In the absence of nuclear pore components, Aurora B delays abscission to ensure that daughter cells separate only when pores are fully formed.
Abstract
Correct assembly of nuclear pore complexes (NPCs), which directly and indirectly control nuclear environment and architecture, is vital to genomic regulation. We previously found that nucleoporin 153 (Nup153) is required for timely progression through late mitosis. In this study, we report that disruption of Nup153 function by either small interfering RNA–mediated depletion or expression of a dominant-interfering Nup153 fragment results in dramatic mistargeting of the pore basket components Tpr and Nup50 in midbody-stage cells. We find a concomitant appearance of aberrantly localized active Aurora B and an Aurora B–dependent delay in abscission. Depletion of Nup50 is also sufficient to increase the number of midbody-stage cells and, likewise, triggers distinctive mislocalization of Aurora B. Together, our results suggest that defects in nuclear pore assembly, and specifically the basket structure, at this time of the cell cycle activate an Aurora B–mediated abscission checkpoint, thereby ensuring that daughter cells are generated only when fully formed NPCs are present.
Introduction
Cell division requires a highly coordinated series of events to precisely partition DNA between two daughter cells. Accurate capture of chromosomes by mitotic spindles is critical to faithful inheritance of DNA and is closely monitored by the spindle assembly checkpoint (Musacchio and Salmon, 2007). Cytokinesis, the final stage of cell division, must then occur coordinately with chromosome segregation to ensure that cells do not physically separate with an incorrect composition of DNA. A protein-rich structure termed the midbody stabilizes an intercellular bridge that is ultimately resolved to create two daughter cells through the process of membrane abscission (Steigemann and Gerlich, 2009). Aurora B kinase activity is essential for maintenance of midbody structure and prolonged midbody stabilization triggered by the presence of chromatin bridges, which can result from improperly segregated chromosomes (Steigemann et al., 2009). Such a delay allows time to resolve the chromatin bridge, thereby preventing aneuploidy in the daughter cells or relapse to a tetraploid state because of cleavage furrow regression. This phenomenon, termed the Aurora B–mediated abscission checkpoint, acts as a final quality control mechanism to ensure that cells do not divide aberrantly (Chen and Doxsey, 2009). Many other organelle segregation and reassembly steps are important for establishing normal function in the newly formed daughter cells. However, thus far, a link between errors in such processes and activation of the Aurora B abscission checkpoint has not been identified.
Within a matter of minutes after anaphase onset, membranes and nucleoporins (Nups) are recruited in concert to the surface of decondensing chromatin and rapidly form two concentric membrane bilayers that enclose the DNA and contain nuclear pore complexes (NPCs; Dultz et al., 2008). NPCs are large, eightfold symmetric macromolecular complexes comprised of ∼30 different proteins, each protein present in multiple copies (D’Angelo and Hetzer, 2008). When completely assembled, NPC architecture includes a central core region spanning the nuclear envelope, filaments emanating into the cytoplasm, and fibers extending ∼50 nm into the nucleoplasm, where they connect to a distal ring to form what is known as the NPC basket (D’Angelo and Hetzer, 2008). NPCs serve both as the gateway for nucleocytoplasmic communication and also as an integral feature of nuclear architecture, helping to regulate chromatin structure and function (Strambio-De-Castillia et al., 2010). We previously obtained the surprising result that Nup153, a key component of the NPC basket (Hase and Cordes, 2003; Ball and Ullman, 2005), is required for timely completion of cytokinesis (Mackay et al., 2009). By further probing this late mitotic function, we have discovered a new link between rapid rebuilding of NPC architecture at the end of mitosis and Aurora B–mediated control over abscission timing.
