Cdc7 phosphorylates Rad18 to integrate S phase progression with postreplication DNA repair, ensuring genome stability.
Abstract
The E3 ubiquitin ligase Rad18 guides DNA Polymerase eta (Polη) to sites of replication fork stalling and mono-ubiquitinates proliferating cell nuclear antigen (PCNA) to facilitate binding of Y family trans-lesion synthesis (TLS) DNA polymerases during TLS. However, it is unclear exactly how Rad18 is regulated in response to DNA damage and how Rad18 activity is coordinated with progression through different phases of the cell cycle. Here we identify Rad18 as a novel substrate of the essential protein kinase Cdc7 (also termed Dbf4/Drf1-dependent Cdc7 kinase [DDK]). A serine cluster in the Polη-binding motif of Rad18 is phosphorylated by DDK. Efficient association of Rad18 with Polη is dependent on DDK and is necessary for redistribution of Polη to sites of replication fork stalling. This is the first demonstration of Rad18 regulation by direct phosphorylation and provides a novel mechanism for integration of S phase progression with postreplication DNA repair to maintain genome stability.
Introduction
Appropriate replication and repair of DNA is important for maintenance of genomic stability. Encounters between replication forks and bulky DNA lesions can lead to stalling of DNA polymerases and incomplete DNA replication or vulnerability to replication fork collapse, both potential sources of genomic instability. Therefore, cells have evolved DNA damage tolerance mechanisms such as trans-lesion synthesis (TLS) for maintaining replication fork progression on damaged templates.
TLS is mediated by the action of specialized DNA polymerases (termed “TLS polymerases”). Collectively, TLS polymerases perform replicative bypass of various DNA lesions by virtue of their flexible active sites (Trincao et al., 2001). In contrast with replicative DNA polymerases, TLS polymerases have low processivity and are highly error-prone on undamaged templates. Therefore, TLS polymerase activities must be strictly regulated to prevent mutagenesis.
Mammalian TLS DNA polymerases include Y family members DNA Polymerase eta (Polη), DNA Polymerase κ (Polκ), DNA Polymerase ι (Polι), Rev1, and DNA Polymerase ζ (Polζ; comprised of the B family DNA Polymerase Rev3L and a regulatory subunit Rev7; Prakash et al., 2005). Each TLS polymerase exhibits a preference for bypass of specific types of DNA damage. For example, Polη is specialized to perform bypass of solar UV radiation–induced cis–syn Thymine–Thymine cyclobutane–pyrimidine dimers (CPDs; Masutani et al., 1999).
The importance of TLS for genome maintenance is demonstrated by individuals with a variant form of the autosomal recessive disease xeroderma pigmentosum (XPV), conferred as a result of Polη deficiency (Masutani et al., 1999). XPV patients exhibit acute sensitivity to sunlight and a high risk of developing skin malignancies, and their Polη-deficient cells are UV sensitive (Maher et al., 1976; Laposa et al., 2003), often exhibiting increased rates of sister chromatid exchange (SCE) relative to wild-type (WT) cells (Cleaver et al., 1999).
Studies with knockout mice have also demonstrated a key role for Polη in tumor suppression. After UV radiation, 100% of polh−/− mice developed epithelial tumors (Lin et al., 2006), and even the loss of a single allele of polh rendered the mice more susceptible to UV-induced tumors (Lin et al., 2006; Ohkumo et al., 2006). The molecular phenotypes of Polη deficiency most likely reflect error-prone bypass of CPD, as polh−/− cells exhibited fivefold higher rates of UV-induced mutagenesis compared with WT cells (Busuttil et al., 2008).
Mechanisms that recruit Polη and other TLS polymerases to sites of DNA damage have been studied extensively. In UV-irradiated cells, Polη redistributes to form nuclear foci that colocalize with proliferating cell nuclear antigen (PCNA) at sites of BrdU incorporation and CPD lesions (Kannouche et al., 2003). Recruitment of Polη and other TLS polymerases to stalled replication forks is mediated in part via mono-ubiquitination of PCNA (Kannouche et al., 2004). Rad18 is a highly conserved ubiquitin E3 ligase that mono-ubiquitinates PCNA in response to DNA damage (Kannouche et al., 2004). Y family polymerases possess Ub-binding domains (termed UBM and ubiquitin-binding zinc finger [UBZ] motifs), and direct interactions between mono-Ub-PCNA and TLS polymerases facilitate their recruitment to replication forks (Bienko et al., 2005). However, additional mechanisms appear to contribute to TLS polymerase recruitment at sites of DNA damage. Rad18 associates directly with Polη and guides the polymerase to sites of DNA damage (Watanabe et al., 2004). Consistent with a key role for Rad18 in TLS, Rad18 deficiency causes sensitivity to DNA-damaging agents (Yamashita et al., 2002; Tateishi et al., 2003; Nakajima et al., 2006). Despite the key roles of Rad18 in TLS, precise mechanisms by which Rad18 effects recruitment of TLS polymerases to sites of DNA damage and mechanisms integrating Rad18 activity with other elements of cell cycle control and various DNA damage responses are not fully understood.
Cdc7 is the catalytic subunit of a protein kinase that was first characterized as a temperature-sensitive mutant essential for the initiation of DNA replication in budding yeast (Hartwell, 1973; Newlon and Fangman, 1975). In higher eukaryotes, Cdc7 has two activating subunits, Dbf4 and ASKL1/Drf1 (Kumagai et al., 1999; Montagnoli et al., 2002), and is essential for initiation of DNA replication (Yamashita et al., 2005). Dbf4/Drf1-Cdc7 (also termed Dbf4/Drf1-dependent kinase [DDK]) phosphorylates components of the MCM2-7 complex and promotes loading of the initiation factor CDC45 (Masai et al., 2006).
Cdc7 mutants are hypersensitive to DNA-damaging agents (Njagi and Kilbey, 1982), which suggests a possible role for DDK in the cellular response to DNA damage. Indeed, in human cells, Dbf4 activates Cdc7, accumulates on chromatin (Tenca et al., 2007), and phosphorylates Claspin (Kim et al., 2008) in response to DNA damage. In budding yeast, Cdc7 prevents UV sensitivity and promotes UV-induced mutagenesis (Njagi and Kilbey, 1982; Kilbey, 1986). Fission yeast mutants of hsk1 and dfp1 (encoding DDK) are also defective for methyl methanesulfonate–induced mutagenesis (Dolan et al., 2010). Conversely, yeast strains engineered to express high levels of Cdc7 show hypermutability in response to UV (Sclafani et al., 1988). Therefore, genetic analyses are consistent with a role for DDK in DNA damage tolerance via TLS. Cdc7 was placed in the Rad6 epistasis group because of phenotypic similarities between cdc7 and rad6 mutants (Njagi and Kilbey, 1982).
Genetic interactions between Cdc7 and members of the TLS pathway have been described in Saccharomyces cerevisiae (Pessoa-Brandão and Sclafani, 2004). Moreover, it is suggested that BRCT domains of the TLS protein Rev1 and Dbf4 domains interact with a common protein or structure (Harkins et al., 2009). Collectively, these studies led us to hypothesize that Cdc7-Dbf4 might participate in TLS by directly phosphorylating a component of this pathway.
Here we have investigated the biochemical relationship between Cdc7 and Rad18, the central regulator of the TLS pathway. We demonstrate that Rad18 is phosphorylated in a Cdc7-mediated manner in vitro and in intact human cells. Moreover we show that Cdc7-dependent Rad18 phosphorylation promotes efficient recruitment of Polη to stalled replication forks after UV treatment. Collectively, these data support a model in which Cdc7 phosphorylation of Rad18 provides a novel mechanism for the regulation of Rad18-mediated TLS.
Results
In vitro phosphorylation of Rad18 by Cdc7
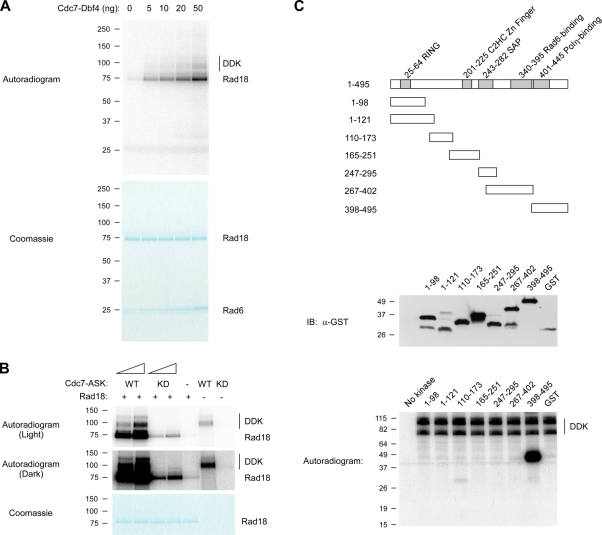
We tested full-length recombinant Rad18 (in complex with its physiological binding partner Rad6) as a potential in vitro substrate for Cdc7-Dbf4. As shown in Fig. 1 A, Rad18 was specifically phosphorylated by Cdc7-Dbf4. Under these reaction conditions, approximately one phosphate was incorporated per molecule of Rad18. This level of incorporation is quite significant and is close to half the level of phosphorylation we typically observed with Mcm2 protein, which is believed to contain multiple phosphorylation sites. Mutant kinase–inactive recombinant DDK failed to phosphorylate Rad18 (Fig. 1 B). Therefore, the Cdc7-dependent Rad18 phosphorylation we observed was not caused by contaminating kinase activities in our DDK preparations.
