Abstract
Purpose of review
The identification of transcriptional activators and repressors of hair cell fates has recently been augmented by the discovery of microRNAs (miRNAs) that can function as post-transcriptional repressors in sensory hair cells.
Recent findings
miRNAs are ~21 nucleotide single-stranded ribonucleic acids that can each repress protein synthesis of many target genes by interacting with messenger RNA transcripts. A triplet of these miRNAs, the miR-183 family, are highly expressed in vertebrate hair cells, as well as a variety of other peripheral neurosensory cells. Point mutations in one member of this family, miR-96, underlie DFNA50 autosomal deafness in humans and lead to abnormal hair cell development and survival in mice. In zebrafish, overexpression of the miR-183 family induces extra and ectopic hair cells, while knockdown reduces hair cell numbers. Genetically-engineered mice with a block in miRNA biosynthesis during early ear development, or during hair cell differentiation, reveal the necessity of miRNAs at these crucial time points.
Summary
Because miRNAs can simultaneously down-regulate dozens to perhaps hundreds of transcripts, they will soon be explored as potential therapeutic agents to repair or regenerate hair cells in animal models.
Keywords: microRNA, hair cells, DFNA50, miR-96, miR-182, miR-183
Introduction
The damage or destruction of sensory hair cells can have a variety of underlying etiologies, including acoustic trauma, genetic disorders, infectious diseases, environmental agents or aging (presbycusis). Presbycusis is by far the most prevalent cause of hearing loss in human populations, with relatively limited options available for therapeutic treatments. One can imagine that an aging hair cell struggles to maintain its stereociliary bundle, mechanosensory channels, tip links, ribbon synapses, apical tight junctions, and/or all the other specialized components needed for detection and transmission at maximal sensitivity. Furthermore, aging could reduce the efficacy of subcellular repair following damage due to oxidative stress or ionic imbalance. The task of rejuvenating a geriatric cell seems formidable. Ideally, the aging cell needs a magic bullet, a hair-cell-specific “fountain of youth” that could regulate the levels of a huge suite of proteins that will, in toto, return the hair cell to a more youthful state. Such is the potential impact of the newly-emerging field of microRNA therapeutics, although at this stage it is but a twinkle in the eye of the prescient optimists among us. This article will summarize recent progress in the study of microRNAs that are associated with the vertebrate inner ear, with a special emphasis on those that are present in sensory hair cells and thus may offer a strategy for hair cell repair or regeneration.
Gene regulation by transcription factors and microRNAs
An exploration of the molecular factors that regulate inner ear development and hair cell specification has yielded considerable insights in the past decade. One of the most exciting discoveries came in 1999, with the observation that the helix-loop-helix transcription factor, Atonal-1 (Atoh1, formerly Math-1), is absolutely necessary for hair cell development in mice [1]. In its absence, cochlear and vestibular hair cells were missing and even supporting cell development failed to progress. In subsequent studies, Atoh1 transduction was also found to be sufficient to specify hair cells ectopically in mammalian hearing organs in organ culture [2, 3] and in vivo [4, 5]. In guinea pigs, virus-mediated delivery of Atoh1 following treatment with ototoxic drugs can either rescue or prevent hair cell destruction [6]. Although Atoh1 is expressed in selected other cell types elsewhere in the body, its role in the inner ear is particularly provocative for the hair cell.
