdh19ΔCP were subcloned into pEGFP-N1 (Takara Bio Inc.) using XhoI and BamHI restriction sites. Both the Ncad-GFP and NcadΔCP-GFP fusions have been described previously (Jontes et al., 2004). Myc-tagged versions of each fusion protein were generated by replacing GFP with a short myc sequence designed to be compatible with the N1 reading frame.
Human CD4 (provided by C. Gu , Ohio State University, Columbus, OH) was amplified by PCR to have an XhoI site at the 5′ end and an EcoRI site at the 3′ end. The cytoplasmic domains of Pcdh19 and Ncad were amplified to have 5′ EcoRI sites and 3′ BamHI and AgeI sites, respectively. These were subcloned behind CD4 to generate CD4-Pcdh19CP-GFP and CD4-NcadCP-GFP.
To generate Venus fusions for BiFC, both the full-length and cytoplasmic-deleted versions of Pcdh19 and Ncad were fused to an N-terminal fragment of Venus (1–172) or a C-terminal fragment of Venus (155–240). For the full-length fusions, we generated Ncad-VenN, Ncad-VenC, Pcdh19-VenN, and Pcdh19-VenC. Equivalent fusions were obtained for NcadΔCP and Pcdh19ΔCP and for CD4-Pcdh19CP and CD4-NcadCP.
To generate Fc fusions, the sequences coding the extracellular domains of pcdh19 and ncad were amplified by PCR and subcloned into a vector encoding the Fc region of human IgG (provided by W.J. Nelson, Stanford University, Palo Alto, CA). These Fc fusions contained the hinge region, resulting in dimeric forms of Ncad-Fc and Pcdh19-Fc.
Zem2S cell and bead aggregation assays
Bead aggregations were performed essentially as described previously (Sivasankar et al., 2009). Pcdh19-Fc and Ncad-Fc were independently transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 h, the growth medium was replaced with serum-free medium. After an additional 48 h, the culture medium containing the secreted Fc fusion proteins was collected. The media were concentrated using Ultracel (Millipore) and incubated with 5 µl protein A Dynabeads (Invitrogen) for 2 h with gentle agitation at 4°C. The beads were washed extensively in binding buffer (50 mM Tris, 100 mM NaCl, 10 mM KCl, and 0.2% BSA, pH 7.4) and sonicated. The beads were split into two tubes, and either 2 mM EDTA or 2 mM CaCl2 was added to each tube. Beads were allowed to aggregate with gentle rocking for 1 h. To document bead aggregation, 5–10-µl drops were spotted in glass depression slides, and images were collected on a microscope (Axiostar) using a 10× objective. Assays were performed three times using three independent protein preps.
Zem2S cell aggregation experiments were performed essentially as described previously (Takeichi, 1977). Zem2S cells were scrape loaded in the absence or presence of 20 µM morpholinos according to manufacturer’s instructions (Gene Tools, LLC). The cells were replated and allowed to grow overnight. Scraped cells were trypsinized in the presence of 2 mM CaCl2, pelleted, and resuspended in HCMF buffer (137 mM NaCl, 5.4 mM KCl, 0.63 mM Na2HPO4, 5.5 mM glucose, and 10 mM Hepes, pH 7.4). For each condition, cells were split into two 2% agar-coated wells in a 6-well plate, and either 2 mM EDTA or 2 mM CaCl2 was added. Cells were shaken for 1 h at 80 rpm at 28.5°C, and images of random fields were collected on a microscope (Axiostar) using a 10× objective.
Quantification of cell or bead aggregation was performed using ImageJ (National Institutes of Health). In brief, images were thresholded, and the size of aggregates was approximated as the area of detected particles in units of pixels. The distribution of aggregate sizes was determined and plotted with Igor Pro. Zem2S cell aggregation experiments were performed three independent times.
CoIP and Western blotting
COS cells were transiently transfected with cDNAs encoding GFP- or myc-tagged Ncad and Pcdh19 (full-length or mutants) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 h, cells were scraped into PBS, pelleted, and lysed on ice in cell lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 0.5% Triton X-100, 1 mM PMSF, and Complete protease inhibitor cocktail [Roche]) and microcentrifuged at 4°C for 10 min. Supernatants were incubated with 2 µg anti-GFP (Invitrogen) or anti-myc (Sigma-Aldrich) primary antibody for 2 h at 4°C before the addition of protein A–Sepharose (GE Healthcare), and the coIP was allowed to incubate overnight at 4°C. The beads were washed five times in cell lysis buffer, resuspended in loading buffer, and boiled for 5 min. Samples were loaded onto 10% Bis-Tris NuPAGE gels (Invitrogen) and subjected to electrophoresis. Proteins were then transferred (Bio-Rad Laboratories) to nitrocellulose (GE Healthcare), blocked with 5% nonfat milk in TBST, and incubated overnight with primary antibody (anti-GFP, 1:1,000; anti-myc, 1:5,000). HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:5,000, and the chemiluminescent signal was amplified using Western Lightning Ultra (PerkinElmer). Blots were imaged on a molecular imaging system (Omega 12iC; UltraLum, Inc.). For coIP of endogenous protein from zebrafish embryos or Zem2S cells, lysates were prepared by homogenizing five-somite embryos or by scraping cultured cells in cell lysis buffer. The immunoprecipitations were performed using anti-Pcdh19 primary antibody, and the Western blots were performed using anti–pan-cadherin primary antibody (Cell Signaling Technology).