Results and discussion
Because Nup153 has been shown to be important for NPC basket assembly (Hase and Cordes, 2003) and because its disruption leads to a delay in late mitotic progression (Mackay et al., 2009; Lussi et al., 2010), we looked closely at NPC assembly in midbody-stage cells after reduction of Nup153 protein levels. To do so, we chose knockdown conditions in which low levels of Nup153 remain and global transport function of the NPC is intact (Mackay et al., 2009). At this level of Nup153 depletion (Fig. S1), recruitment of Tpr, a major component of the NPC basket (Krull et al., 2004), was strikingly impaired in midbody-stage cells, with little to no incorporation of Tpr at the newly forming nuclear rim and aberrant localization to cytoplasmic foci (Fig. 1, A and C; and Fig. S1 B). Nup50/Npap60, a dynamic component of the NPC basket (Guan et al., 2000; Rabut et al., 2004), similarly displayed a dramatic NPC recruitment defect in midbody-stage cells depleted of Nup153 (Fig. 1, B and C; and Fig. S1 C). We observed reduced recruitment of both Tpr and Nup50 to the nuclear rim in interphase cells no longer linked by midbodies, which is consistent with a previous study (Hase and Cordes, 2003), but no corresponding mislocalization to cytoplasmic puncta (Fig. 1, A and B).
Figure 1.
Nup153 depletion leads to distinctive defects in nuclear pore basket assembly in midbody-stage cells. (A and B) HeLa cells transfected with either control or Nup153-specific siRNA oligos were analyzed by immunofluorescence using antibodies directed against the NPC basket components Tpr (A) and Nup50 (B). α-Tubulin was used to identify midbody-stage cells (brackets). (C) Quantification of the prevalence of the mislocalized Nup phenotypes in the cell population (Ø indicates a mean of 0%; n = 3). (D) Montages of time-lapse imaging illustrating the recruitment of POM121-3GFP to chromatin (H2B-mCherry) immediately after anaphase onset (t = 0:00) in siControl or siNup153-1–treated cells. (E) Quantification of POM121-3GFP recruitment to chromatin shows similar kinetics between control and Nup153-depleted cells (n = 3). (F–H) siRNA-treated cells stained for the central NPC component Nup133 (F), the NPC basket–associated protein Mad1 (G), and Lamin A/C (H). Error bars indicate mean ± SD. Bars, 10 µm.
Nup153 translocates to chromatin early during nuclear reassembly (Bodoor et al., 1999; Dultz et al., 2008), raising the question of whether its depletion might result in a global defect in either nuclear envelope formation or NPC assembly. To address these possibilities, we first analyzed recruitment of the integral membrane Nup, POM121, to chromatin in newly forming nuclei by live imaging (Fig. 1 D). Quantification of POM121-GFP fluorescence at the chromatin surface indicated that the kinetics of membrane recruitment and the NPC component POM121 in particular are not affected by reduced levels of Nup153 (Fig. 1 E). Incorporation of Nup133, a central NPC component (Strambio-De-Castillia et al., 2010), was also not noticeably affected in either interphase- or midbody-stage cells depleted of Nup153 (Fig. 1 F). Together, these results indicate that NPC basket assembly is particularly sensitive to reduced levels of Nup153 in midbody-stage cells, resulting in diversion of basket components to ectopic sites. Eventually, mislocalized NPC basket components may turn over or accumulate slowly at the NPC basket along with residual Nup153. Sensitivity of pore basket structure to perturbation during late stages of cell division may be because of differential requirements for efficient postmitotic and interphase NPC assembly (Doucet et al., 2010).
In addition to serving as a docking site for transport factors, the NPC basket also provides a scaffold required for binding and sequestration of the spindle checkpoint regulatory proteins Mad1 and Mad2 (Lee et al., 2008) and for anchoring the nuclear lamina to the NPC (Smythe et al., 2000; Walther et al., 2001). Incorporation of both Mad1 and Lamin A at the nuclear rim was reduced in midbody-stage cells depleted of Nup153, and these proteins instead displayed cytoplasmic mislocalization similar to that seen for Tpr and Nup50 (Fig. 1, G and H). Although multiple basket components and associated factors are found in cytoplasmic foci in midbody-stage cells upon Nup153 depletion, these sites do not represent a single entity nor do they represent fully assembled NPCs in the cytoplasm. Foci containing Tpr and Mad1 often colocalized (Fig. S2 A), which is consistent with findings that Tpr directly binds Mad1 (Lee et al., 2008); however, those containing Mad1 (and Tpr) were never found overlapping with either Nup50 or Lamin A/C (Fig. S2 B and not depicted). Other Nups, such as Nup133 (Fig. 1 F), were not detected in the cytoplasm. Most importantly, mislocalization of basket Nups and associated factors reflects a disruption of organization normally established at this stage of nuclear reassembly.