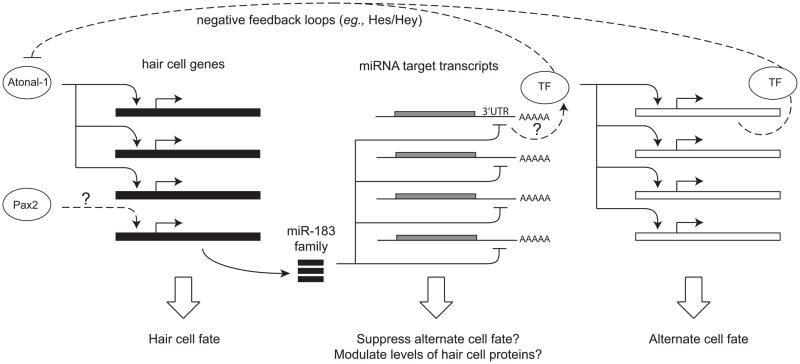
Transcriptional activators such as the Atoh1 protein can be powerful mediators of cell fate specification because of their ability to upregulate multiple gene products needed for cell-specific differentiation (Figure 1). Transcriptional repressors can be equally important, playing a role in transitioning cells from cycling progenitors into committed post-mitotic precursors, or in suppressing cells from acquiring alternative fates. In the mammalian inner ear, members of the Hes/Hey family of transcription factors act downstream of Notch signaling to mediate lateral inhibition during development and are implicated in the repression of hair cell fates (Figure 1). Furthermore, their expression in supporting cells suggests that they may directly or indirectly promote these phenotypes as an alternative to the hair cell fate, sometimes via a Notch-independent pathway [7].
Figure 1. Regulatory factors that can influence cell fates.
Cell fate specification relies on different types of regulatory factors that can each control multiple downstream targets. Shown here are examples of transcription factors (TF, Atonal, Pax2) that can modulate several genes at once by binding to the regulatory regions on the gene that lie upstream of the coding sequence to turn on (as shown) or off (not shown) the transcription of messenger RNA (mRNA). Transcription factors play a key role in the progenitor cells assuming one or another cell fate (such as hair cell vs. supporting cell). Small non-coding microRNAs, such as the miR-183 family, can down-regulate several genes at once by binding to the 3′ untranslated region (3′UTR) of their mRNAs. Some of transcripts that are targeted by the miR-183 family might encode inhibitors of the hair cell fate (such as the Hes/Hey transcription), positive transcriptional regulators of alternate fates, or proteins required by hair cells whose levels must be precisely modulated.
A relatively new class of negative regulators has been discovered that act not at the level of transcription but at the level of protein translation. These are the microRNAs (miRNAs), small single-stranded RNAs typically ~21 nucleotides in length in their mature, cytosolic forms. Like transcriptional repressors, miRNAs can act on many genes simultaneously (Figure 1). These small regulators base pair with target sequences on the 3′ untranslated regions (3′UTRs) of messenger RNA (mRNA) transcripts and block protein synthesis. The binding of miRNAs to their target transcripts is imperfect in animals, which significantly challenges our ability to predict which transcripts are bone fide targets for particular miRNAs. Several different algorithms are currently in use, most of which show that individual mRNAs often have many potential miRNA binding sites, and conversely each miRNA is predicted to have up to hundreds of distinct target transcripts [8]. Verification of the target genes is an important and active area of research. For a small subset of miRNAs, whole-scale proteomics approaches have indeed confirmed that hundreds of transcripts can be repressed in response to the forced expression of a single miRNA [9, 10].
The miRNA-183 family in sensory cells
Expression of the miRNAs in the mouse inner ear was initially studied with microarray analyses, in which over 100 miRNAs were detected in postnatal ear samples [11]. A few of these were verified by in situ hybridization (Figure 2). The miR-183 family, consisting of the miR-96, miR-182 and miR-183 triplet, is of special interest due to its strong and specific expression in hair cells and slightly less intense staining in the spiral and vestibular ganglia of the inner ear [11]. These genes are also expressed in retina, nose, tongue and dorsal root ganglion. The miR-183 family is highly conserved: it shares nearly the same sequence from human to zebrafish. Similarly, in the zebrafish, expression of the miR-183 family in hair cells and the statoacoustic ganglia is specific and abundant [14, 15*]. Interestingly, expression of the miR-183 family in ciliated neurosensory organs is conserved among both vertebrates and invertebrates. In Drosophila and C. elegans, homologs to miR-183 (miR-263b and miR-228, respectively) are expressed in sensory organs [16*]. Such conserved expression of miR-183 in highly-diverged peripheral sensory organs might be critical to tissue identity during evolution [17*].