Figure 1.
DDK phosphorylates the C terminus of Rad18. (A) Recombinant Rad18–Rad6 complex was incubated with purified Cdc7-Dbf4 in the presence of [32P]γATP. Reaction products were resolved by SDS-PAGE and analyzed by autoradiography (top) and Coomassie staining (bottom). The Rad18 and Rad6 bands were excised from the dried gel, and the radioactivity of each band was measured by scintillation counter. The incorporation of phosphate per molecule of Rad18 protein was calculated on the basis of the specific radioactivity of [32P]γATP in the kinase reactions. (B) Recombinant Rad18–Rad6 complex was incubated with purified DDK complexes containing WT Cdc7 or a kinase-dead (KD) Cdc7 K90 → E mutant in the presence of [32P]γATP. Reaction products were analyzed as described for A. (C) The indicated regions of hRad18 were expressed as recombinant GST fusion proteins in E. coli. Expression of the GST fusion proteins was validated by immunoblotting with anti-GST. GST fusions of Rad18 were tested for in vitro phosphorylation by Cdc7-Dbf4. Molecular mass is indicated in kilodaltons next to the gel blots.
In previous studies, sequence motifs targeted by DDK have been identified via mutational analyses using recombinant bacterial GST fusion proteins as substrates (Cho et al., 2006; Sheu and Stillman, 2006). To identify the Rad18 regions phosphorylated by Cdc7, we generated a series of partially overlapping GST fusion fragments of Rad18 spanning specific domains of the Rad18 protein (indicated in Fig. 1 C). The GST-Rad18 fragments were tested as possible in vitro substrates for Cdc7-Dbf4. As shown in Fig. 1 C, recombinant Dbf4-Cdc7 specifically phosphorylated a C-terminal Rad18 fragment spanning amino acids 398–495.
The C terminus of Rad18 contains three clusters of serine residues representing potential Cdc7 phosphorylation sites: cluster 1 comprises serines 403, 405, and 409; cluster 2 comprises serines 421–423; and cluster 3 (which we designated the “S box”) comprises serines 432–434, 436, 438, and 441–444 (Fig. 2 A). To identify the specific Cdc7 phospho-acceptor sites, serines within clusters 1–3 were mutated to alanines, and mutant GST-Rad18 fragments harboring those S-to-A substitutions were tested as Cdc7 substrates. A GST-Rad18 398–495 fusion with alanine substitutions at residues 403, 405, and 409 was phosphorylated as efficiently as GST-Rad18 398–495 (WT), thereby excluding cluster 1 serines as Cdc7 targets (unpublished data). Although there is no specific consensus sequence present in known DDK substrates, Cdc7 preferentially phosphorylates clusters of serines flanked by acidic amino acids (Cho et al., 2006). The Rad18 S box contains nine serines preceding a glutamate-rich region, similar to the sequence of Mcm2, the best-characterized Cdc7 substrate (Cho et al., 2006). Therefore, we asked whether serines in the S box were necessary for Rad18 phosphorylation by Cdc7. First, we deleted the entire S box and tested the resulting protein (GST-Rad18 398–495 ΔS-Box) for in vitro phosphorylation by Cdc7. As shown in Fig. 2 B, deletion of the S box reduced phosphorylation of GST-Rad18 398–495 by 90%, which suggests that amino acids phosphorylated by Cdc7 lie within the S box.
Figure 2.
Identification of Cdc7 target residues in the C-terminal domain of Rad18. (A) Amino acid sequence of the Rad18 C terminus. A serine-rich region within the Polη-binding domain is highlighted in blue. Serines representing potential DDK targets are underlined. (B) Mcm2, GST-Rad18 398–495 (WT), and GST-Rad18 398–495 (ΔS-box) were tested as DDK substrates using in vitro kinase assays. Phosphorylated GST-Rad18 substrates and the closely migrating bands corresponding to phosphorylated Mcm2 (top) and autophosphorylated Cdc7 (bottom) are indicated. Equivalent levels of GST-Rad18 substrates in each reaction were verified by Coomassie staining of the radioactive gels (not depicted). (C) GST-Rad18 398–495 (WT) and GST-Rad18 398–495 3S → A (containing alanine substitution of serine residues 434, 436, and 444) were tested as DDK substrates using in vitro kinase assays. To verify equivalent levels of substrate between reactions, a portion of each GST-Rad18 preparation was analyzed on a separate gel using SDS-PAGE and immunoblotting with anti-GST. (D) GST-Rad18 398–495 (WT) and various derivatives containing serine or alanine substitutions were tested as DDK substrates using in vitro kinase assays. To verify equivalent levels of substrate between reactions, a portion of each GST-Rad18 preparation was analyzed by SDS-PAGE and Coomassie staining on a separate gel. In the sequences below the gel images, residues highlighted in blue represent serines within the Rad18 S box that were mutated to alanine. Residues highlighted in red represent individual S box serines that were available for in vitro phosphorylation by Cdc7 in each GST-Rad18 mutant. Although not shown in this particular experiment, a GST-Rad18 S-Box → A mutant containing only one serine at position 441 was also unable to support phosphorylation by DDK (not depicted). (E) Sequence alignment of Rad18 from various species. Underlined residues represent the S box of human Rad18. Serine residues highlighted in blue indicate potential phosphorylation sites present in human Rad18. The residues highlighted in red indicate a conserved serine corresponding to S434 of human Rad18. The dashed lines represent residues that are absent from mouse, zebrafish, and S. pombe homologues of Rad18. Molecular mass is indicated in kilodaltons next to the gel blots.
Next we performed alanine substitution of S434, S436, and S444, all of which lie within the S box and precede acidic residues, similar to sites preferentially phosphorylated by Cdc7 in Mcm2 (Cho et al., 2006). Again, the resulting GST-Rad18 398–495 (3S → A) protein was tested as a Cdc7 substrate using in vitro kinase assays. As shown in Fig. 2 C, the level of in vitro phosphorylation of GST-Rad18 398–495 (3S → A) was reduced by 60% relative to WT. Therefore, S434, S436, and S444 are likely Cdc7 targets. However, Cdc7-dependent phosphorylation of GST-Rad18 398–495 (3S → A) was not completely abrogated by mutating S434, S436, and S444, which indicates that Cdc7 phosphorylates additional sites within the S box.
To identify the serines most efficiently phosphorylated by Cdc7-Dbf4, all serines in the S box were mutated to alanine (indicated in blue in Fig. 2 D), and individual serines were sequentially added back (indicated in red in Fig. 2 D) to detect sites conferring phosphorylation. Using substrates in which only one serine was available to Cdc7, S434 was preferentially phosphorylated by Cdc7 (Fig. 2 D). GST-Rad18 398–495 (434S) was only phosphorylated ∼10% as efficiently as GST-Rad18 398–495 WT. Therefore, consistent with results of Fig. 2 C, S434 is most probably not the only residue phosphorylated by Cdc7. Similar to Mcm2, the pattern of Rad18 phosphorylation is likely to be complex, with multiple residues serving as potential Cdc7 targets. Alignment of the C terminus of hRad18 shows that S434 and many elements of the S box are highly conserved from chicken to humans (Fig. 2 E), which may suggest conservation of putative Cdc7-based Rad18 phosphorylation between some species.
Rad18 S434 is phosphorylated in intact cells
To determine whether S434 of Rad18 is phosphorylated in intact cells, we raised a phospho-specific antibody against Rad18 pS434 (described in Materials and methods). Three complementary approaches were used to validate the Rad18 pS434 phospho-specific antibody.
