The precise balance of spindle orientations required for proper epidermal morphogenesis is regulated by mInscuteable expression and NuMA localization.
Abstract
Progenitor cells must balance self-amplification and production of differentiated progeny during development and homeostasis. In the epidermis, progenitors divide symmetrically to increase surface area and asymmetrically to promote stratification. In this study, we show that individual epidermal cells can undergo both types of division, and therefore, the balance is provided by the sum of individual cells’ choices. In addition, we define two control points for determining a cell’s mode of division. First is the expression of the mouse Inscuteable gene, which is sufficient to drive asymmetric cell division (ACD). However, there is robust control of division orientation as excessive ACDs are prevented by a change in the localization of NuMA, an effector of spindle orientation. Finally, we show that p63, a transcriptional regulator of stratification, does not control either of these processes. These data have uncovered two important regulatory points controlling ACD in the epidermis and allow a framework for analysis of how external cues control this important choice.
Introduction
During development of the epidermis, basal progenitor cells undergo both symmetric cell divisions (SCDs) to increase surface area and asymmetric cell divisions (ACDs) to increase thickness of the tissue (Fig. 1 A; Smart, 1970; Lechler and Fuchs, 2005). In SCD, mitotic spindles are oriented parallel to the underlying basement membrane, whereas ACDs have spindles perpendicular to it (Fig. 1 A). A primary role for the perpendicular spindles is to directly drive the morphogenesis (stratification) of this tissue. These divisions are also associated with a change in cell fate, as they produce one daughter cell in the proliferative basal layer of the epidermis and another daughter committed to differentiation in the suprabasal cell layer. Little is known about how the balance of SCD/ACD is controlled to allow proper development.
Figure 1.
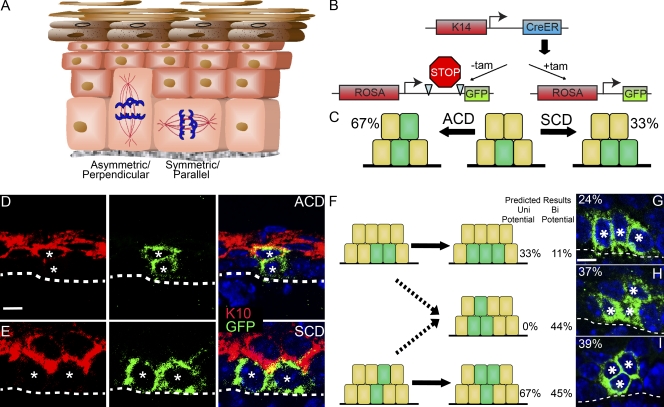
Most epidermal progenitors can divide both symmetrically and asymmetrically. (A) Diagram of the epidermis showing an SCD and an ACD. (B) Genetics of short-term lineage-tracing experiments. K14-CreER mice were mated to Rosa-lox-stop-lox-GFP. (C) A single labeled cell (green) resulting from recombination at the Rosa locus (middle) can divide symmetrically to generate two basal cells or asymmetrically to yield one basal and one suprabasal cell. Quantitation of the percentage of clones 16–20 h after tamoxifen-induced recombination is listed next to the diagram (n = 100). (D and E) Examples of two-cell clones resulting from an ACD (D) and an SCD (E). GFP (green) marks the clones, whereas K10 (red) marks cells committed to differentiation. Hoechst labels nuclei (blue). (F) Resulting three-cell clones and their prevalence if cells are unipotential in their division orientation (solid arrows) or if they are bipotential (solid and dashed arrows). (G–I) Examples of three types of three-cell clones, two SCD (G), one SCD and one ACD (H), and two ACD (I). n = 100 three-cell clones. Asterisks highlight the GFP-labeled cells. Dashed lines indicate the basement membrane separating epidermis from dermis. Bars, 10 µm.
A conserved complex of proteins (including Par3, mouse Inscuteable [Insc; mInsc], Leu-Gly-Asn–enriched protein [LGN], and NuMA) localizes to the apical cell cortex during ACDs (Kraut et al., 1996; Schober et al., 1999; Parmentier et al., 2000; Lechler and Fuchs, 2005; Siller et al., 2006). These proteins are required for orienting the mitotic spindle along the apical–basal axis of the cell. Beyond their cell cycle–dependent expression and localization, we know very little about the control of these proteins in the epidermis and whether their expression and/or activity is regulated to direct ACDs.