Next, we further characterized the late mitotic delay seen upon Nup153 depletion (Mackay et al., 2009; Lussi et al., 2010) by following the kinetics of midbody persistence. HeLa cells stably expressing both GFP-tubulin and H2B-mCherry were treated with either control or Nup153-specific siRNAs for 48 h and then analyzed by live imaging (Fig. 2 A). Although the midbody remained visible for a median time of 1.5 h in control cells, Nup153-depleted cells exhibited a profound delay (median time of 3.25–4 h) in midbody disassembly (Fig. 2, A and B). Therefore, either Nup153 is directly involved in the process of midbody disassembly or it impacts the regulation of cytokinesis progression. To distinguish between these two possibilities, we inhibited the activity of Aurora B, a kinase known to regulate a checkpoint for the timing of abscission (Steigemann et al., 2009). The number of midbody-stage cells dramatically decreased within 60 min in both control and Nup153-depleted cells after the addition of the Aurora B kinase inhibitor ZM447439 (Fig. 2, C and D). The reduced number of Nup153-depleted cells at midbody stage after treatment with inhibitor does not correspond to a similar increase in multinucleate cells (Fig. 2 D), indicating that these cells progressed through abscission. This supports the notion that, rather than having a direct role in the process of abscission, Nup153 function influences the active state of Aurora B specifically at this time of cell division.
Figure 2.
Nup153 depletion leads to an Aurora B–dependent delay in abscission. (A) Montages illustrating the prolonged timing of midbody persistence in Nup153-depleted cells expressing GFP-tubulin and H2B-mCherry, where the first frame in which a fully formed midbody was observed (t = 0:00; closed arrows) until the frame in which the midbody was no longer present (open arrows) was tracked. Bars, 10 µm. (B) Quantification of midbody persistence. Black bars indicate the median time of midbody persistence in cells from two to three independent experiments (siControl = 1.5 h, 238 cells analyzed; siNup153-1 = 3.25 h, 220 cells analyzed; siNup153-2 = 4 h, 87 cells analyzed). (C) Control or Nup153-depleted HeLa cells were briefly treated with the Aurora B inhibitor ZM447439 or DMSO and analyzed after 60 min for the presence of midbody cells (arrowheads) by detection of α-tubulin. Bar, 20 µm. (D) Quantification of midbody-stage cells and cells with more than one nucleus present after treatment with ZM447439 at the indicated time points (n = 3). Error bars indicate mean ± SD.
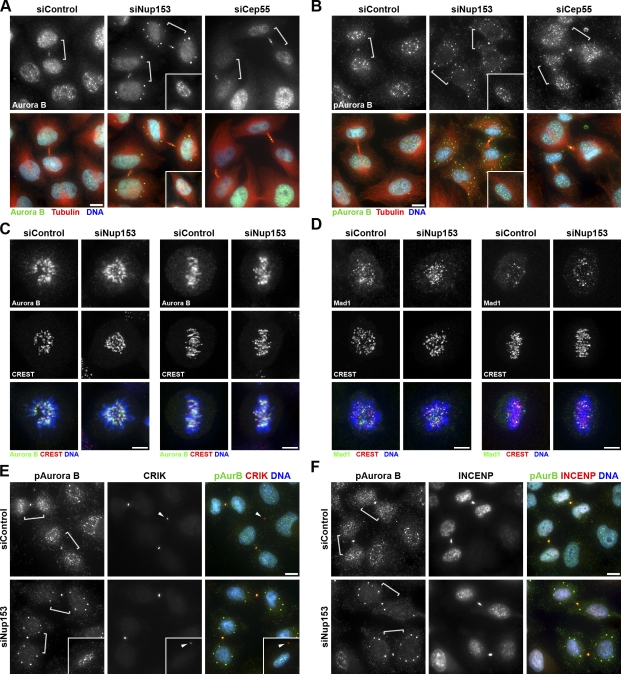
A hallmark of the abscission checkpoint is the sustained presence of active Aurora B at the midbody (Steigemann et al., 2009). Both control and Nup153-depleted cells displayed normal Aurora B staining at the midbody; however, midbody-stage cells depleted of Nup153 showed additional Aurora B localizing to cytoplasmic foci (Fig. 3 A). A phospho-specific antibody that recognizes the active form of Aurora B–phosphorylated at T232 (pAurora B) also decorated cytoplasmic puncta (Fig. 3 B), indicating not only that Aurora B is mislocalized, but also that the active form of Aurora B is stabilized within the cytoplasm. Notably, these foci were distinct from those containing Mad1 (and Tpr) or Lamin A/C (Fig. S2, C and D). Aurora B foci formation is not a general consequence of delayed abscission, as cells depleted of Cep55 showed an increased number of midbody-stage cells as expected (Fabbro et al., 2005) but maintained normal Aurora B localization patterns (Fig. 3, A and B).