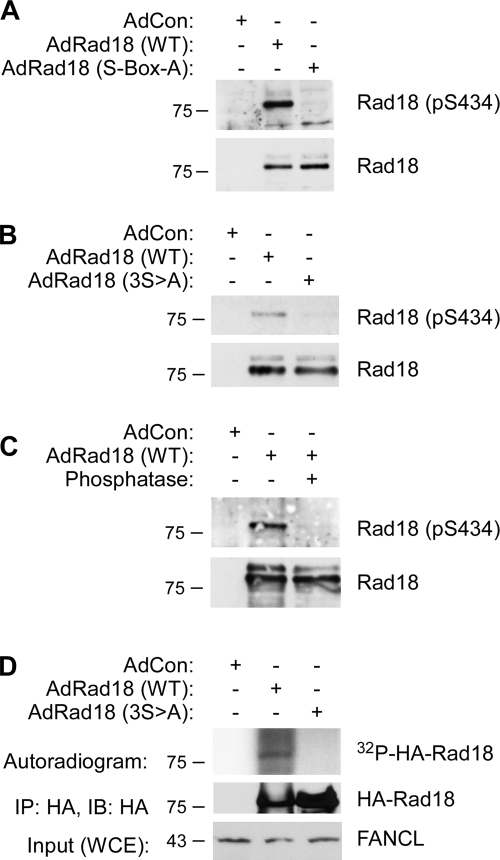
We used adenoviral vectors to ectopically express HA-tagged Rad18 in H1299 cells. The levels of Rad18 achieved using adenovirus were similar to or less than levels of endogenous Rad18 (unpublished data). Using immunoblot analysis of HA-Rad18 immunoprecipitates, we demonstrated that our Rad18 S434 phospho-specific antibody recognized Rad18 WT, but not Rad18 S-Box-A, a mutant in which all of the serines in the S box were substituted with alanines (Fig. 3 A). The phospho-specific antibody detected a Rad18 mutant containing alanine substitutions at residues S434, S436, and S438 (hereafter designated “Rad18 3S → A”) poorly relative to Rad18 WT (Fig. 3 B).
Figure 3.
Rad18 S434 is phosphorylated in intact human cells. (A) Phospho-specificity of Rad18 anti-pS434 antibody demonstrated by analysis of S box mutant Rad18. H1299 cells were infected with adenovirus vectors encoding HA-tagged Rad18 (WT), Rad18 S-Box → A (which harbors alanine substitution of all serines in the S box), or “empty” vector control (AdCon). Extracts from the resulting cells were immunoprecipitated with anti-HA, and the immune complexes were analyzed by SDS-PAGE and immunoblotting. Immunoblots were probed sequentially with anti-Rad18 (pS434) and anti-HA. As with most commonly used phospho-specific antibodies, we routinely observed minor phospho-independent reactivity of anti-Rad18 (pS434) with Rad18. (B) Phospho-specificity of Rad18 anti-pS434 antibody demonstrated by analysis of mutant Rad18. H1299 cells were infected with adenovirus vectors encoding HA-tagged Rad18 (WT), Rad18 3S → A (which harbors alanine substitution of residues 434, 436, and 444 in the S-box), or AdCon. Extracts from the resulting cells were immunoprecipitated with anti-HA and analyzed by SDS-PAGE and immunoblotting with anti-Rad18 (pS434) and anti-HA. (C) Phospho-specificity of Rad18 anti-pS434 antibody demonstrated by phosphatase treatment. H1299 cells were infected with adenovirus vectors encoding HA-tagged Rad18 (WT) or AdCon. Extracts from the resulting cells were immunoprecipitated with anti-HA. The immune complexes were incubated with λ phosphatase (or left untreated for controls) as described in Materials and methods. Control and phosphatase-treated immune complexes were analyzed by SDS-PAGE and immunoblotting with anti-Rad18 (pS434) and anti-HA. (D) The S box is necessary for phosphorylation of full-length Rad18 by DDK. RAD18−/− HCT116 cells were infected with adenovirus vectors encoding HA-tagged Rad18 (WT), Ad-Rad18 S-box-A, or AdCon. Soluble extracts from the resulting cells were immunoprecipitated with anti-HA. The immune complexes were incubated with λ phosphatase (to dephosphorylate any preexisting DDK-mediated phosphorylations) and washed extensively. 90% of each immune complex was subject to in vitro kinase assays using recombinant Cdc7-Dbf4 as described under in Materials and methods. Reaction products were resolved by SDS-PAGE, and phosphorylated species were detected by autoradiography (top). Approximately 10% of each immune complex was analyzed by SDS-PAGE and immunoblotting with anti-HA to test for relative levels of WT and S-Box-A mutant Rad18 (middle). The bottom panel shows immunoblot analysis of the soluble cell extracts probed with antibody against an irrelevant protein (FANCL) as a control for protein normalization in the “input” fractions. Molecular mass is indicated in kilodaltons next to the gel blots.
We consistently observed low-level detection of Rad18 3S → A by our Rad18 S434 phospho-specific antibody, which likely reflects phosphorylation of alternative phosphorylated serines within the S box. This result is expected because many kinases (including Cdc7) phosphorylate amino acid clusters rather than individual residues. Finally, λ phosphatase treatment of immunoprecipitated Rad18 WT eliminated the phospho-S434 signal detected by immunoblotting (Fig. 3 C). Collectively, the results of Fig. 3 validate the phospho-specificity of our Rad18 phospho-S434 antibody and indicate that phosphorylation of the Rad18 S box occurs in intact cells. We tested immunoprecipitated HA-Rad18 (WT) and HA-Rad18 (S-Box-A) as in vitro substrates for Cdc7-Dbf4. As shown in Fig. 3 D, HA-Rad18 (WT), but not HA-Rad18 (S-Box-A), was phosphorylated by Cdc7-Dbf4 in vitro, which is further consistent with phosphorylation of the Rad18 S box by DDK.
S434 phosphorylation of Rad18 in human cells is Cdc7 dependent
We investigated the Cdc7 dependence of Rad18 phosphorylation at S434. Cdc7 was depleted from H1299 cells using siRNA. As shown in Fig. 4 A, we achieved ∼75% depletion of Cdc7 after 24 h. At this time point, S phase populations of Cdc7-depleted cells were similar to control cells, as determined by BrdU labeling (Fig. 4 B). Cdc7 is an essential kinase in S phase for viability and DNA synthesis (Bousset and Diffley, 1998). Therefore, the persistence of BrdU-positive nuclei in siCdc7-transfected cultures is consistent with immunoblots (Fig. 4 A) showing that the depletion of Cdc7 achieved under our standard experimental conditions is partial. Although inhibitory effects of Cdc7 deficiency on the cell cycle of cultured mammalian cells and mouse ES cells have been reported, these are p53 dependent (Kim et al., 2002; Montagnoli et al., 2004). The H1299 cells used in our experiments lack p53 and therefore do not arrest immediately or die in response to acute and partial Cdc7 depletion. Moreover, Woodward et al. (2006) and Ge et al. (2007) have shown that 90% depletion of Mcm2-7 complex proteins (the crucial targets of Cdc7 in the context of DNA synthesis) also has no overt inhibitory effect on DNA synthesis during an unperturbed cell cycle in experimental systems from Xenopus egg extracts, Caenorhabditis elegans, and cultured mammalian cells. Therefore, the persistence of DNA synthesis after acute and partial Cdc7 depletion is fully consistent with published and generally accepted literature.
Figure 4.
DDK is important for mediating Rad18 S434 phosphorylation in intact human cells. (A) Partial knockdown of Cdc7 achieved using siRNA. Replicate plates of H1299 cells were transfected with siRNA against Cdc7 (siCdc7) or nontargeting control RNA duplexes (siCon), or were left untransfected. Cell lysates prepared 24 h and 48 h after transfection were analyzed for Cdc7 expression using SDS-PAGE and immunoblotting. A minor nonspecific band (NS) recognized by the Cdc7 antibody serves as a loading control. (B) Partial Cdc7 depletion does not affect numbers of S phase cells. Replicate plates of H1299 cells were transfected with siCdc7 or siCon RNA duplexes. 24 h and 48 h after transfection, cells were incubated with 10 µM BrdU for 1 h. Levels of BrdU incorporation were determined by anti-BrdU staining and FACS analysis. (C) Effect of partial Cdc7 or Dbf4 depletion of Rad18 S434 phosphorylation. H1299 cells were transfected with siRNA against Cdc7 or Dbf4 (or nontargeting control RNA duplexes). Extracts from the resulting cells were immunoprecipitated with anti-HA and analyzed by SDS-PAGE and immunoblotting with anti-Rad18 (pS434) and anti-HA. Input fractions were normalized for protein and analyzed by SDS-PAGE and immunoblotting with anti-Cdc7 or anti-Dbf4 antibodies to test for knockdown efficiency. Molecular mass is indicated in kilodaltons next to the gel blots.
We measured S434 phosphorylation of HA-Rad18 immunoprecipitated from Cdc7-depleted cells. As shown in Fig. 4 C, depletion of Cdc7 led to reduced phosphorylation of Rad18 at S434. Similar results were obtained with siRNA depletion of Dbf4, the noncatalytic binding partner of Cdc7 (Fig. 4 C). Numbers of BrdU-positive S phase cells in cultures acutely depleted of Cdc7 and Dbf4 were comparable (±8% in three independent experiments). Therefore, consistent with the results of our in vitro kinase assays, S434 of Rad18 is phosphorylated in a DDK-dependent manner.