Results and discussion
To determine whether epidermal progenitors are committed to a single division orientation, we used genetic lineage tracing to mark and follow individual cells after division. We injected pregnant dams (K14-CreER; Rosa-lox-stop-lox-GFP) with a low dose of tamoxifen to achieve limited recombination, allowing expression of GFP in a small subset of epidermal progenitors (∼3%; Fig. 1 B). Embryos were examined at embryonic day (e) 15.5, 16–20 h after injection. Clones of two cells were easily distinguishable and well separated from each other, suggesting they are the result of a single recombination event. If this assumption is true, we would expect to see two types of two-cell clones. One type would consist of two basal cells generated from an SCD of a labeled progenitor. The other would consist of one basal and one suprabasal cell, the result of an ACD (Fig. 1 C). 67% of clones were of the basal/suprabasal type, whereas 33% were composed of two basal cells (n = 100; Fig. 1 C). These results are in close agreement with the ratio of spindles perpendicular or parallel to the basement membrane (∼70% perpendicular/30% parallel; Lechler and Fuchs, 2005). To demonstrate that the ACD results in cell fate changes, we stained cryosections of these embryos for the differentiation marker keratin 10 (K10). K10 is expressed in almost all suprabasal cells (>97%; n = 167) and is excluded from the proliferative basal compartment (Fig. S1, A and B). In the two-cell clones with one basal and one suprabasal cell, the suprabasal cell was always K10 positive, whereas the basal cell was K10 negative (n = 61; Fig. 1 D). In clones of two basal cells (n = 42), all cells were K10 negative (Fig. 1 E). Thus, short-term lineage tracing is a feasible method to track division orientations and cell fates of epidermal progenitors and supports a direct relationship between division orientation and cell fate.
We next examined the organization of cells in three-cell clones, resulting from two progenitor divisions. If cells are committed to their division orientation (unipotent), then clones would consist of three basal cells (for SCD) or one basal cell with two suprabasal cells (for ACD; Fig. 1 F). However, our analysis demonstrated that three-cell clones consisting of two basal cells and one suprabasal cell were frequently seen (37% of clones; n = 100; Fig. 1 H). These results are inconsistent with cells being committed to their division orientations. Only if orientations are relatively independent at each division would so many clones of this type exist (see Materials and methods). These data suggest that the majority of basal cells have the capability to divide both symmetrically and asymmetrically.
The finding that individual cells can divide in either orientation raised an intriguing question about the timing and mechanism of establishment of cell division orientation. In many invertebrate models for ACD, the orientation of the mitotic spindle results from centrosome migration during interphase (Yamashita et al., 2003; Rebollo et al., 2007; Rusan and Peifer, 2007). However, in the studied cell types, cells divide exclusively asymmetrically and do not have the ability to divide symmetrically. We wanted to determine when epidermal cells establish their division orientation and whether the ability to divide in both orientations results in differences in the mechanism of spindle positioning.
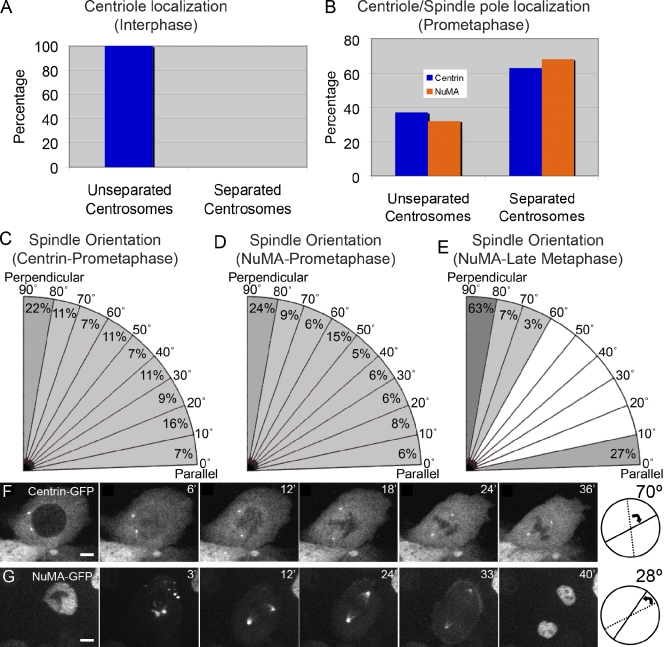
To determine whether centrosome splitting and migration occur during interphase in epidermal progenitors, we examined the localization of centrioles in these cells using centrin-GFP–expressing mice (Lechler and Fuchs, 2005). We examined >1,000 interphase cells in e14.5 epidermis and did not find any in which centrosomes have migrated away from each other (Fig. 2 A). Additionally, in 37% of prometaphase cells, centrosomes have not yet separated (n = 98 cells; Fig. 2 B). Without a bipolar spindle, these cells, by definition, cannot have established their division orientation.