Figure 3.
Aurora B is mislocalized and aberrantly active in midbody-stage cells depleted of Nup153. (A and B) siRNA-treated cells were stained for Aurora B (A) and pAurora B (B). Note that Aurora B (and pAurora B) mislocalizes in cytoplasmic foci upon Nup153 depletion. Insets show normal Aurora B localization in interphase cells. (C and D) Localization of Aurora B (C) and the spindle checkpoint protein Mad1 (D) during prometaphase (left) and metaphase (right). CREST antiserum was used to indicate centromeres. (E) Detection of pAurora B and CRIK illustrates that mislocalized pAurora B foci are neither ectopic nor residual midbodies (arrowheads). Insets show a cell with what is likely a residual midbody. (F) Cells stained for pAurora B and INCENP. Brackets indicate midbody-stage cells. Bars: (A, B, E, and F) 10 µm; (C and D) 5 µm.
In addition to being critical for cytokinesis, Aurora B activity is also required during mitotic spindle formation, where it functions in the correction of erroneous kinetochore–microtubule attachments, thus guarding against improper chromosome segregation (Ditchfield et al., 2003; Hauf et al., 2003). Interestingly, the Nup107–160 complex was recently implicated in recruiting Aurora B to centromeres associated with aligned kinetochores (Platani et al., 2009). However, in Nup153-depleted cells, Aurora B localization was comparable with control cells during both prometaphase and metaphase (Fig. 3 C), indicating that the influence of Nup153 on Aurora B is restricted temporally to abscission. Although Mad1 localization is affected in midbody-stage cells (Fig. 1 F), Mad1 kinetochore localization was unchanged in early mitosis after Nup153 depletion (Fig. 3 D), suggesting that these conditions do not impair or hyperactivate the spindle assembly checkpoint. Furthermore, we did not observe an increase in hallmarks of chromosome missegregation, such as the presence of chromatin bridges or multilobed nuclei. Thus, mislocalized Aurora B and delayed abscission observed under these conditions of Nup153 depletion are unlikely caused by disruption of events earlier in mitosis.
To address whether Aurora B foci seen upon Nup153 depletion represent ectopic midbodies in the cytoplasm, we tracked the midbody-associated protein citron Rho-interacting kinase (CRIK; Madaule et al., 1998). CRIK colocalized with pAurora B at the midbody, as expected, but was not found in pAurora B foci (Fig. 3 E). Furthermore, pAurora B did not localize to what are likely residual midbodies (Fig. 3 E, arrowheads) in recently divided cells. pAurora B is typically present in conjunction with the chromosomal passenger complex, a complex consisting of Aurora B, INCENP, Survivin, and Borealin (Ruchaud et al., 2007). Surprisingly, although INCENP was robustly detected at midbodies upon depletion of Nup153, it was not present in the cytoplasmic Aurora B foci (Fig. 3 F). Thus, stabilization of pAurora B triggered by depletion of Nup153 has a unique signature (the appearance of cytoplasmic foci and a population uncoupled from the chromosomal passenger complex) that distinguishes it from that seen upon chromosome missegregation (Steigemann et al., 2009). However, in both cases, abscission is delayed in an Aurora B–dependent manner.
To identify the Nup153 domain requirements for rescue of the mislocalized Aurora B phenotype, we expressed different fragments of Nup153 fused to GFP in cells depleted of Nup153 (Fig. 4 A; Mackay et al., 2009). Consistent with our previous data, only full-length or N + C constructs rescued the number of midbody-stage cells (Fig. 4 B; Mackay et al., 2009). Mislocalized Aurora B was similarly rescued under the same conditions (Fig. 4 C). Thus, the increase in midbody-containing cells upon Nup153 depletion tightly correlates with formation of Aurora B foci. Moreover, Nup153 function at this time in the cell cycle requires simultaneous and perhaps cooperative function of the N- and C-terminal domains.