Online supplemental material
Fig. S1 shows that Zem2S cells express both pcdh19 and ncad. Fig. S2 shows the expression of Pcdh19EC-Fc and NcadEC-Fc in HEK293 cells, which was secreted into the culture medium and purified on magnetic protein A beads. Video 1 shows convergence movements in a control transgenic embryo expressing Histone H2A–GFP to label nuclei. Video 2 shows cell movements in an embryo injected with morpholinos directed against pcdh19. Video 3 shows cell movements in a Histone H2A–GFP transgenic embryo that was injected with morpholinos against ncad. Video 4 shows cell movements in a Histone H2A–GFP transgenic embryo that was coinjected with subthreshold doses of morpholinos directed against both pcdh19 and ncad. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007008/DC1.
Acknowledgments
We would like to thank James Nelson for the human IgG Fc, Chen Gu for the human CD4, and Christine Beattie for comments on the manuscript.
This work was supported by the National Science Foundation (grant ARRA/IOS 0920357 to J.D. Jontes), a neurosciences core grant (P30-NS045758), and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences (to J.D. Jontes).
Footnotes
Abbreviations used in this paper:
- BiFC
- bimolecular fluorescence complementation
- coIP
- coimmunoprecipitation
- hpf
- h postfertilization
- Ncad
- N-cadherin
- PAPC
- paraxial protocadherin
References
- Aamar E., Dawid I.B. 2008. Protocadherin-18a has a role in cell adhesion, behavior and migration in zebrafish development. Dev. Biol. 318:335–346 10.1016/j.ydbio.2008.03.040 [DOI] [PubMed] [Google Scholar]
- Chen X., Gumbiner B.M. 2006. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J. Cell Biol. 174:301–313 10.1083/jcb.200602062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Koh E., Yoder M., Gumbiner B.M. 2009. A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS One. 4:e8411 10.1371/journal.pone.0008411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collodi P., Kamei Y., Ernst T., Miranda C., Buhler D.R., Barnes D.W. 1992. Culture of cells from zebrafish (Brachydanio rerio) embryo and adult tissues. Cell Biol. Toxicol. 8:43–61 10.1007/BF00119294 [DOI] [PubMed] [Google Scholar]
- Crocker J.C., Grier D.G. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Depienne C., Bouteiller D., Keren B., Cheuret E., Poirier K., Trouillard O., Benyahia B., Quelin C., Carpentier W., Julia S., et al. 2009. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 5:e1000381 10.1371/journal.pgen.1000381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derycke L.D.M., Bracke M.E. 2004. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int. J. Dev. Biol. 48:463–476 10.1387/ijdb.041793ld [DOI] [PubMed] [Google Scholar]
- Dibbens L.M., Tarpey P.S., Hynes K., Bayly M.A., Scheffer I.E., Smith R., Bomar J., Sutton E., Vandeleur L., Shoubridge C., et al. 2008. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 40:776–781 10.1038/ng.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond M.R., Jontes J.D. 2008. Inhibition of protocadherin-alpha function results in neuronal death in the developing zebrafish. Dev. Biol. 321:175–187 10.1016/j.ydbio.2008.06.011 [DOI] [PubMed] [Google Scholar]
- Emond M.R., Biswas S., Jontes J.D. 2009. Protocadherin-19 is essential for early steps in brain morphogenesis. Dev. Biol. 334:72–83 10.1016/j.ydbio.2009.07.008 [DOI] [PubMed] [Google Scholar]
- England S.J., Blanchard G.B., Mahadevan L., Adams R.J. 2006. A dynamic fate map of the forebrain shows how vertebrate eyes form and explains two causes of cyclopia. Development. 133:4613–4617 10.1242/dev.02678 [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B., Reugels A.M., Pauls S., Campos-Ortega J.A. 2003. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development. 130:3767–3780 10.1242/dev.00603 [DOI] [PubMed] [Google Scholar]
- Ghosh C., Collodi P. 1994. Culture of cells from zebrafish (Brachydanio rerio) blastula-stage embryos. Cytotechnology. 14:21–26 10.1007/BF00772192 [DOI] [PubMed] [Google Scholar]
- Hong E., Brewster R. 2006. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 133:3895–3905 10.1242/dev.02560 [DOI] [PubMed] [Google Scholar]
- Hulpiau P., van Roy F. 2009. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 41:349–369 10.1016/j.biocel.2008.09.027 [DOI] [PubMed] [Google Scholar]
- Jontes J.D., Emond M.R., Smith S.J. 2004. In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J. Neurosci. 24:9027–9034 10.1523/JNEUROSCI.5399-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449–456 10.1038/nrm1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37:465–487 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Yamamoto A., Bouwmeester T., Agius E., Robertis E.M. 1998. The role of paraxial protocadherin in selective adhesion and cell movements of the mesoderm during Xenopus gastrulation. Development. 125:4681–4690 [DOI] [PubMed] [Google Scholar]