Figure 2.
Cell division orientation is established during metaphase. (A) Quantification of basal epidermal cells in interphase with unseparated and separated centrosomes as marked by centrin (n = 1,012 cells). (B) Quantification of basal epidermal cells in prometaphase with unseparated and separated centrosomes/spindle poles. Centrosomes were identified by centrin-GFP localization, and spindle poles were identified by NuMA-GFP localization (n = 98 cells for centrin-GFP; n = 132 for NuMA-GFP). (C–E) Spindle orientation was determined in cells with bipolar spindles as visualized with centrin-GFP in prometaphase (C), with NuMA-GFP in prometaphase (D), and with NuMA-GFP at late metaphase (E). Angles are relative to the basement membrane. (F and G) Rotation of mitotic spindles in cultured keratinocytes as visualized with centrin-GFP (F) and NuMA-GFP (G). (right) Diagrams indicate the angle of rotation of the spindle. Bars, 2.5 µm.
To determine whether the same behavior is seen when visualizing a spindle pole marker, we generated mice expressing NuMA-GFP under the control of K14 promoter (Fig. S2 A). NuMA-GFP localizes to the nucleus in interphase cells and to spindle poles during mitosis (Fig. S2, B–D). Importantly, NuMA-GFP mice displayed wild-type ratios of spindle orientations (Fig. 2 E). 32% of prometaphase cells (n = 132) have monopolar NuMA-GFP localization, demonstrating that a bipolar mitotic spindle has not yet formed (Fig. 2 B). Similar localization was found when staining for endogenous NuMA (unpublished data).
Together, these data demonstrate that spindle orientation is not predetermined by interphase centrosome positioning but rather is established during mitosis. To determine whether spindles elongate along their future axis or rotate into it, we examined the orientation of mitotic spindles in the population of prometaphase cells that had already formed bipolar spindles. Using both centrin-GFP and NuMA-GFP as markers, we found that spindle orientation was essentially random in cells during prometaphase (Fig. 2, C and D). However, during late metaphase/anaphase, the ratio of perpendicular and parallel spindles was similar to those previously published, 73%/27% (n = 106 cells; Fig. 2 E). These data suggest that mitotic spindles form and then rotate into their final position during metaphase.
To determine whether spindle rotation also occurs in cultured keratinocytes, we performed time-lapse imaging of centrin-GFP– and NuMA-GFP–expressing cells. Cultured keratinocytes behave somewhat differently from their in vivo counterparts. Although centrioles remain closely apposed in vivo, they do not do so in culture. Despite this difference, there is a pronounced rotation of the mitotic spindle in some dividing cells, which occurs after nuclear envelope breakdown and spindle elongation (Fig. 2, F and G). Therefore, the dynamics of spindle orientation in epidermal progenitors is mechanistically distinct from what has been reported in several invertebrate model systems. Altogether, the aforementioned data demonstrate that individual progenitor cells can divide either asymmetrically or symmetrically and that this orientation is achieved in metaphase through spindle rotation.
What determines whether a cell divides symmetrically or asymmetrically? We hypothesized that either the expression or localization/activity of the machinery that directs spindle reorientation regulates this decision. Although Par3 is expressed and apically localized in most if not all polarized epithelia, mInsc is much more restricted in its expression pattern. Loss of Insc results in loss of ACD in Drosophila neuroblasts, whereas misexpression of Insc in symmetrically dividing neuroepithelial cells forces these cells to reorient their mitotic spindles along the apical–basal axis (Kraut et al., 1996).