Relationship between Rad18 and Cdc7 in UV-treated cells
To test the hypothesis that Cdc7 might play a role in the TLS pathway after acquisition of DNA damage, we investigated a potential relationship between Rad18 and Cdc7. After UV treatment, we observed an increase in the amount of Rad18-associated Cdc7 as determined by coimmunoprecipitation (Fig. 5 A). The association of Rad18 and Cdc7 was robust over a range of UV doses (including 100 J/m2 and 10 J/m2, as shown in Fig. 5, A and B, respectively). Therefore, UV treatment induces the formation of a complex containing Rad18 and Cdc7. In siRNA experiments, Chk1 depletion reduced both basal and UV-induced association of Rad18 with Cdc7 (Fig. 5 B).
Figure 5.
Rad18 S box phosphorylation is UV-inducible and promotes interactions with Polη. (A) H1299 cells expressing HA-Rad18 were treated with UV (100 J/m2). After 1 h, extracts from the resulting cells were immunoprecipitated with anti-HA and analyzed by SDS-PAGE and immunoblotting with anti-HA and anti-Cdc7 antibodies. (B) HA-Rad18–expressing H1299 cells were transfected with siChk1 (to ablate Chk1 expression) or with siCon for controls. 24 h after transfection, cells were treated with UV (10 J/m2), and 1 h later, extracts were immunoprecipitated and analyzed as described for A. (C) H1299 cells were infected with adenovirus vectors encoding HA-tagged Rad18 (WT) or AdCon. Cells were treated optionally with UV (100 J/m2). After 1 h, extracts from the resulting cells were immunoprecipitated with anti-HA, and the immune complexes were analyzed by SDS-PAGE and immunoblotting. Immunoblots were probed sequentially with anti-Rad18 (pS434) and anti-HA. Input fractions were probed with a Chk1 (p317) antibody. (D) H1299 cells were coinfected with adenoviruses encoding HA-Rad18 (WT) and YFP-Polη, then transfected with siRNA against Cdc7 (or with nontargeting siCon RNA duplexes). Some cultures were treated with 100 J/m2 UV as indicated. One h after UV treatment, cells were harvested and the resulting extracts were immunoprecipitated with anti-HA. Immune complexes and input fractions were analyzed by SDS-PAGE and immunoblotting with anti-GFP (to detect YFP-Polη), anti-Rad18, and anti-Cdc7, as indicated. (E) H1299 cells were infected with adenoviruses encoding HA-Rad18 (WT), HA-Rad18 S-Box-A, or HA-Rad18 S-Box-D, or with AdCon. 48 h after infection, cells were harvested and the resulting extracts were immunoprecipitated with anti-HA. Immune complexes and input fractions were analyzed by SDS-PAGE and immunoblotting with anti-Polη, anti-Rad18, and anti-Rad6 as indicated. (F) H1299 cells were infected with adenoviruses encoding HA-Rad18 (WT) or HA-Rad18 S-Box-A, or with AdCon. Some cultures were treated with 100 J/m2 UV as indicated. 1 h after UV treatment, cells were harvested and the resulting extracts were immunoprecipitated with anti-HA. Immune complexes were analyzed by SDS-PAGE and immunoblotting with anti-Polη, anti-Rad18, and anti-Rad6 as indicated. (G) HCT116 RAD18−/− cells were infected with adenoviruses encoding HA-Rad18 (WT) or HA-Rad18 S-Box-A, or with AdCon. The resulting cells were treated with UV (0, 10, and 100 J/m2) as indicated. 1 h after UV treatment, cells were harvested and the resulting cell extracts were analyzed by SDS-PAGE and immunoblotting with anti-HA and anti-PCNA as indicated. Molecular mass is indicated in kilodaltons next to the gel blots.
Because Cdc7 and Rad18 formed a complex in response to UV irradiation (Fig. 5, A and B), we asked whether phosphorylation of Rad18 at S434 was similarly genotoxin inducible.
Chk1 phosphorylation at S317 was measured as a known UV-inducible event. As shown in Fig. 5 C, Chk1 pS317 and Rad18 pS434 were induced by treatment with UV radiation. Collectively, these data suggest that phosphorylation of Rad18 at S434 is Cdc7-Dbf4 dependent, which is further consistent with Rad18 being a novel target of DDK. Furthermore, similar to Rad18–Cdc7 association, Rad18 phosphorylation at S434 occurs both basally and in a DNA damage–inducible manner.
Acute depletion of Cdc7 or mutation of the S box affects Polη binding
The C-terminal amino acid region of Rad18 participates in Polη binding (Watanabe et al., 2004). Because we demonstrated that the Polη-binding domain of Rad18 is phosphorylated by DDK, we hypothesized that this phosphorylation regulates Rad18–Polη interactions. To test this hypothesis, we assessed the effect of partial Cdc7 ablation (using siRNA) on the interaction between Rad18 and YFP-tagged Polη, as determined by coimmunoprecipitation. As shown in Fig. 5 D, Cdc7 ablation attenuated the UV-inducible interaction between Rad18 and YFP-Polη, which is consistent with the hypothesis that Cdc7 regulates association of Rad18 with Polη.
As an alternative approach to test the importance of Rad18 S box phosphorylation for Polη binding, we determined levels of Polη present in immunoprecipitated complexes of WT Rad18 or a Rad18 phosphorylation-resistant S box mutant (designated “Rad18 S-Box-A”) from H1299 cells. For comparison, we also measured association of Polη with a Rad18 mutant in which all S box serines were substituted with Asp (S-Box-D) in anticipation of a possible phosphomimetic effect. Similar amounts of Rad18 were immunoprecipitated from cells expressing Rad18 WT, Rad18 S-Box-A, and Rad18 S-Box-D (Fig. 5 E). In the absence of DNA damage, Polη was detectable in the Rad18 (WT) immune complexes, but under the same experimental conditions we did not detect Polη in Rad18 S-Box-A immunoprecipitates (Fig. 5 E). Rad18 S-Box-D substitutions did not promote increased Polη binding (Fig. 5 E). Similarly, glutamate substitution of serines in the Rad18 S box failed to confer Polη binding, which indicates that there was no phosphomimetic activity of S → D or S → E substitutions (unpublished data). For phosphorylation-dependent protein–protein interactions, the geometry of phosphate binding is critical, and phosphomimics often do not fully recapitulate the phosphorylated amino acid. In many phosphoproteins, the phosphate can hydrogen bond with more geometries and provides hydrogen bonding or salt bridging combinations that Glu and Asp cannot (Williams et al., 2010), which most likely explains the lack of phosphomimetic phenotypes of Rad18 S → D and S → E mutations in the S box. Nevertheless, our analysis of Rad18 mutants harboring S → A substitutions indicates that Cdc7-mediated phosphorylation promotes basal association with Polη.
As shown in Fig. 5 F, UV treatment induced an ∼3.5-fold increase in the association of Polη with WT Rad18. However, in UV-treated cells, the association of Polη with Rad18 S-Box-A was reduced by >80% relative to its interaction with Rad18 WT.
The defective binding of Polη to Rad18 S-Box-A was specific because levels of Rad6 (another Rad18 binding partner) in WT Rad18 and S-Box-A immunoprecipitates were similar (Fig. 5 E, and F). Therefore, both basal and UV-induced Rad18–Polη associations are compromised in the Rad18 S-Box-A mutant.
S box mutation did not influence PCNA-directed E3 ubiquitin ligase activity because both Rad18 WT and Rad18 S-Box-A promoted similar levels of PCNA mono-ubiquitination when expressed in RAD18−/− HCT 116 cells (Fig. 5 G). The chromatin association of Rad18 was unaffected by S box mutations as determined by immunoblotting of chromatin fractions (Fig. 5, E and F) and by fluorescence microscopy experiments in which we examined the subcellular distribution of ectopically expressed CFP-Rad18 fusion proteins (not depicted).
Polη regulation is compromised by Cdc7 depletion and Rad18 S box mutations
The interaction between Rad18 and Polη is thought to be essential for guiding the TLS polymerase to sites of replication stalling. Rad18-dependent chaperoning of Polη is evident from the formation of Polη nuclear foci, microscopic structures likely representing sites of ongoing TLS (Watanabe et al., 2004).