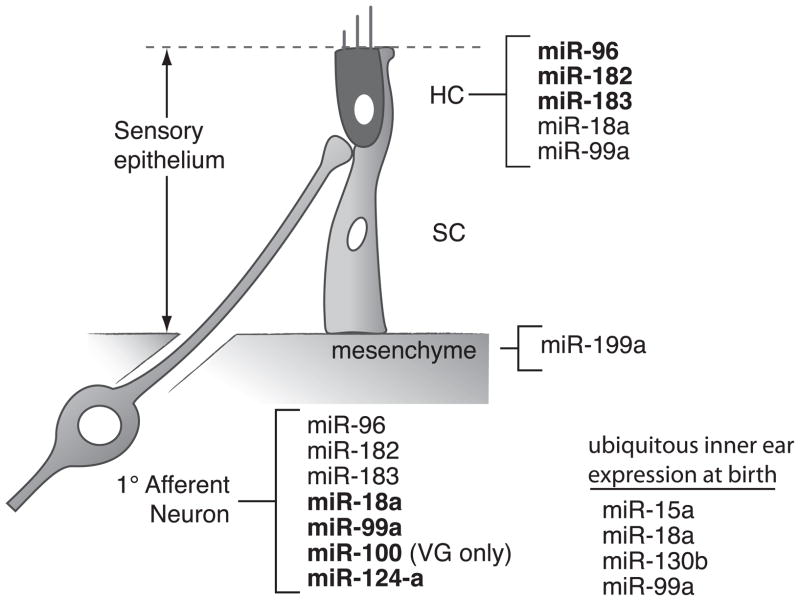
Figure 2. Many microRNAs are expressed in the mammalian cochlea.
Schematic showing miRNA gene expression in the mouse inner ear at birth based on in situ hybridization. Bold denotes the location showing relatively stronger expression. Data are based on [11–13*].
Expression of the miR-183 family is dynamic and closely related to specification of sensory cells in the vertebrate inner ear. During embryonic development of the mouse, the miR-183 triplet is first expressed in the entire otic vesicle and spiral and vestibular ganglion neurons, and then becomes restricted to hair cells and their associated neurons. The family is also observed in the greater epithelial ridge at embryonic days 15.5–17.5, and in the inner sulcus and the spiral limbus from postnatal days 4–11 [13*]. Although the miR-183 family members are detected in adult mouse inner ear using microarray data, detailed in situ hybridization shows their disappearance from hair cells and the spiral ganglion during the second and third postnatal weeks, respectively [11, 13*]. In contrast to the mouse, in zebrafish the three miRNAs are not detected in sensory regions prior to hair cell differentiation [15*]; like the mouse, zebrafish also express these genes in the otic ganglion.
Mutations in miR-96, a member of the miRNA-183 family, underlie deafness
The specific expression and conservation of the miR-183 family predicts their potential importance in the inner ear. Binding between miRNAs and their targets largely relies on a perfect match to the 2nd to 8th nucleotides of the miRNA, named the seed region [18]. Recently, mutations in the seed region of miR-96 were found to cosegregate with inherited sensorineural hearing loss at the DFNA50 locus in human families [19**]. One family shows a G>A change of the fifth nucleotide of mature miR-96. A second family has a C>A change in the sixth nucleotide of the seed (also indicated as position 14 of the gene). In parallel, a mutant mouse called diminuendo has an A>T change of the seventh nucleotide of mature miR-96. Diminuendo mice have progressive hearing loss in heterozygotes. The homozygotes have irregular hair cell bundles and hair cells degenerate rapidly after birth [20**].
To find out how mutations in miR-96 are related to these disorders, both groups investigated whether mutant miR-96 loses its ability to downregulate original target genes of miR-96. Interactions between human miR-96/mutant miR-96 and five potential target genes, chosen due to their association with the ear in animal models, were tested in vitro using luciferase assays. Indeed, mutant miR-96 is less effective at repressing the target genes compared to the wildtype miR-96 [19**]. In addition, the expression levels of the transcripts in the mouse ear were also quantified, and two of them are increased slightly in diminuendo [20**]. Results from human and mouse agree that certain target genes are de-repressed (i.e. up-regulated) in conjunction with miR-96 seed mutations (Figure 3A). However, the impact on the translation of individual targets is surprisingly subtle, and could not be confirmed by visualizing the levels of the encoded proteins in immunohistochemistry of tissue sections.