To determine whether the presence of mInsc obligately leads to ACD in the epidermis, we generated mice that allow inducible expression of mInsc from a tetracycline/doxycycline-responsive promoter (TRE). The TRE-mInsc-HA mice include an epitope tag to visualize the resulting protein. We mated these mice to a K14-promoter-rtTA mouse line to allow for controlled expression in the epidermis (Fig. 3 A). Transgene expression was dependent on both the TRE-mInsc-HA and K14-rtTA alleles as well as doxycycline (unpublished data).
Figure 3.
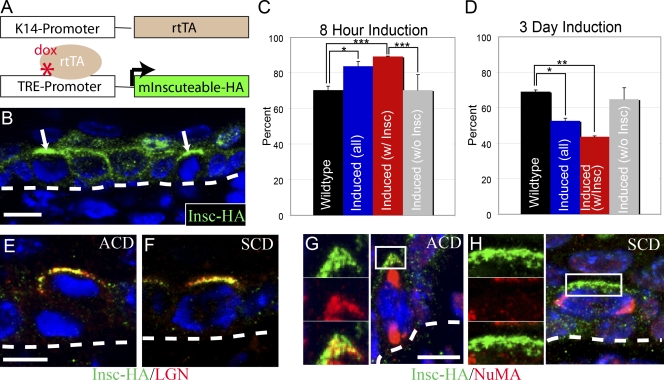
mInsc expression is sufficient for transient but not long-term increases in ACD. (A) Diagram of genetic induction system. K14-rtTA mice were mated to mice expressing mInsc-HA under the control of TRE. (B) With both alleles and doxycycline present, mInsc-HA (green) is expressed in the epidermis. Arrows indicate apical accumulation of mInsc-HA in mitotic cells. Hoeschst labels nuclei (blue). (C) Quantification of mitotic spindles oriented perpendicular to the basement membrane in embryos induced to express mInsc-HA for 8 h. Expression of mInsc-HA was mosaic so data are presented for all cells (induced(all)), only cells with detectable apical mInsc-HA (induced(w/Insc)), and cells without detectable mInsc-HA (induced(w/o Insc)). P-value for wild-type versus induced (all) cells is 0.017, wild-type versus induced (w/mInsc) is 0.0002, and induced (w/mInsc) versus induced (w/o mInsc) is 0.0005. (D) Quantification of mitotic spindles oriented perpendicular to the basement membrane in embryos induced to express mInsc-HA for 3 d. P-values are 0.017 for wild-type versus induced (all) cells and 0.003 for wild-type versus induced (w/ mInsc) cells. There was no statistically significant difference between either induced (w/o mInsc) and induced (w/mInsc) or wild-type cells. (C and D) Error bars indicate mean ± SEM. (E and F) Colocalization of mInsc-HA (green) and LGN (red) at the apical cortex of mitotic cells dividing either asymmetrically (E) or symmetrically (F). (G) mInsc-HA (green) colocalizes with NuMA (red) at the apical cortex of an asymmetrically dividing cell. (H) Lack of colocalization of mInsc-HA (green) and NuMA (red) in a symmetrically dividing cell. (G and H, left) Higher magnification views of boxed areas are shown. Dashed lines indicate the basement membrane. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 10 µm.
K14-rtTA;TRE-mInsc-HA (mInscIND) mice were injected with a single dose of doxycycline and embryos harvested 8 h later at e14.5. mInsc-HA was expressed in 50–70% of cells of the epidermis, depending on the specific embryo. We found that mInsc-HA localizes normally and tightly to the apical region of mitotic basal cells (Fig. 3 B). Importantly, mInsc-HA expression affected spindle orientation. Although 70% of divisions were perpendicular in control mice, 80–89% were perpendicular in mInscIND mice (Fig. 3 C). Because there was mosaicism in mInsc-HA expression, we determined the frequency of division orientation in cells expressing the transgene. After the 8-h induction, the cells not expressing mInsc-HA showed a normal 70% perpendicular/30% parallel ratio (n = 50 cells). In cells expressing mInsc-HA, 90% of divisions were perpendicular (n = 102 from three embryos; Fig. 3 C). This change is highly statistically significant (P = 0.0002). Thus, transient forced expression of mInsc-HA is sufficient to increase perpendicular spindle orientation in the epidermis and suggests that regulation of mInsc expression is a key control point in the decision between SCD/ACD. Consistent with this, in mInscIND embryos, 78% of lineage-traced two-cell clones were composed of one basal and one suprabasal cell (n = 115; compared with 67% in control mice; P < 0.05). We did not find any perturbation in the pattern of K10 expression in these mice. Therefore, both by analysis of spindle orientation and resulting cell clones, mInsc-HA resulted in an increase in ACDs.