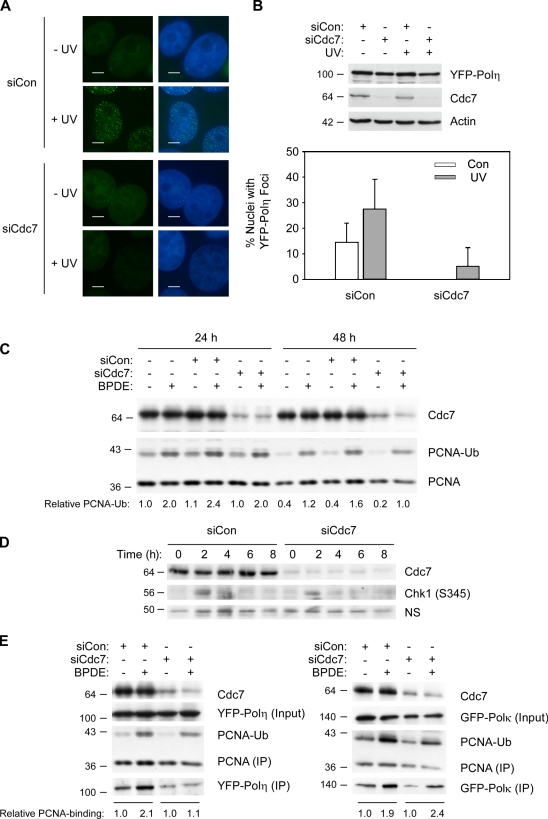
We determined the subcellular distribution of YFP-Polη before and after UV treatment in control and Cdc7-depleted H1299 cells. In nonirradiated control (Cdc7-expressing) cells, we observed a basal level of nuclear YFP-Polη foci corresponding to the reported association of Polη with the replication machinery (Kannouche et al., 2003). As expected, in Cdc7-replete cells, UV irradiation led to a twofold increase in YFP-Polη foci (Fig. 6 B; P = 0.0006). In contrast, after Cdc7 ablation, numbers of cells containing basal YFP-Polη nuclei foci were reduced ∼14-fold (Fig. 6 B; P = 0.0001). Moreover, Cdc7 depletion led to an 80% decrease in the number of cells containing UV-induced nuclear YFP-Polη foci Fig. 6 B; P = 0.0001). The efficiency of Cdc7 knockdown and similar expression levels of YFP-Polη in this experiment were confirmed by immunoblotting (Fig. 6 B). These results suggest that Cdc7 is important for the formation of Polη foci both in the absence of exogenous DNA damage and after UV radiation.
Figure 6.
Partial Cdc7 depletion inhibits formation of YFP-Polη nuclear foci. (A) Effect of partial Cdc7 depletion on subcellular distribution of YFP-Polη. YFP-Polη–expressing H1299 cells were transfected with siCdc7 (or siCon) oligos. Some cultures were treated with 100 J/m2 UV. 6 h after UV treatment, cells were fixed and stained with DAPI. Nuclear YFP-Polη foci were visualized by fluorescence microscopy as described in Materials and methods. Bars, 5 µm. (B) Validation of Polη expression and Cdc7 depletion (top). Cell extracts from the experiment described in A and B were analyzed by SDS-PAGE and immunoblotting with anti-GFP (to detect YFP-Polη), anti-Cdc7, and β-actin. Nuclear foci observed in the experiment described in A were enumerated as described in Materials and methods (bottom). To calculate the percentage of cells with Polη foci, ∼250 nuclei were scored for each condition. Data points represent the mean for 250 cells, with error bars representing the range. (C) Effect of Cdc7 depletion on PCNA mono-ubiquitination. H1299 cells were transfected with siCon or siCdc7, or were left untransfected. After 24 and 48 h, the resulting cells were treated with 500 nM BPDE for 1 h. After harvest, cell extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (D) Effect of Cdc7 depletion on kinetics of Chk1 phosphorylation. H1299 cells were transfected with siCon or siCdc7. After 24 h and 48 h, the resulting cells were treated with 500 nM BPDE and harvested at the indicated times. Cell extracts were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (E) Effect of Cdc7 depletion on PCNA binding of Polη and Polκ. YFP-Polη– or GFP-Polκ–expressing H1299 cells were transfected with siCon or siCdc7. 24 h later, cells were treated with BPDE for 4 h or left untreated for controls. Chromatin fractions from the resulting cells were immunoprecipitated with PCNA antibodies, and the resulting immune complexes were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Relative levels of YFP-Polη and GFP-Polκ in the PCNA immune complexes were quantified by densitometry and are expressed as fold change relative to controls (that received no BPDE). Molecular mass is indicated in kilodaltons next to the gel blots.
We considered the possibility that partial Cdc7 depletion might affect Polη recruitment secondarily because of changes in PCNA mono-ubiquitylation. However, basal and DNA damage–inducible PCNA mono-ubiquitylation were unaffected by partial Cdc7 depletion (Fig. 6 C). We also examined DNA damage–induced Chk1 phosphorylation in Cdc7-depleted cells. As shown in Fig. 6 D, the kinetics and magnitude of DNA damage–induced Chk1 phosphorylation were also unaffected by Cdc7 depletion. Therefore, two key S phase–coupled events (DNA damage–induced PCNA monoubiquitylation and Chk1 phosphorylation) were not perturbed by partial and acute Cdc7 depletion. These results are consistent with published work indicating that Mcm proteins (crucial targets of Cdc7) can be depleted substantially without affecting S phase progression (Woodward et al., 2006; Ge et al., 2007).
Consistent with the results of Fig. 6 A, in co-immunoprecipitation experiments we found that the genotoxin-inducible association of YFP-Polη with PCNA is sensitive to partial Cdc7 depletion (Fig. 6 E, left), whereas associations of PCNA with GFP-Polκ were relatively unaffected (Fig. 6 E, right). Collectively, these results indicate that partial Cdc7 depletion specifically impairs the association of Polη with stalled replication forks.
As a complementary approach to investigate the roles of Cdc7 and Rad18 S box phosphorylation in Polη recruitment, we compared the basal and UV-induced levels of YFP-Polη foci in response to ectopic expression of different Rad18 mutants. Immunofluorescence images of representative nuclei from this experiment are shown in Fig. 7 A. Although expression of WT Rad18 led to a statistically significant increase in the number of basal foci relative to AdCon (P < 0.0001 as calculated using Fisher’s exact test), expression of S-Box-A Rad18 did not (Fig. 7 B; P = 0.53). Moreover, after UV irradiation, cells expressing Rad18 S-Box-A exhibited almost 50% fewer foci than those expressing WT Rad18 (Fig. 7 B; P < 0.0001).
Figure 7.
Effect S box phosphorylation site mutation on Rad18-induced formation of YFP-Polη foci. (A) H1299 cells were coinfected with adenovirus vectors encoding YFP-Polη and HA-Rad18 (WT) or HA-Rad18 S-Box-A, or AdCon for controls. Some cultures were treated with 100 J/m2 UV. 6 h after UV treatment, cells were fixed and stained with DAPI. Nuclear YFP-Polη foci were visualized by fluorescence microscopy. Bars, 5 µm. (B) Cell extracts from the experiment described in A were analyzed by SDS-PAGE and immunoblotting with anti-HA to confirm equivalent expression of Rad18 (WT) and Rad18 S-Box-A (left). Nuclear foci observed in the experiment were enumerated (right). To calculate the percentage of cells with Polη foci, ∼250 nuclei were scored for each condition. Data points represent the mean for 250 cells, with error bars representing the range. (C) H1299 cells were transfected with siRNA directed against the 3′ UTR of endogenous Rad18 mRNA or with a nontargeting control siRNA, siCon. The resulting cells were transfected with CMV-driven mammalian expression plasmids encoding HA-Rad18 (WT), HA-Rad18 S-Box-A, or empty vector for control. Control and Rad18-complemented cells were trypsinized and re-plated. 24 h later, cells were harvested for SDS-PAGE and immunoblotting analysis (left) or were treated with varying doses of UV and analyzed for clonogenic survival (right). Molecular mass is indicated in kilodaltons next to the gel blots.
Even in the presence of endogenous Rad18 in H1299 cells, expression of the S box Rad18 mutant had a slight dominant-negative effect and impaired the formation of UV damage–induced YFP-Polη foci. Rad18 exists as a dimer in vivo, and formation of Polη interaction–defective heterodimers (comprising endogenous Rad18 WT and ectopically expressed Rad18 S-Box-A) likely accounts for the apparent dominant-negative activity of Rad18 S-Box-A in YFP-Polη focus-forming assays.