Figure 4.
Rescue of both delayed abscission and aberrant Aurora B localization in midbody-stage cells requires the same domains of Nup153. (A) Schematic drawing of the domain structure of Nup153, including the N terminus (N), zinc finger region (Z), and FG-rich C terminus (C), and the different constructs used to determine the domains of Nup153 required to rescue function after Nup153 depletion. (B) Rescue of midbody accumulation after depletion of endogenous Nup153 is seen with expression of full-length and N + C constructs. Brackets indicate a nonsignificant difference between control and Nup153 siRNA treatment (n = 3). (C) Significant reduction in Aurora B foci is seen after Nup153 depletion when cells express full-length or N + C constructs compared with GFP (*, P < 0.005; n = 3). Control siRNA-treated cells never displayed mislocalized Aurora B (Ø indicates a mean of 0%). Error bars indicate mean ± SD.
We used these observations to take an independent tactic to interfere with Nup153 mitotic function. Specifically, we made cell lines that express high levels of the Nup153 C-terminal domain (Nup153-C) or GFP alone upon tetracycline induction (Fig. S3 A). The overexpressed C-terminal fragment would be predicted to interfere with the role of Nup153 during abscission if coupling between the N- and C-terminal domains is important for this function. At the same time, other functions of Nup153 would be expected to be preserved, such as its incorporation into the NPC, which is solely dependent on the N-terminal region (Enarson et al., 1998). Overexpression of the Nup153-C fragment indeed resulted in an approximately threefold increase in the number of midbody-stage cells (Fig. 5, A and C), which is similar to that seen upon reduction of Nup153 (Fig. 1 C), indicating that Nup153-C can dominantly interfere with the late mitotic function of Nup153 and lead to delayed abscission.
Figure 5.
Induced expression of Nup153-C fragment dominantly interferes with NPC basket assembly and results in mislocalized active Aurora B in midbody-stage cells. (A) HeLa-T-REx cell lines without (−DOX) and with (+DOX) induced expression of GFP or a Nup153 C-terminal fragment were analyzed for the presence of midbody-stage cells (arrowheads). (B) Immunostaining of GFP or Nup153-C–GFP-expressing cells shows that localization of both Nup153 and Nup133 is unaffected, whereas Tpr and Nup50 mislocalize to cytoplasmic foci in midbody-stage cells expressing Nup153-C–GFP. Detection of α-tubulin (not depicted) was used to identify midbody-stage cells. (C) Quantification of the incidence of midbody-stage cells and the mislocalized Nup phenotypes within the cell population (Ø indicates a mean of 0%; n = 3). (D and E) Staining for both Aurora B (D) and pAurora B (E) indicates that expression of Nup153-C–GFP leads to mislocalized and aberrantly active Aurora B in midbody-stage cells. (F–H) HeLa cells treated with Nup50-specific siRNA oligos display normal localization of both Nup153 (F) and Tpr (G) but display increased numbers of midbody-stage cells, many of which contain mislocalized pAurora B (H). (I) Quantification of the incidence of pAurora B foci in a midbody-stage cell after either siRNA treatment or fragment expression as indicated (n = 3). (J) Model for cross talk between nuclear pore assembly and the abscission checkpoint. Under normal conditions, resolution of cytokinesis ensues after proper nuclear pore assembly and Aurora B dephosphorylation (dashed red circle). pAurora B (solid red circles) is stabilized throughout the cytoplasm, including at the midbody, when assembly of the nuclear basket is disrupted, whether by a discrete change, such as the depletion of Nup50 (purple ovals), or a more potent disruption, as results from either interfering with Nup153 function (but not localization) by dominant interference (C terminus) or depleting Nup153. Activated Aurora B delays abscission, but the specific contributions of cytoplasmic versus midbody pools of Aurora B remain to be elucidated. (A, B, and D–H) Brackets indicate midbody-stage cells. Error bars indicate mean ± SD. Bars, 10 µm.