To determine the longer-term effects of misexpression, we collected embryos after 3 d of induction of mInsc-HA. To our surprise, these mice did not demonstrate changes in epidermal architecture as compared with littermate controls. Thus, we determined the percentage of perpendicular spindles and found them to be significantly decreased to 53% (n = 95 cells from two embryos; Fig. 3 D). Thus, although short-term induction of mInsc-HA results in increased ACDs, this is not sustained, demonstrating a robust control of division orientation.
To determine why there is a decrease in perpendicular spindles, we analyzed the expression and localization of mInsc-HA. As shown in Fig. 3 E, mInsc-HA was present at levels comparable with the 8-h induction, and it still localized to the apical region of mitotic cells. Furthermore, LGN clearly colocalized with mInsc-HA at the apical membrane of mitotic cells (Fig. 3 E). Unexpectedly, we also observed mInsc-HA and LGN colocalization in some cells dividing symmetrically (Fig. 3 F). Thus, although mInsc–LGN complexes have been a useful marker for perpendicular spindles, their localization can clearly be uncoupled from spindle orientation. LGN directly binds to NuMA, which is an important effector for spindle orientation (Du et al., 2001; Srinivasan et al., 2003; Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). NuMA localizes to spindle poles in all mitotic cells and to the apical cell cortex in cells with spindles perpendicular to the basement membrane (Fig. 3 G; Lechler and Fuchs, 2005). However, NuMA did not colocalize with mInsc-HA in symmetrically dividing cells (Fig. 3 H). These data reveal a control point downstream of mInsc expression, which regulates the localization of the machinery driving spindle reorientation. In addition to the uncoupling of mInsc from spindle orientation, we also saw an increase in cells dividing at oblique angles relative to the basement membrane. This effect was much more common in cells expressing mInsc-HA (21%; n = 48) than surrounding cells, which did not (4%; n = 47). Although further investigation into these divisions is required, these results are consistent with competition between machinery for SCD and ACD or with external tissue forces altering division orientation.
Uncoupling of Insc from spindle orientation was also found in transgenic embryos constitutively expressing GFP-mInsc from the K14 promoter (Fig. S3, A–C). During SCDs, GFP-mInsc colocalized with LGN but not with NuMA (Fig. S3, C and E).
To determine whether mInsc–LGN can be uncoupled from division orientation in wild-type embryos, we examined cells with parallel spindles to determine whether any of them had apically localized LGN. A small percentage of these cells (∼5% of cells with parallel spindles; n = 41) had apical LGN, suggesting that mInsc–LGN can be functionally uncoupled from the mitotic spindle under normal conditions. Thus, uncoupling increases from 5% in wild-type embryos to 9% after 8-h mInsc induction to almost 40% at 3-d induction.
The aforementioned data implicate a regulated recruitment of NuMA by mInsc–LGN in controlling spindle orientation and reveal that events downstream of mInsc transcription can alter division orientation. We suggest that cells use expression of mInsc as a set point for ACDs but can quickly alter division orientation by controlling NuMA recruitment. This may be important to allow cells to quickly respond to environmental changes, allowing them to choose the correct division orientation as circumstances change.
Because mInsc expression controls division orientation later in development, we wanted to determine whether its expression is regulated in the commitment to stratification that occurs around e13.5. Before this time, the majority of cell divisions occur parallel to the basement membrane (∼75%). We prepared epidermal extracts from e11.5 and e13.5 mice and compared levels of Par3, mInsc, and LGN by Western blotting (Lechler and Fuchs, 2005). Levels of Par3 and LGN did not vary at the two time points. In contrast, mInsc levels were low in e11.5 epidermis and increased markedly by e13.5 (Fig. 4 A). The temporal expression pattern as well as the inducible mInsc-HA expression results suggest that the control of mInsc expression is one important factor contributing to commitment to stratification.