Figure 3. The human miR-96 gene has been linked with human deafness and efforts to understand the etiology have focused on its interactions with predicted target transcripts.
(A) The 3′ untranslated region (3′UTR) of adenylate cyclase 6 (ACDY6) messenger RNA has a perfect match to the seed region (positions 2–8) of miR-96 (and miR-182) and is negatively regulated by miR-96 in an in vitro luciferase assay [21**]. The ADCY6 protein is expressed at the base of the stereociliary bundle in the mouse cochlea [22], as noted by Xu and collaborators [21**]. (B) Mutations in the seed region of miR-96 causes autosomal dominant hearing loss in humans [19**]. The 14C>A mutation (boxed) is predicted to acquire many new targets for miR-96, including the SLC19A2 gene shown here. (C) The search for mutations in predicted binding sites of the miR-183 family that segregates with human deafness revealed a C>A mutation (boxed) in the 3′UTR of the RDX gene in one human family [23*]. This mutation should interfere with binding between the 3′UTR and the seed region of miR-96, although this could not be verified by an in vitro luciferase assay.
Importantly, the impact of the miR-96 diminuendo mutation is not limited to the de-repression of target genes. A subset of genes is more downregulated in diminuendo as compared to wildtype and these often contain a sequence complementary to the mutant miR-96 seed region (Figure 3B). Thus, the mutant miRNA acquires new targets whose translational repression could be responsible, in whole or in part, for the mutant phenotypes in mouse and human [19**, 20**]. Interestingly, one such potential new target gene, SLC19A2, causes hair cell loss and auditory neuropathy when it is deleted in knockout mice [24]. Genome-wide profiling of gene expression in diminuendo also shows large-scale up- and down-regulation of gene transcripts, either of which could reflect an indirect impact of the miR-96 mutation [20**].
If interactions between miRNAs and their targets depend on a precise sequence match of the seed regions, then mutations in the 3′UTR of target genes should also compromise the fine regulation of gene expression. Such target mutations might also lead to hearing loss. Additional screening for mutations in the miR-183 family or in their predicted target genes identified a C>A mutation in a predicted binding site of miR-96/182 in the RDX 3′UTR in a human family with autosomal recessive non-syndromic deafness (Figure 3C) [23*]. Although in vitro luciferase assays fail to show an interaction between RDX and miR-96/182, RDX’s potential as a target of other miRNAs remains. Therefore, besides a gene’s protein coding region, mutations in the non-coding 3′UTR, where miRNAs typically bind, can be important sites for disruption of gene function (also shown first in worms and flies).
Using zebrafish to reveal the functions of the miR-183 family in the inner ear
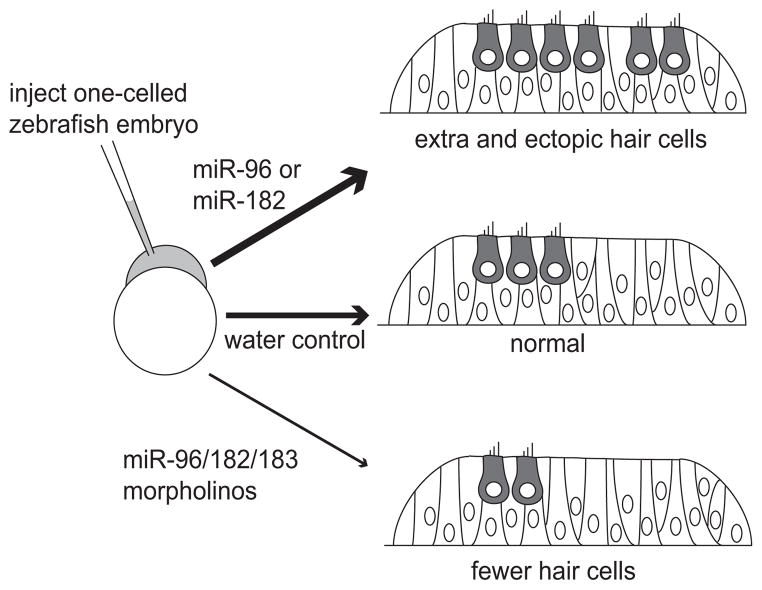
Our understanding of the miR-183 family in the inner ear was further advanced through direct manipulation of their levels in the zebrafish. Data show that individual miRNAs are capable of influencing the developing sensory epithelium to promote the hair cell fate [15*]. Synthesized miRNAs are injected into 1-celled embryos, and are overexpressed ubiquitously during the first few days of development. Overexpression of miR-182 or miR-96 induces duplicated otic vesicles, ectopic sensory patches and excess hair cells at stages when the earliest hair cells first appear (Figure 4). As more hair cells are added to sensory patches, expanded sensory patches and excess hair cells are found in miR-182-injected embryos. In contrast, miR-96-injected embryos discontinue the production of extra hair cells at this later stage, while miR-183 injection never produces ectopic or excess hair cells [15*]. This study broadened our mind to consider that hair cell fate can be influenced not only by transcriptional activators like the Atoh1 protein, but also repressors like the miRNAs which can function post-transcriptionally. Specifically, overexpression of miRNAs is a valid test for the sufficiency of miRNAs in regulating cell fates.