Endogenous Nup153, as predicted, targeted normally to the NPC in both interphase and midbody-stage cells expressing the Nup153-C fragment, as did the core component Nup133 (Fig. 5 B). However, incorporation of the NPC basket components Tpr and Nup50 during late mitotic NPC assembly was disrupted in a manner similar to that observed after Nup153 depletion (Fig. 5 B). In fact, this defect in NPC basket assembly was unexpected, as the determinants for Nup153 interaction with both Tpr (Hase and Cordes, 2003) and Nup50 (Kosako et al., 2009) map to the Nup153 N-terminal region. Thus, although the C-terminal domain of Nup153 does not directly interact with these NPC basket components, our results suggest that it plays a role in their incorporation at this specific stage of the cell cycle. This also extended to both Mad1 and Lamin A/C (Fig. S3, D and E). Moreover, midbody-stage cells expressing Nup153-C displayed mislocalized and aberrantly active Aurora B (Fig. 5, D and E), supporting the notion that NPC basket defects activate an Aurora B abscission checkpoint. As a final test of whether NPC basket assembly disruption can lead to activation of an abscission checkpoint, we depleted cells of Nup50 (Fig. S3, F and G). Although this treatment results in discrete disruption of the NPC basket, with localization of Nup153 and Tpr unaffected (Fig. 5, F and G), it was accompanied by an approximately twofold increase in the number of midbody-stage cells (siControl = 5.3 ± 0.4%; siNup50 = 9.5 ± 0.3%; n = 3). Furthermore, pAurora B was found in ectopic cytoplasmic foci in 28% of midbody-stage cells (Fig. 5, H and I). Consistent with causing modest alteration of the NPC basket, Nup50 depletion elicits a less-robust phenotype than larger-scale alterations (>60% of midbody stage cells have Aurora B foci after Nup153 depletion or Nup153-C expression; Fig. 5 I). Nonetheless, the absence of Nup50 appears sufficient to similarly trigger the abscission checkpoint.
Collectively, our data indicate that an Aurora B checkpoint at cytokinesis is linked to Nup function and support a model in which the status of postmitotic NPC assembly is under surveillance (Fig. 5 J). These results point to a new mode by which Nups contribute to the orchestration of cell division and, to our knowledge, are only the second example in mammalian cells of a defect that can trigger such a checkpoint at cytokinesis. Significantly, this model suggests that mechanisms are in place to ensure not only that DNA content is accurately inherited but also that it is packaged into nuclei in which key elements, such as the NPC basket structure, are properly poised for their functional role.
Materials and methods
Cell culture, siRNA transfection, and ZM447439 treatment
HeLa cells grown in DME supplemented with 10% FBS were transfected with control or Nup153-specific siRNA oligos as described previously (Mackay et al., 2009) using Lipofectamine RNAiMAX (Invitrogen) and final concentrations of 10 nM siRNA for siControl (a scrambled version of siNup153-1) and siNup153-2 and 170 pM for siNup153-1. All Nup153 siRNA experiments, except for those in Figs. 1 (D and E) and 4 (B and C), were performed using both Nup153-specific siRNA oligos and produced essentially identical results, although results from only one siRNA oligo are depicted. Other siRNA oligos used in this study are used as described previously: Nup50 (Ogawa et al., 2010) and Cep55 (Morita et al., 2007) oligos were both used at 10 nM. Cells were harvested for analysis 48 h after siRNA transfection. For Aurora B inhibition experiments, ZM447439 was added at a final concentration of 2 µM to siRNA-treated cells 48 h after transfection. Cells were harvested after 60–120-min incubation.
Plasmids and stable cell lines
H2B-mCherry plasmid was generated by cloning an H2B-mCherry fragment in place of the EGFP fragment in pEGFP-N2. The pgk-puromycin cassette from the pRetroQ-AcGFP1-C1 plasmid (Takara Bio Inc.) was then introduced into the H2B-mCherry vector to allow double selection. The G418-resistance cassette was disrupted by restriction digestion with NaeI, and the resulting linearized vector was used for transfection in subsequent steps. Stable HeLa cell lines used in live imaging expressing rat POM121-3GFP (provided by E. Hallberg, Södertörn University College, Södertörn, Sweden; Kihlmark et al., 2001). H2B-mCherry or GFP-tubulin (Takara Bio Inc.)/H2B-mCherry were generated by sequentially transfecting the appropriate plasmids into HeLa cells using Lipofectamine LTX (Invitrogen) and selecting in 1 mg/ml G418 + 0.5 µg/ml puromycin. Stable lines used in the phenotype rescue analysis have been previously described (Mackay et al., 2009). To generate inducible stable cell lines, we used the Flp-In T-REx system (Invitrogen). In brief, GFP or Nup153-C–GFP fragments were cloned into the pcDNA5/FRT/TO plasmid, transfected into the T-REx–HeLa cell line along with the pOG44 Flp recombinase plasmid, and selected in 100 µg/ml hygromycin. Resistant cells were pooled and expanded. Protein expression was induced by addition of 0.3 µg/ml doxycycline for 24 h before analysis.