Figure 4.
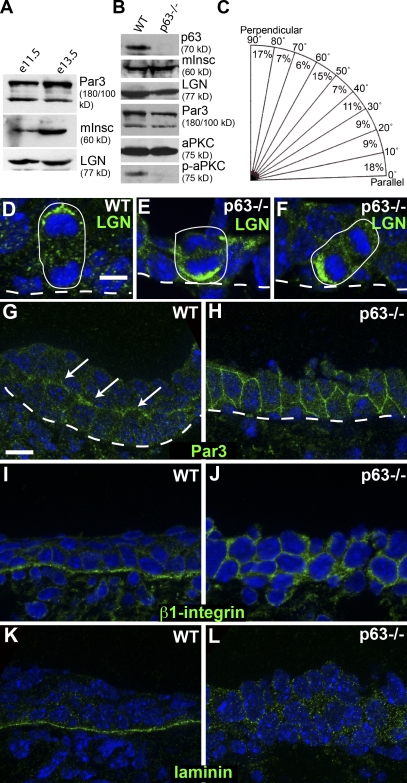
p63 does not control mInsc expression but is required for proper cell division orientation. (A) Western blot analysis of e11.5 and e13.5 epidermis. (B) Western blot of e14.5 wild-type (WT) and p63-null epidermis. (C) Mitotic spindle orientations in e14.5 p63-null epidermis. (D–F) Localization of LGN (green) in anaphase wild-type (D) and p63-null epidermal cells (E and F). (G and H) Localization of Par3 (green) in wild-type (G) and p63-null (H) epidermis. Arrows denote apical accumulation of Par3. (I and J) Localization of β1-integrin (green) in wild-type (I) and p63-null (J) epidermis. (K and L) Localization of laminin (green) in wild-type (K) and p63-null (L) epidermis. Dashed lines indicate the basement membrane. Circles indicate boundaries of mitotic cells. Bars, 10 µm.
An excellent candidate for a regulator of mInsc expression is p63, a transcription factor required for epidermal stratification (Mills et al., 1999; Yang et al., 1999). To determine whether p63-null mice are able to express mInsc, we prepared protein extracts from wild-type and littermate p63-null embryos at e14.5. These samples had equivalent expression of mInsc, demonstrating that p63 is not essential for mInsc expression (Fig. 4 B). However, e14.5 p63-null embryos displayed random division orientation (Fig. 4 C). Although LGN remains polarized at the cell cortex, it is no longer exclusively found apically (Fig. 4, D–F). Phenotypically, this is very similar to mutations in β1-integrin, which lead to loss of cell–matrix interactions and cell polarity (Lechler and Fuchs, 2005). To directly determine whether polarity markers are altered, we examined the localization of Par3. In wild-type epidermis, Par3 was weakly enriched at the apical end of the basal cells (Fig. 4 G). In p63-null mice, Par3 was present at cell–cell junctions all around the cell (Fig. 4 H). Although levels of Par3 and aPKC were normal, phosphorylation of aPKC, reflective of its activation, was considerably diminished in p63-null epidermis (Fig. 4 B).
A previous study demonstrated that p63 controls cell–matrix interactions by regulating the expression of several integrins and extracellular matrix components (Carroll et al., 2006). In support of this, β1-integrin was mislocalized around the cell, and there was no enrichment for laminin in the basement membrane of the p63-null epidermis (Fig. 4, I–L). These data are consistent with polarity defects in p63-null epidermis being secondary to defects in cell–substratum adhesion. However, we cannot rule out the possibility that p63 plays more direct roles in controlling epithelial polarity. Regardless, these data strongly suggest that p63 does not directly control mInsc expression.
We have demonstrated that epidermal progenitors choose between SCD and ACD. As division orientation directly controls morphogenesis, this decision is tightly regulated. Two control points have been identified: expression of mInsc and recruitment of NuMA to the apical cell cortex. The physiological factors that control these are likely to be key regulators of epidermal development. In addition, understanding how environmental signals impinge on these control points will be an important area of future investigation.