Figure 4. The miRNA-183 family regulates hair cell numbers in the developing zebrafish.
Injection of reagents into the one-celled embryos is followed by analysis of their inner ears at 1–2 days post-fertilization. Excessive amounts of miR-96 or miR-182 (but not miR-183) generate too many hair cells one day later (thick arrow). Reduced levels of each of the family members resulting from antisense morpholino injections lead to the generation of too few hair cells two days later (thin arrow). Results are based on [15*].
Knockout of the miRNAs is a test of their necessity during development. In zebrafish, we explored the necessity of the miR-183 family in inner ear development by knockdown of the miRNAs using antisense morpholinos. Morpholinos complementary to each miRNA were designed and injected into 1-celled zebrafish embryos (Figure 4). Knockdown of miR-183/96/182 individually or in combinations results in fewer hair cells, smaller otic ganglia, and defects in semicircular canals in the inner ear [15*]. Loss of hair cells in the sensory organ is proportional to the amount of morpholinos injected, and is more severe following combined miRNA knockdown than with individual knockdown. Therefore, the absolute levels of the miR-183 family members are critical to obtain a normal number of hair cells. Although the impact of the miR-183 family on hair cell number is profound, its role in the sensory epithelia is more of a fine tuner than a switch of hair cell fate: even in the apparent absence of all three miRNAs, about half the normal number of hair cells are still present [15*].
If the total level of the miR-183 family remains constant, another interesting question is whether members of the miR-183 family can substitute for each other. By overexpressing miR-182 in embryos with miR-96 knockdown, we found that hair cells loss is partially rescued (i.e. increased more than miR-96 knockdown embryos, but decreased relative to normal embryos) [15*]. We speculate that by sharing redundant functions, the three miRNAs may provide better capability to buffer environmental fluctuations and to ensure a precise number of hair cells is produced.
Using mouse models to explore the necessity of all miRNAs in hair cells
Others have used a knockdown approach to test the necessity of all miRNAs in the development of hair cells. Results show that a total elimination of miRNAs in hair cells yields more a severe phenotype when compared to knockdown of the miR-183 family alone. Mutant mice are generated through conditional Cre-mediated knockout of Dicer specifically in Pou4f3-expressing cells [12*]. The Dicer protein is a type III ribonuclease required for miRNA biogenesis, and Pou4f3 is expressed in hair cells of both cochlea and vestibule. Thus, miRNA production is interrupted in newly-generated hair cells of the mutants. At postnatal day 38, mutant mice are deaf, and hair cells in the cochlea are abnormally shaped and missing stereocilia. Vestibular hair cells are less disrupted, showing only disorganized stereocilia [12*]. These results reveal the importance of miRNAs in maintaining specialized structures and survival of hair cells. This study suggests that the combined impact of all miRNAs is essential and substantial, while phenotypes resulting from knockdown of individual miRNAs might be more moderate. Nevertheless, it is worth noting a surprising exception: the knockout of miR-9a in Drosophila wing development causes more profound defects than a Dicer-mediated knockout of all miRNAs in this organ [25]. This implies that individual miRNAs may have opposing effects under wildtype conditions, which are effectively cancelled out when both functions are eliminated but not when only one is absent.