- Kim S.H., Jen W.C., De Robertis E.M., Kintner C. 2000. The protocadherin PAPC establishes segmental boundaries during somitogenesis in xenopus embryos. Curr. Biol. 10:821–830 10.1016/S0960-9822(00)00580-7 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. 1991. Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature. 353:267–270 10.1038/353267a0 [DOI] [PubMed] [Google Scholar]
- Lele Z., Folchert A., Concha M., Rauch G.J., Geisler R., Rosa F., Wilson S.W., Hammerschmidt M., Bally-Cuif L. 2002. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development. 129:3281–3294 [DOI] [PubMed] [Google Scholar]
- Marini C., Mei D., Parmeggiani L., Norci V., Calado E., Ferrari A., Moreira A., Pisano T., Specchio N., Vigevano F., et al. 2010. Protocadherin 19 mutations in girls with infantile-onset epilepsy. Neurology. 75:646–653 10.1212/WNL.0b013e3181ed9e67 [DOI] [PubMed] [Google Scholar]
- McMahon A., Supatto W., Fraser S.E., Stathopoulos A. 2008. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science. 322:1546–1550 10.1126/science.1167094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina A., Swain R.K., Kuerner K.M., Steinbeisser H. 2004. Xenopus paraxial protocadherin has signaling functions and is involved in tissue separation. EMBO J. 23:3249–3258 10.1038/sj.emboj.7600329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H., Nandadasa S., Yamamoto T.S., Terasaka-Iioka C., Wylie C., Ueno N. 2010. Nectin-2 and N-cadherin interact through extracellular domains and induce apical accumulation of F-actin in apical constriction of Xenopus neural tube morphogenesis. Development. 137:1315–1325 10.1242/dev.043190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S., Tao Q., Menon N.R., Heasman J., Wylie C. 2009. N- and E-cadherins in Xenopus are specifically required in the neural and non-neural ectoderm, respectively, for F-actin assembly and morphogenetic movements. Development. 136:1327–1338 10.1242/dev.031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F., Kools P., van Roy F. 2000. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 299:551–572 10.1006/jmbi.2000.3777 [DOI] [PubMed] [Google Scholar]
- Pauls S., Geldmacher-Voss B., Campos-Ortega J.A. 2001. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev. Genes Evol. 211:603–610 10.1007/s00427-001-0196-x [DOI] [PubMed] [Google Scholar]
- Redies C. 2000. Cadherins in the central nervous system. Prog. Neurobiol. 61:611–648 10.1016/S0301-0082(99)00070-2 [DOI] [PubMed] [Google Scholar]
- Sivasankar S., Zhang Y., Nelson W.J., Chu S. 2009. Characterizing the initial encounter complex in cadherin adhesion. Structure. 17:1075–1081 10.1016/j.str.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai K., Kubota M., Shiono K., Tokutsu H., Suzuki S.T. 2010. Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 1344:13–24 10.1016/j.brainres.2010.04.065 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 1977. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 75:464–474 10.1083/jcb.75.2.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 1988. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 102:639–655 [DOI] [PubMed] [Google Scholar]
- Unterseher F., Hefele J.A., Giehl K., De Robertis E.M., Wedlich D., Schambony A. 2004. Paraxial protocadherin coordinates cell polarity during convergent extension via Rho A and JNK. EMBO J. 23:3259–3269 10.1038/sj.emboj.7600332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Janicki P., Köster I., Berger C.D., Wenzl C., Grosshans J., Steinbeisser H. 2008. Xenopus paraxial protocadherin regulates morphogenesis by antagonizing Sprouty. Genes Dev. 22:878–883 10.1101/gad.452908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. 1995. The Zebrafish Book: a guide for the laboratory use of zebrafish (Brachydanio rerio). University of Oregon Press, Eugene, OR. 300 pp [Google Scholar]
- Wheelock M.J., Shintani Y., Maeda M., Fukumoto Y., Johnson K.R. 2008. Cadherin switching. J. Cell Sci. 121:727–735 10.1242/jcs.000455 [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G., Bhatt S., Chavrier P., Bravo R., Charnay P. 1989. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature. 337:461–464 10.1038/337461a0 [DOI] [PubMed] [Google Scholar]
- Wu Q., Maniatis T. 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 97:779–790 10.1016/S0092-8674(00)80789-8 [DOI] [PubMed] [Google Scholar]
- Yagi T. 2003. Diversity of the cadherin-related neuronal receptor/protocadherin family and possible DNA rearrangement in the brain. Genes Cells. 8:1–8 10.1046/j.1365-2443.2003.00614.x [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Amacher S.L., Kim S.H., Geissert D., Kimmel C.B., De Robertis E.M. 1998. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 125:3389–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Tanaka H., Sugiura H., Okamura K., Sakaguchi T., Tran U., Takemiya T., Mizoguchi A., Yagita Y., Sakurai T., et al. 2007. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 56:456–471 10.1016/j.neuron.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]