Materials and methods
Lineage-tracing experiments
K14-CreER mice (provided by E. Fuchs, the Rockefeller University, New York, NY; Vasioukhin et al., 1999) were mated with either Rosa-lox-stop-lox-GFP (provided by F. Wang, Duke University, Durham, NC) or Rosa mT/mG mice (Jackson ImmunoResearch Laboratories, Inc.; Muzumdar et al., 2007). When embryos were e14.5, the pregnant dams were injected intraperitoneally with tamoxifen. For Rosa-lox-stop-lox-GFP mice, a dose of 10 µg/g of the dam was used. For Rosa mT/mG mice, a dose of 5 µg/g of the dam was used. After 16–20 h, embryos were harvested, cryoembedded, sectioned, and analyzed via immunofluorescence.
Analysis of mitotic spindle orientation
Centrin-GFP and NuMA-GFP embryos were sacrificed at e14.5, cryoembedded, and sectioned for analysis. After staining, sections were covered with coverslips (no. 1.5; VWR International). Images were collected using a microscope (AxioImager Z1) with an ApoTome attachment, a 63× 1.4 NA Plan Apochromat objective, and AxioVision software (all obtained from Carl Zeiss, Inc.). Immersion oil was used (Immersol 518F; Carl Zeiss, Inc.). Images were collected at room temperature using a camera (AxioCam MRm; Carl Zeiss, Inc.). Maximum intensity projections are presented for tissue images. Photoshop (Adobe) was used for postacquisition processing of brightness and contrast. Spindle angles were measured with respect to the basement membrane using ImageJ (version 1.40G; National Institutes of Health). Angles within 30° of the basement membrane were counted as symmetric divisions. Similarly, angles >60° with respect to the basement membrane were counted as asymmetric divisions.
mInsc induction
K14-rtTA mice (provided by E. Fuchs) were mated with TRE-mInsc-HA mice (Nguyen et al., 2006). When embryos were at e14.5, pregnant dams were injected with 200 µg doxycycline. For short-term induction of mInsc-HA, a single injection was given, and the embryos were taken 8 h later. For long-term induction, the pregnant dam was injected daily in addition to being fed doxycycline chow (PicoLab rodent diet [LabDiet] with 0.1% doxycycline). Timing of induction did not alter results, as an 8-h induction at e16.5 led to an increase in ACDs (86%; n = 50). In addition, 3-d treatment with doxycycline did not cause changes in proliferation rate or apoptosis.
Time-lapse imaging
Cultured centrin-GFP– or NuMA-GFP–expressing cells were grown in glass-bottom dishes (MatTek) with cover glass (no. 1.5). They were imaged with a swept-field confocal (VT-Infinity; Visitech) mounted on a microscope (DMI6000; Leica) with a 63× 1.4 NA Plan Apo objective. Immersion oil (Leica) and a camera (OrcaER; Hamamatsu Photonics) were also used. Cells were maintained at 37°C and 6% CO2. Simple software was used for image acquisition (PCI eCommerce Solutions). Postacquisition modification was performed with Photoshop.
Immunofluorescence and Western blot analysis
The following primary antibodies were used in this study: rabbit anti-GFP (Invitrogen), mouse anti–keratin10 (Thermo Fisher Scientific), rabbit anti-NuMA (Abcam), rat anti-HA (Roche), rabbit anti-LGN (provided by W. Chia, National University of Singapore, Singapore), rabbit anti-Par3 (Millipore), rabbit anti–β1-integrin (Millipore), rabbit anti-laminin, rabbit anti-aPKC (Santa Cruz Biotechnology, Inc.), rabbit anti–phospho-aPKC (Cell Signaling Technology), mouse anti-p63 (Santa Cruz Biotechnology, Inc.), and rabbit anti-Insc (laboratory generated). Secondary antibodies included Alexa Fluor 488–conjugated antibodies (Invitrogen) and Rhodamine red– and horseradish peroxidase–conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc.). TIF images were imported into Photoshop, where they were modified for brightness and contrast.