Differences in hair cell phenotypes between the cochlea and vestibule of Pou4f3-Cre:Dicer knockout mouse might be explained by their composition of miRNAs. Surprisingly, comparison of miRNA expression profiles in the wildtype mouse cochlea and vestibule at P0 reveals limited differences [12*]. Potential hearing-related miRNAs were identified by matching their chromosomal loci with known deafness loci and several were chosen for further analysis. Examples include miR-15a, miR-99a and miR-18a that are expressed in hair cells of both the cochlea and the vestibule (Figure 2). Homologs of miR-15a and miR-18a in zebrafish are shown to affect sensory organs and morphology of the inner ear [12*]. Moreover, absolute levels of miRNAs and their target genes might be different in cochlear and vestibular hair cells, revealing distinct degrees of repression by miRNAs. Claudin12 is a predicted target gene of miR-15a, and its protein is detected in cochlear hair cells but not vestibular hair cells [12*]. It is possible that Claudin12 translation is repressed completely by miR-15a in the vestibule, but only moderately in the cochlea.
Using mouse models to explore the necessity of all miRNAs in the entire inner ear
Just as Cre-mediated excision of Dicer was used to test the necessity of all miRNAs in developing hair cells, this approach was also used to ask about miRNA function throughout the entire inner ear. Over 100 miRNAs are detected in the inner ear and many of them are abundant in sensory patches or ganglia, such as miR-100, miR-125b (Figure 2) and miR-133a [11]. Furthermore, a few are differentially expressed in auditory versus vestibular sensory organs during development [12*]. Conditional knockout of Dicer at early stages was accomplished by putting the Cre recombinase under the control of the Pax2 promoter, which is widely expressed in the otic placode. In this way, global miRNA processing is interrupted before specification of sensory patches. By embryonic day 17.5, depletion of miRNAs adversely affects the morphogenesis of the entire ear, including nearly all the sensory organs. In the abnormally uncoiled cochlea, the prosensory region shrinks and hair cells are disarranged. Additionally, neurons in the ganglia are sparse, and afferent innervation to sensory patches is largely reduced. In contrast, the posterior crista is the only intact sensory patch, and displays normal hair cell stereocilia. Interestingly, residual expression of mature miR-183 is found in the posterior crista, and correlates well with the formation of those sensory patches and their innervation [26**].
The search for miRNA target genes
The functions of miRNAs should be closely related to their targets. Currently, it is believed that miRNAs change a cell’s gene expression profile on multiple levels, including direct repression of target genes, indirect influence on transcripts, and alternative splicing of mRNAs [27]. The global impact of miRNAs can modulate cell fates, and might be utilized for repair and regeneration of specific cell types. For example, abundant expression of the miR-183 family in hair cells might help differentiate and maintain their apical structures. During aging, hair cells gradually lose their apical structures and hearing threshold increases. Through boosting levels of the miR-183 family in hair cells, those apical structures and hearing ability might be restored. In addition, overexpression of miR-182 yields extra and ectopic hair cells in the inner ear. One possibility is that ectopic expression of miR-182 in supporting cells could repress supporting cell genes and convert them into hair cells. Thus, for hearing loss caused by missing hair cells, directional delivery of miR-182 into supporting cells might facilitate the regeneration of hair cells.
Previous authors have suggested that the miR-183 family may repress either prosensory genes or supporting cell genes to help specify hair cell fate, and predicted Sox2, a prosensory marker, as a target of miR-182 [28]. However, there was no visible up-regulation of Sox2 revealed by antibody in the zebrafish inner ear with miR-182 knockdown [15*]. miRNAs can also promote de-differentiation of somatic cells into stem cells, which can be induced into desired cell types. Let-7 is shown to repress self-renewal of embryonic stem cells, and inhibition of it helps switch somatic cells into pluripotent stem cells [29]. Two major approaches of miRNA therapy include inhibition of undesired miRNAs and delivery of desired miRNAs. For knocking down undesired miRNAs, systematic delivery of modified short RNAs with complementary sequence to target miRNAs can be effective. In addition, constructs designed to contain multiple binding sites of related miRNAs inhibit their function by absorbing them, and can be delivered through viral infection. Increase of desired miRNAs has been shown in mice through viral delivery of miRNA expressing constructs [30].