Phosphorylation-resistant Rad18 S-Box-A fails to complement UV sensitivity of Rad18-depleted cells
TLS-deficient cells sometimes exhibit sensitivity to genotoxic agents, and we predicted that Rad18 S-Box-A mutation might compromise cellular sensitivity to UV. For reasons that are unclear, HCT 116 cells are extraordinarily resistant to UV and other genotoxins, even in a RAD18−/− background (Shiomi et al., 2007), and we could not determine the effect of mutant forms of Rad18 on UV sensitivity using these cells. Therefore, as an alternative to using HCT116 cells, we depleted endogenous Rad18 from H1299 cells. The resulting Rad18-depleted cells were complemented using expression vectors encoding siRNA-resistant forms of Rad18 (WT), Rad18 S-Box-A, or with “empty” vector for control. Equivalent expression levels of Rad18 (WT) and Rad18 S-Box-A were confirmed by immunoblotting. As expected, both CMV-Rad18 WT and S-Box-A expression plasmids induced a similar level of PCNA mono-ubiquitination (Fig. 7 C). The resulting cells were irradiated with various doses of UV, and survival was determined using colony formation assays. As expected, UV sensitivity of Rad18-depleted H1299 cells was almost fully corrected by CMV-Rad18 WT. In contrast, the Rad18 S-Box-A mutant only conferred partial complementation of UV sensitivity. In the experiment shown in Fig. 7 C, irradiation with 10 J/m2 UV electromagnetic radiation subtype C (UVC) reduced survival to 79 ± 6% and 36% ± 6% in Rad18-expressing and Rad18-depleted cells, respectively. CMV-Rad18 (WT) expression corrected the UV sensitivity of Rad18-depleted cells by 82 ± 4.5%, whereas the Rad18 S-Box-A mutant only resulted in 38 ± 6% correction of UV sensitivity. These results further indicate that Rad18 S box phosphorylation is important for appropriate tolerance of UV-induced DNA damage.
Discussion
It has long been accepted that the cell cycle kinase Cdc7 is essential for the initiation phase of DNA synthesis. However, recent work also supports an active role for Cdc7 in the DNA damage response. For example, in human cells and Xenopus extracts, Cdc7 and Dbf4 form a stable complex and accumulate on chromatin after DNA damage or replication stress (Tenca et al., 2007; Tsuji et al., 2008). Claspin and AND-1 were recently characterized as DNA damage–induced Cdc7 substrates (Kim et al., 2008; Yoshizawa-Sugata and Masai, 2009), which supports a model in which Cdc7 maintains its kinase activity in the context of cellular response to DNA damage. It has been proposed that Cdc7 is important for replication restart after DNA damage–induced stalling of replication forks (Takeda et al., 1999; Tsuji et al., 2008). A role for Cdc7 in replication restart that implies a delayed activation of Cdc7 could potentially reconcile the apparent contradiction between the early findings that Cdc7 is inhibited and the more recent findings, including ours, that it is active in the context of the checkpoint.
Studies in yeast have demonstrated that Cdc7 and Dbf4 are required for induced mutagenesis (Ostroff and Sclafani, 1995). Moreover, ectopic expression of Cdc7 induced hypermutability in response to UV treatment (Sclafani et al., 1988). These data suggest that Cdc7 plays a role in the DNA damage tolerance TLS pathway. Indeed, a genetic study demonstrating that Cdc7 is epistatic to members of the Rad6 pathway (Pessoa-Brandão and Sclafani, 2004) led the authors to hypothesize that Cdc7 phosphorylates some member of the TLS pathway.
Our study identifies hRad18 as a novel DDK substrate in human cells and provides a potential molecular explanation for the observed genetic interaction between the Cdc7 and TLS pathways in yeast. We note, however, that the S box region of Rad18 is not conserved in S. cerevisiae or Schizosaccharomyces pombe. Therefore, DDK may regulate TLS by phosphorylating alternative sites in yeast Rad18 or via regulation of other TLS components.
Similar to phosphorylated motifs of other DDK substrates, the human Rad18 S box is both serine-rich and extremely acidic owing to a high density of aspartic and glutamic acid residues. Cdc7 is known to be an acidophilic kinase (Masai et al., 2006). We draw a useful analogy between Rad18 and Mcm2, a well-characterized substrate of Cdc7. Studies performed in vitro and using whole cells have shown that Cdc7 phosphorylates multiple residues in the acidic, serine-rich N terminus of Mcm2 (Cho et al., 2006; Charych et al., 2008), a region similar to the S box of Rad18. One detailed biochemical study demonstrated that, in vitro, the minimal requirement to support Cdc7 phosphorylation is an adjacent acidic amino acid (Cho et al., 2006). We noted that S434 is adjacent to a glutamic acid. The requirement for an adjacent acidic amino acid also invites the question of whether an adjacent phosphorylation might equally fulfill this condition. Indeed, Cho et al. (2006) found a peptide substrate that was only phosphorylated by Cdc7 after it was prephosphorylated on an adjacent serine, which suggests the possibility of a priming mechanism. Several studies have postulated models in which phosphorylation by a Cdk is a prerequisite for Cdc7 activity (Masai et al., 2000; Nougarède et al., 2000; Montagnoli et al., 2006; Wan et al., 2008), and a priming mechanism offers an attractive biochemical explanation for this relationship.
Although the Cdc7/Rad18 interaction and phosphorylation at S434 are induced by DNA damage, S434 was also observed to be phosphorylated basally. Therefore, Cdc7-mediated recruitment of Polη may be important for S phase progression even in the absence of exogenously induced DNA damage. Polη is localized to replication factories during an unperturbed S phase (Kannouche et al., 2003), which suggests a role for Polη in normal DNA replication. A recent study demonstrated that depletion of Polη from human cells affects cell cycle progression and rates of proliferation (Rey et al., 2009). In addition, Rey et al. (2009) described spontaneous chromosome instability, characterized by chromatid breaks, in Polη-depleted cells. Interestingly, Polη depletion led to a significant increase in chromatid aberrations at fragile sites, which suggests a role for Polη in replicating breakage-prone DNA regions. Polη has also been implicated in replication of another endogenous, aberrant DNA structure that can lead to fork pausing in a normal S phase: G4 quartet DNA is a four-stranded DNA structure that can form in guanine-rich nucleotide sequences and presents a challenge to the normal replicative polymerase. Depletion of Polη in human cells reduced their ability to replicate G4 quartet DNA (Bétous et al., 2009). These findings, combined with the knowledge that Cdc7 is a well-characterized cell cycle kinase, suggest that Cdc7 activity could be important for recruiting Polη during normal replication to aid in bypass of endogenous DNA structures.
Until recently, it has been accepted that Polη recruitment in response to exogenous DNA damage relies primarily on PCNA mono-ubiquitination and the subsequent binding of mono-ubiquitinated PCNA to the Polη UBZ motif (Bienko et al., 2005). However, the UV sensitivity of XPV cells can be complemented by Polη UBZ mutants (Acharya et al., 2008), which suggests that alternate mechanisms exist for the recruitment of Polη. Watanabe et al., (2004) demonstrated that Rad18 interacts with Polη and chaperones the polymerase to stalled replication forks. We observed that although the S box mutant does not interact efficiently with Polη or support the formation of Polη foci, it is competent to mono-ubiquitinate PCNA. Therefore, our findings are consistent with the work of Acharya et al. (2008) and we validate the model proposed by Watanabe et al. (2004), demonstrating a separation of Rad18’s primary TLS functions: modifier of PCNA and chaperone of Polη. We extend the results of those workers by demonstrating that the Rad18–Polη interaction is subject to regulation by phosphorylation.
Further studies are necessary to determine the mechanism by which Chk1 promotes phosphorylation of Rad18 by Cdc7. Rad18 does not appear to be a Chk1 substrate (unpublished data). The role of Chk1 in stimulating Rad18–Cdc7 binding could be caused by Chk1-dependent stabilization of stalled forks, potentially creating or maintaining replication intermediates that facilitate Rad18–Cdc7 interactions. Alternatively, Chk1 might target DDK (directly or indirectly), and there is evidence that Dbf4 is phosphorylated by Chk1 at multiple sites (Kim et al., 2003).
In conclusion, our study identifies a novel biochemical relationship between the S-phase kinase Cdc7 and the TLS regulator Rad18. Our work is the first demonstration of Rad18 regulation via phosphorylation. The finding that Cdc7 regulates recruitment of Polη by modification of Rad18 suggests a novel biochemical basis for a genetic relationship that was previously recognized in yeast. Our findings provide important new mechanistic details regarding the regulation of the DNA damage tolerance pathway and how postreplication repair is integrated with other elements of cell cycle control and the response to either exogenously induced or spontaneously arising forms of DNA damage in human cells.
Materials and methods
Adenovirus construction and infection
Adenovirus construction and infection were performed as described previously (Bi et al., 2005, 2006). H1299 and HCT116 cells were routinely infected with at 4 × 109 pfu/ml and 109 pfu/ml of adenovirus, respectively.