Immunofluorescence analysis
Cells were fixed for immunofluorescence analysis using one of the following methods: (a) incubation in −20°C methanol for 10 min, (b) incubation in RT methanol for either 10 or 30 min, or (c) incubation in 4% paraformaldehyde for 10 min at RT followed by 10 min in −20°C methanol. The following antibodies were used in this study: α-tubulin (YL1/2 [Accurate Chemical & Scientific Corp.] and ab18251 [Abcam]), Nup153 (SA1; provided by B. Burke, Institute of Medical Biology, Singapore), Nup153-Z (Liu et al., 2003), Tpr (IHC-00099-1; Bethyl Laboratories, Inc.), Nup50 (a custom antibody raised against a peptide, including the C-terminal 18 amino acids of Nup50), Nup133 (provided by D. Forbes, University of California, San Diego, La Jolla, CA), Mad1 (9B10; Santa Cruz Biotechnology, Inc.), Lamin A/C (ab20740; Abcam), Aurora B (3094 [Cell Signaling Technology] and ab2254 [Abcam]), Aurora B pT232 (600-401-677; Rockland), CREST (HCT-0100; Immunovision), GFP (JL8 [Takara Bio Inc.] and ab290 [Abcam]). DNA was stained with Hoechst 33258 included in the final wash. Phenotype quantification in Figs. 1 C, 2 D, 4 (B and C), and 5 (C and I) represents the mean and SD of at least three independent experiments, where >900 cells were counted per experiment.
Live cell time-lapse imaging and image analysis
Time-lapse imaging was performed essentially as described previously (Mackay et al., 2009, 2010) with a few modifications. To image POM121-3GFP recruitment to chromatin, stable cells expressing both POM121-3GFP and H2B-mCherry were plated on a 4-well chambered cover glass (LabTek), treated with siRNA for 48 h, and imaged using a 40× NA 0.9 Plan Apo objective at several stage positions every 2 min for 3 h. The microscope used (IX81; Olympus) was equipped with a stage-top incubator (OKO Laboratory), a motorized XY stage, and a camera (Orca ER; Hamamatsu Photonics) all controlled using MetaMorph software (MDS Analytical Technologies). Image analysis and intensity measurements were performed using ImageJ (National Institutes of Health). Specifically, background-subtracted images in the H2B-mCherry channel were used to set a thresholded area, which was used to measure the mean pixel intensity in the POM121-3GFP channel. Mean pixel intensities were normalized according to the maximum value for individual cells. Data points in Fig. 1 E represent the mean and standard error from three independent experiments in which ∼20 nuclei were analyzed per experiment. To image midbody persistence, cells expressing both GFP-tubulin and H2B-mCherry were treated with siRNA for 48 h and imaged at several stage positions every 15 min for 18 h. Quantification was performed manually, and statistical analysis was performed using Prism software (GraphPad Software, Inc.).
Online supplemental material
Fig. S1 shows Western blot analysis of the levels of Nup153 and other proteins analyzed in this study after Nup153 depletion by two independent siRNA oligonucleotides and that mislocalization of Tpr and Nup50 only occurs in midbody-stage cells depleted of Nup153. Fig. S2 illustrates that mislocalized Nups, Nup-associated factors, and Aurora B in cytoplasmic foci do not represent a single entity. Fig. S3 shows induced expression of GFP and Nup153-C–GFP, that mislocalization of Tpr, Nup50, Mad1, and Lamin A/C only occurs in midbody-stage cells expressing Nup153-C–GFP, and the levels of depletion of Nup50. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007124/DC1.