Predicting ratios of three-cell clones in lineage-tracing experiments
Of the two-cell clones analyzed, 67% were asymmetric, whereas 33% were symmetric. If all cells are unipotential in their division orientation, we would expect to see no clones consisting of two basal cells and one suprabasal cell. If division orientation is essentially random at each division, than we would expect the following: 0.33 × 0.33 × 100% = 11% of clones as three basal cells (two SCDs); 0.67 × 0.67 × 100% = 45% of clones with one basal cell/two suprabasal cells (two ACDs); and 2(0.67 × 0.33) × 100% = 44% of clones with two basal cells/one suprabasal. For one ACD/one SCD, this population occurs not only from an ACD followed by an SCD, but also the converse. For clarity, four cell clones were not quantitated. Although this may bias the results, it does not alter the basic conclusions of this experiment.
Initially, a Pearson χ2 goodness of fit test was used to test whether the observed data deviated from the predicted. A one-sample z test was performed and indicated that there is no evidence for a statistically significant difference between the observed and predicted numbers of clones resulting from two ACDs or from one ACD and one SCD (based on randomness in division orientation). However, a statistically significant increase in clones resulting from two SCDs (P = 0.00003) was noted. This may reflect either a preference for a cell that has divided symmetrically to divide symmetrically again or the presence of a small population of cells that divide exclusively symmetrically. However, it most likely results at least in part from a small percentage of cells in which recombination occurred in adjacent cells.
Online supplemental material
Fig. S1 shows keratin 10 expression in basal and suprabasal epidermis. Fig. S2 shows characterization of a NuMA-GFP transgenic mouse model. Fig. S3 shows characterization of mInsc-GFP transgenic embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008001/DC1.
Acknowledgments
We thank Matthew Heaton for help with statistical analysis, Fan Wang, Elaine Fuchs, and Alea Mills for mouse strains, Cheryl Bock and the Duke Transgenic Core facility for microinjection, and Amanda Snyder and Julie Underwood for mouse care. Kaelyn Sumigray provided valuable technical/intellectual input, and Adelheid Soubry contributed to initial experiments. We thank Michel Bagnat, Scott Soderling, Fan Wang, and members of the Lechler Laboratory for comments on the manuscript.
This work was supported by the National Institutes of Health (grant R03-AR056045 to T. Lechler), the Sidney Kimmel Foundation, the Alexander and Margaret Stewart Trust, and the Duke Cancer Center.
Footnotes
Abbreviations used in this paper:
- ACD
- asymmetric cell division
- Insc
- Inscuteable
- LGN
- Leu-Gly-Asn–enriched protein
- mInsc
- mouse Insc
- SCD
- symmetric cell division
- TRE
- tetracycline/doxycycline-responsive promoter
References
- Bowman S.K., Neumüller R.A., Novatchkova M., Du Q., Knoblich J.A. 2006. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 10:731–742 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Carroll D.K., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W. 2006. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8:551–561 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- Du Q., Stukenberg P.T., Macara I.G. 2001. A mammalian partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3:1069–1075 10.1038/ncb1201-1069 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Hisata K., Raabe T., Matsuzaki F. 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8:586–593 10.1038/ncb1409 [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L.Y., Jan Y.N., Knoblich J.A. 1996. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 383:50–55 10.1038/383050a0 [DOI] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 398:708–713 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., Fuchs E. 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 127:171–183 10.1016/j.cell.2006.07.036 [DOI] [PubMed] [Google Scholar]
- Parmentier M.L., Woods D., Greig S., Phan P.G., Radovic A., Bryant P., O’Kane C.J. 2000. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20:RC84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., González C. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Rusan N.M., Peifer M. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M., Schaefer M., Knoblich J.A. 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 402:548–551 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Cabernard C., Doe C.Q. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- Smart I.H. 1970. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br. J. Dermatol. 82:276–282 10.1111/j.1365-2133.1970.tb12437.x [DOI] [PubMed] [Google Scholar]
- Srinivasan D.G., Fisk R.M., Xu H., van den Heuvel S. 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 17:1225–1239 10.1101/gad.1081203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein L., Wise B., Fuchs E. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA. 96:8551–8556 10.1073/pnas.96.15.8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Jones D.L., Fuller M.T. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 301:1547–1550 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 398:714–718 10.1038/19539 [DOI] [PubMed] [Google Scholar]