Cell lines and reagents
Human lung carcinoma H1299 cells, 293T cells, and RAD18−/− HCT116 cells (Shiomi et al., 2007) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and 100 µg/ml streptomycin sulfate and 100 U/ml penicillin. The antibodies were purchased as indicated: anti-Polη (A301-231A) and anti-Rad18 (A301-340A) were from Bethyl Laboratories, Inc.; anti–HA-tag (Y11, sc-805), anti-CHEK1 (sc-8408), and anti-PCNA (sc-56) were from Santa Cruz Biotechnology, Inc.; anti–phospho-Chk1 Ser-317 (2344) was from Cell Signaling Technology; anti-Dbf4 (MBS120101) was from MyBiosource.com; anti-ACTB (A5441) was from Sigma Aldrich; anti-GFP (A11122) was from Invitrogen; and anti-Cdc7 (K0070-3) was from the Marine Biological Laboratory. The Rad18 S434 phospho-specific antibody was generated by immunizing rabbits with the peptide IQEVLSS[pS]ESDS. To ensure phospho-specificity, the resulting areas were depleted using unmodified immunogen and purified using the phosphopeptide. Peptide synthesis, immunizations, and antibody purification steps were performed by 21st Century Biochemicals, Inc.
Genotoxin treatment
For UVC treatment, the growth medium was removed from the cells and replaced with PBS. The plates were transferred to a UV cross-linker (Agilent Technologies) and irradiated. The UVC dose delivered to the cells was confirmed with a UV radiometer (UVP, Inc.). The cells were re-fed with complete growth medium and returned to the incubator. Benzo(a)pyrene diolepoxide (BPDE; National Cancer Institute Carcinogen Repository) was dissolved in anhydrous DMSO and added directly to the growth medium as a 1,000× stock to give final a concentration of 500 nM.
RNAi interference
Cells were plated to achieve 75% confluence in 6-cm tissue culture dishes. Transfection of siRNA constructs was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each plate of cells was transfected with 20 µl of Lipofectamine reagent and contained a final siRNA concentration of 100 nM in a total volume of 3 ml. Sequences for siRNA are as follows. Cdc7 siRNA, 5′-GCUCAGCAGGAAAGGAGUUdTdT-3′, originally described by Montagnoli et al. (2004); Rad18 3′ untranslated region (UTR) siRNA, 5′-UUAUAAAUGCCCAAGGAAAUU-3′; Dbf4 siRNA, 5′-CAGUAUCAAGUUGUUGAUGAU-3′; and control nontargeting siRNA, 5′-UAGCGACUAAACACAUCAA-3′ (Thermo Fisher Scientific).
Fluorescence microscopy
H1299 cells expressing WT and mutant forms of Rad18 and YFP-Polη were treated with UVC as described above and analyzed exactly as described previously (Bi et al., 2006). In brief, to visualize YFP-Polη fluorescence, the cells were fixed with 4% paraformaldehyde for 10 min, then permeabilized with 0.2% Triton X-100 for 5 min. After washing the slides with PBS, cells were DAPI-stained and mounted with Vectashield solution (Vector Laboratories). Images were acquired at room temperature on an inverted microscope (IX-70; Olympus) attached to a restoration imaging system (Deltavision Spectris; Applied Precision, Inc.) equipped with a charge-coupled device camera (Photometrics CoolSNAP HQ). Images were captured using the SoftWoRx Acquire 3D software using 40× (NA. 1.35), 60× (NA 1.42), or 100× (NA 1.40) oil objective lenses (Olympus). Images were collected as z stacks of 0.1–0.5-µm increments (1–15 sections total). Image stacks were deconvolved using the SoftWoRx conservative algorithm with 10 iterations, then collapsed into single-plane images using the Quick Projection option. Multidimensional analysis of image stacks was done using the SoftWoRx software and displayed as either solid or wireframe models. 3D projections and models were rotated and saved in file formats that were exported to Photoshop (Adobe) for viewing and analysis.
Immunoprecipitations and immunoblotting
To prepare extracts containing soluble and chromatin-associated proteins, monolayers of cultured cells (typically in 60-mm or 100-mm dishes) were washed three times in ice-cold PBS and scraped into 500 µl of ice-cold cytoskeleton buffer (CSK buffer; 10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF, and 0.1% Triton X-100) freshly supplemented with 0.2 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche). Lysates were transferred to microcentrifuge tubes and incubated on ice for 5 min, then centrifuged at 1,000 g for 2 min. The supernatants were removed and further clarified by centrifugation at 10,000 g for 10 min to obtain Triton X-100–soluble proteins. The Triton X-100–extracted insoluble nuclear fractions were washed once with 1 ml of CSK buffer and then resuspended in a minimal volume of CSK. In some experiments, chromatin-bound proteins were released by digestion of nuclei in CSK with 1,000 U/ml of RNase-free DNase I (Roche) at 25°C for 30 min. After DNase I digestion, soluble and insoluble materials were separated by centrifugation at 10,000 g for 10 min. For immunoprecipitation, samples were normalized for protein concentration. Primary antibody incubations were performed overnight at 4°C on rotating racks. Immune complexes were recovered by the addition of secondary antibody and 20 µl of protein G–Sepharose 4 Fast Flow beads (GE Healthcare) and further incubation for 4–6 h. The beads were recovered by brief centrifugation and washed four times with 1 ml CSK (5–10 min per wash). The washed immune complexes were boiled in protein loading buffer for 10 min to release and denature immunoprecipitated proteins before separation on SDS-PAGE.
Expression of recombinant Rad18 GST fusion fragments
A series of cDNAs spanning the length of Rad18 and mutational variants of fragment 7 were cloned into the pGEX 2T vector (GE Healthcare). All cloning was verified by sequencing (Dana-Farber/Harvard Cancer Center Sequence Core). GST fusion proteins were expressed in One Shot BL21 (DE3) Escherichia coli (Invitrogen) and purified according to the manufacturer’s instructions.
Survival assays
Endogenous Rad18 was depleted using siRNA, and complementation with WT and mutant Rad18 was achieved using transfected expression plasmids, as described previously (Song et al., 2010). The resulting cultures were reseeded at low density in replicate and irradiated with various doses of UVC. Control and irradiated plates were returned to the incubator and maintained until colonies comprising ∼50 cells were evident (typically 10–12 d). The resulting cells were fixed (using 40% methanol and 10% acetic acid) and stained with crystal violet, and colonies were enumerated as described previously (Ohashi et al., 2009).
In vitro Cdc7 kinase assays
Recombinant GST-Cdc7–Dbf4 complexes were produced in baculovirus and used to phosphorylate various substrates in vitro as described previously (Masai et al., 2006).
In brief, kinase assays were conducted in 25-µl reactions containing 40 mM Hepes-KOH, pH 7.6, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM β-glycerophosphate, 1 mM NaF, 2 mM DTT, 8 mM MgOAC, 0.1 mM ATP, 10 µCi [32P]γATP, and substrate proteins indicated in the figure legends. Reactions were incubated at 30°C for 45 min, followed by the addition of SDS-PAGE sample buffer. After boiling for 3 min, half of each sample was analyzed by SDS-PAGE. Radioactive proteins were detected by autoradiography of stained and dried gels.
Reproducibility
All data shown are representative of experiments that were repeated at least three times with similar results on each separate occasion.
Acknowledgments
We are grateful to Haruo Ohmori, Chikahide Masutani, and Scott Williams for helpful discussions during these studies. We thank Jordan Fishman and Pam Crowley for synthesizing peptides, generating phosphospecific antibodies, and providing valuable advice on immunological methods. We thank Beth Sullivan for help with microscopy.
This work was supported by National Institute of Environmental Health Sciences grants ES09558 and ES016280 (to C. Vaziri).