Acknowledgments
We thank D. Ayer, B. Burke, D. Forbes, E. Hallberg, B. Liebold, J. Rosenblatt, and W. Sundquist for reagents, C. Rodesch and the University of Utah Fluorescence Microscopy Core Facility for assistance with time-lapse imaging and analysis, J. Rosenblatt and W. Sundquist for critical reading of this manuscript, and D. Lim for help with graphic illustration.
D.R. Mackay was supported by the American Cancer Society (Michael Schmidt Postdoctoral Fellowship PF-07-103-01-CSM). Core facilities were supported in part by a grant (P30 CA042014) awarded to the Huntsman Cancer Institute. This work was funded by the National Institutes of Health (grant R01 GM61275), a scholar award from the Leukemia and Lymphoma Society, and the Huntsman Cancer Foundation (to K.S. Ullman).
Footnotes
Abbreviations used in this paper:
- CRIK
- citron Rho-interacting kinase
- NPC
- nuclear pore complex
- Nup
- nucleoporin
References
- Ball J.R., Ullman K.S. 2005. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 114:319–330 10.1007/s00412-005-0019-3 [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264 [DOI] [PubMed] [Google Scholar]
- Chen C.T., Doxsey S. 2009. A last-minute rescue of trapped chromatin. Cell. 136:397–399 10.1016/j.cell.2009.01.028 [DOI] [PubMed] [Google Scholar]
- D’Angelo M.A., Hetzer M.W. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456–466 10.1016/j.tcb.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C.M., Talamas J.A., Hetzer M.W. 2010. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 141:1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P., Enarson M., Bastos R., Burke B. 1998. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 107:228–236 10.1007/s004120050301 [DOI] [PubMed] [Google Scholar]
- Fabbro M., Zhou B.B., Takahashi M., Sarcevic B., Lal P., Graham M.E., Gabrielli B.G., Robinson P.J., Nigg E.A., Ono Y., Khanna K.K. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell. 9:477–488 10.1016/j.devcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Guan T., Kehlenbach R.H., Schirmer E.C., Kehlenbach A., Fan F., Clurman B.E., Arnheim N., Gerace L. 2000. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20:5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase M.E., Cordes V.C. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 14:1923–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihlmark M., Imreh G., Hallberg E. 2001. Sequential degradation of proteins from the nuclear envelope during apoptosis. J. Cell Sci. 114:3643–3653 [DOI] [PubMed] [Google Scholar]
- Kosako H., Yamaguchi N., Aranami C., Ushiyama M., Kose S., Imamoto N., Taniguchi H., Nishida E., Hattori S. 2009. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16:1026–1035 10.1038/nsmb.1656 [DOI] [PubMed] [Google Scholar]
- Krull S., Thyberg J., Björkroth B., Rackwitz H.R., Cordes V.C. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., McCormick F. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Prunuske A.J., Fager A.M., Ullman K.S. 2003. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev. Cell. 5:487–498 10.1016/S1534-5807(03)00262-4 [DOI] [PubMed] [Google Scholar]
- Lussi Y.C., Shumaker D.C., Shimi T., Fahrenkrog B. 2010. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 1:71–84 10.4161/nucl.1.1.10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D.R., Elgort S.W., Ullman K.S. 2009. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol. Biol. Cell. 20:1652–1660 10.1091/mbc.E08-08-0883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D.R., Ullman K.S., Rodesch C.K. 2010. Time-lapse imaging of mitosis after siRNA transfection. J. Vis. Exp. 40:1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 394:491–494 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., Sundquist W.I. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Miyamoto Y., Asally M., Oka M., Yasuda Y., Yoneda Y. 2010. Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol. Biol. Cell. 21:630–638 10.1091/mbc.E09-05-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M., Santarella-Mellwig R., Posch M., Walczak R., Swedlow J.R., Mattaj I.W. 2009. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell. 20:5260–5275 10.1091/mbc.E09-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 [DOI] [PubMed] [Google Scholar]
- Smythe C., Jenkins H.E., Hutchison C.J. 2000. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19:3918–3931 10.1093/emboj/19.15.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P., Gerlich D.W. 2009. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 19:606–616 10.1016/j.tcb.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., Gerlich D.W. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Fornerod M., Pickersgill H., Goldberg M., Allen T.D., Mattaj I.W. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]