Footnotes
Abbreviations used in this paper:
- BPDE
- benzo(a)pyrene diol epoxide
- CPD
- cyclobutane–pyrimidine dimer
- DDK
- Dbf4/Drf1-dependent kinase
- PCNA
- proliferating cell nuclear antigen
- TLS
- trans-lesion synthesis
- XPV
- xeroderma pigmentosum variant
- WT
- wild type
References
- Acharya N., Yoon J.H., Gali H., Unk I., Haracska L., Johnson R.E., Hurwitz J., Prakash L., Prakash S. 2008. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 105:17724–17729 10.1073/pnas.0809844105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Rey L., Wang G., Pillaire M.J., Puget N., Selves J., Biard D.S., Shin-ya K., Vasquez K.M., Cazaux C., Hoffmann J.S. 2009. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 48:369–378 10.1002/mc.20509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Slater D.M., Ohmori H., Vaziri C. 2005. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 280:22343–22355 10.1074/jbc.M501562200 [DOI] [PubMed] [Google Scholar]
- Bi X., Barkley L.R., Slater D.M., Tateishi S., Yamaizumi M., Ohmori H., Vaziri C. 2006. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell. Biol. 26:3527–3540 10.1128/MCB.26.9.3527-3540.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 310:1821–1824 [DOI] [PubMed] [Google Scholar]
- Bousset K., Diffley J.F. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12:480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil R.A., Lin Q., Stambrook P.J., Kucherlapati R., Vijg J. 2008. Mutation frequencies and spectra in DNA polymerase eta-deficient mice. Cancer Res. 68:2081–2084 10.1158/0008-5472.CAN-07-6274 [DOI] [PubMed] [Google Scholar]
- Charych D.H., Coyne M., Yabannavar A., Narberes J., Chow S., Wallroth M., Shafer C., Walter A.O. 2008. Inhibition of Cdc7/Dbf4 kinase activity affects specific phosphorylation sites on MCM2 in cancer cells. J. Cell. Biochem. 104:1075–1086 10.1002/jcb.21698 [DOI] [PubMed] [Google Scholar]
- Cho W.H., Lee Y.J., Kong S.I., Hurwitz J., Lee J.K. 2006. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc. Natl. Acad. Sci. USA. 103:11521–11526 10.1073/pnas.0604990103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J.E., Afzal V., Feeney L., McDowell M., Sadinski W., Volpe J.P., Busch D.B., Coleman D.M., Ziffer D.W., Yu Y., et al. 1999. Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 59:1102–1108 [PubMed] [Google Scholar]
- Dolan W.P., Le A.H., Schmidt H., Yuan J.P., Green M., Forsburg S.L. 2010. Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage. Genetics. 185:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Q., Jackson D.A., Blow J.J. 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21:3331–3341 10.1101/gad.457807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins V., Gabrielse C., Haste L., Weinreich M. 2009. Budding yeast Dbf4 sequences required for Cdc7 kinase activation and identification of a functional relationship between the Dbf4 and Rev1 BRCT domains. Genetics. 183:1269–1282 10.1534/genetics.109.110155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. 1973. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J. Bacteriol. 115:966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P., Fernández de Henestrosa A.R., Coull B., Vidal A.E., Gray C., Zicha D., Woodgate R., Lehmann A.R. 2003. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 22:1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500 [DOI] [PubMed] [Google Scholar]
- Kilbey B.J. 1986. cdc7 alleles and the control of induced mutagenesis in yeast. Mutagenesis. 1:29–31 10.1093/mutage/1.1.29 [DOI] [PubMed] [Google Scholar]
- Kim J.M., Nakao K., Nakamura K., Saito I., Katsuki M., Arai K., Masai H. 2002. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 21:2168–2179 10.1093/emboj/21.9.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Yamada M., Masai H. 2003. Functions of mammalian Cdc7 kinase in initiation/monitoring of DNA replication and development. Mutat. Res. 532:29–40 [DOI] [PubMed] [Google Scholar]
- Kim J.M., Kakusho N., Yamada M., Kanoh Y., Takemoto N., Masai H. 2008. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 27:3475–3482 [DOI] [PubMed] [Google Scholar]
- Kumagai H., Sato N., Yamada M., Mahony D., Seghezzi W., Lees E., Arai K., Masai H. 1999. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol. 19:5083–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposa R.R., Feeney L., Cleaver J.E. 2003. Recapitulation of the cellular xeroderma pigmentosum-variant phenotypes using short interfering RNA for DNA polymerase H. Cancer Res. 63:3909–3912 [PubMed] [Google Scholar]
- Lin Q., Clark A.B., McCulloch S.D., Yuan T., Bronson R.T., Kunkel T.A., Kucherlapati R. 2006. Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res. 66:87–94 10.1158/0008-5472.CAN-05-1862 [DOI] [PubMed] [Google Scholar]
- Maher V.M., Ouellette L.M., Curren R.D., McCormick J.J. 1976. Caffeine enhancement of the cytotoxic and mutagenic effect of ultraviolet irradiation in a xeroderma pigmentosum variant strain of human cells. Biochem. Biophys. Res. Commun. 71:228–234 10.1016/0006-291X(76)90272-2 [DOI] [PubMed] [Google Scholar]
- Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J. Biol. Chem. 275:29042–29052 [DOI] [PubMed] [Google Scholar]
- Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J.M., Ishii A., Tanaka T., Kobayashi T., et al. 2006. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281:39249–39261 [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki M., Yamada A., Kusumoto R., Nogimori T., Maekawa T., Iwai S., Hanaoka F. 1999. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18:3491–3501 10.1093/emboj/18.12.3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Bosotti R., Villa F., Rialland M., Brotherton D., Mercurio C., Berthelsen J., Santocanale C. 2002. Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 21:3171–3181 10.1093/emboj/cdf290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Tenca P., Sola F., Carpani D., Brotherton D., Albanese C., Santocanale C. 2004. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 64:7110–7116 10.1158/0008-5472.CAN-04-1547 [DOI] [PubMed] [Google Scholar]
- Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. 2006. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281:10281–10290 [DOI] [PubMed] [Google Scholar]
- Nakajima S., Lan L., Kanno S., Usami N., Kobayashi K., Mori M., Shiomi T., Yasui A. 2006. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 281:34687–34695 [DOI] [PubMed] [Google Scholar]
- Newlon C.S., Fangman W.L. 1975. Mitochondrial DNA synthesis in cell cycle mutants of Saccharomyces cerevisiae. Cell. 5:423–428 10.1016/0092-8674(75)90029-X [DOI] [PubMed] [Google Scholar]
- Njagi G.D., Kilbey B.J. 1982. cdc7-1 a temperature sensitive cell-cycle mutant which interferes with induced mutagenesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 186:478–481 10.1007/BF00337951 [DOI] [PubMed] [Google Scholar]
- Nougarède R., Della Seta F., Zarzov P., Schwob E. 2000. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 20:3795–3806 10.1128/MCB.20.11.3795-3806.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E., Hanafusa T., Kamei K., Song I., Tomida J., Hashimoto H., Vaziri C., Ohmori H. 2009. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 14:101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkumo T., Kondo Y., Yokoi M., Tsukamoto T., Yamada A., Sugimoto T., Kanao R., Higashi Y., Kondoh H., Tatematsu M., et al. 2006. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol. Cell. Biol. 26:7696–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff R.M., Sclafani R.A. 1995. Cell cycle regulation of induced mutagenesis in yeast. Mutat. Res. 329:143–152 [DOI] [PubMed] [Google Scholar]
- Pessoa-Brandão L., Sclafani R.A. 2004. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 167:1597–1610 10.1534/genetics.103.021675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Rey L., Sidorova J.M., Puget N., Boudsocq F., Biard D.S., Monnat R.J., Jr., Cazaux C., Hoffmann J.S. 2009. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol. Cell. Biol. 29:3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R.A., Patterson M., Rosamond J., Fangman W.L. 1988. Differential regulation of the yeast CDC7 gene during mitosis and meiosis. Mol. Cell. Biol. 8:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.J., Stillman B. 2006. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 24:101–113 10.1016/j.molcel.2006.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N., Mori M., Tsuji H., Imai T., Inoue H., Tateishi S., Yamaizumi M., Shiomi T. 2007. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 35:e9 10.1093/nar/gkl979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I.Y., Palle K., Gurkar A., Tateishi S., Kupfer G.M., Vaziri C. 2010. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J. Biol. Chem. 285:31525–31536 10.1074/jbc.M110.138206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Ogino K., Matsui E., Cho M.K., Kumagai H., Miyake T., Arai K., Masai H. 1999. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol. 19:5535–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. 2003. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23:474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenca P., Brotherton D., Montagnoli A., Rainoldi S., Albanese C., Santocanale C. 2007. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J. Biol. Chem. 282:208–215 10.1074/jbc.M604457200 [DOI] [PubMed] [Google Scholar]
- Trincao J., Johnson R.E., Escalante C.R., Prakash S., Prakash L., Aggarwal A.K. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell. 8:417–426 10.1016/S1097-2765(01)00306-9 [DOI] [PubMed] [Google Scholar]
- Tsuji T., Lau E., Chiang G.G., Jiang W. 2008. The role of Dbf4/Drf1-dependent kinase Cdc7 in DNA-damage checkpoint control. Mol. Cell. 32:862–869 10.1016/j.molcel.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K.M., Boulton S.J., Hollingsworth N.M. 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22:386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.S., Williams R.S., Dovey C.L., Guenther G., Tainer J.A., Russell P. 2010. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 29:1136–1148 10.1038/emboj.2009.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.M., Göhler T., Luciani M.G., Oehlmann M., Ge X., Gartner A., Jackson D.A., Blow J.J. 2006. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 173:673–683 10.1083/jcb.200602108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Okada T., Matsusaka T., Sonoda E., Zhao G.Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. 2002. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 21:5558–5566 10.1093/emboj/cdf534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N., Kim J.M., Koiwai O., Arai K., Masai H. 2005. Functional analyses of mouse ASK, an activation subunit for Cdc7 kinase, using conditional ASK knockout ES cells. Genes Cells. 10:551–563 [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N., Masai H. 2009. Roles of human AND-1 in chromosome transactions in S phase. J. Biol. Chem. 284:20718–20728 [DOI] [PMC free article] [PubMed] [Google Scholar]