Conclusion
There is palpable excitement about recent new discoveries that implicate miRNAs in hearing and deafness. A considerable amount of work lies ahead to reveal how these negative regulators function in the mammalian inner ear. Future studies will also include designing vectors to deliver miRNAs to residual hair cells or supporting cells in a therapeutic context, with initial testing in animal models.
Acknowledgments
Funding Sources: National Institutes of Health R21DC008997 and R01DC002756
This work was supported by NIH R01DC002756 and R21DC008997 (to D.M.F.). We thank Rodney McPhail for help with the figures.
Reference Section
(*) of special interest
(**) of outstanding interest
- 1.Bermingham NA, Hassan BA, Price SD, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 2.Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 3.Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 4.Kawamoto K, Ishimoto S, Minoda R, et al. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubbels SP, Woessner DW, Mitchell JC, et al. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumikawa M, Minoda R, Kawamoto K, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 7.Doetzlhofer A, Basch ML, Ohyama T, et al. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37:2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 11.Weston MD, Pierce ML, Rocha-Sanchez S, et al. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 12 *.Friedman LM, Dror AA, Mor E, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. Large-scale microarray screening was combined with bioinformatics to identify microRNAs that are expressed in auditory vs. vestibular sensory epithelium in mice. Genetic knockdown of Dicer1, a microRNA-processing enzyme, shortly after hair cells are specified showed that microRNAs are required for hair cell survival postnatally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 *.Sacheli R, Nguyen L, Borgs L, et al. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9:364–370. doi: 10.1016/j.gep.2009.01.003. This study provides a detailed time course of expression of the miRNA-183 family in various cell types of the mouse inner ear through postnatal day 11 using in situ hybridization techniques. It maps the appearance of the miRNAs in the embryonic inner ear and their disappearance from hair cells during the second postnatal week even as they persist in certain non-sensory cells of the cochlea. [DOI] [PubMed] [Google Scholar]
- 14.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 15 *.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30:3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. Three microRNAs highly expressed in hair cells and moderately expressed in the statoacoustic ganglion were evaluated for their roles in the developing inner ear of the zebrafish. Overexpression of the microRNAs generated extra hair cells in the inner ear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16 *.Pierce ML, Weston MD, Fritzsch B, et al. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. Orthologs of miR-183/96 in representative deuterostomes and protostomes were predicted, and their expression in ciliated sensory and neural organs were compared. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 *.Christodoulou F, Raible F, Tomer R, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. Two of the genes in the miR-183 cluster belong to a small group of microRNAs that show conservation of expression in peripheral sensory organs from both protostomes (zebrafish, mouse) and a deuterostome (Platynereis). This ancient evolutionary conservation in bilateria argues for an essential role for miRNA-182 and -183 in sensory cell specification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19 **.Mencia A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. Published simultaneously with Lewis et al., (2009) as the first papers to link microRNAs with deafness. Point mutations in miR-96 were associated with autosomal dominant, progressive, non-syndromic hearing loss in humans that mapped to the DFNA50 locus. [DOI] [PubMed] [Google Scholar]
- 20 **.Lewis MA, Quint E, Glazier AM, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. Published simultaneously with Mencia et al., 2009, with both reports showing for the first time that a microRNA point mutation can be an underlying cause of deafness, in this case in the diminuendo mouse model. Hair cells developed abnormally in both heterozygotes and homozygous mutants. A thorough microarray analysis of the changes in mRNA transcripts in mutants compared to controls showed that hundreds of genes were de-repressed, a few hair cell genes were upregulated, and many new genes were repressed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21 **.Xu S, Witmer PD, Lumayag S, et al. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. Three of the microRNAs that are highly expressed in hair cells are also important in retinal development, as indicated by this large-scale screen for miRNAs expressed in retina. This gene cluster includes miR-183, -96 and -182. The transcripts for Mitf and adenylate-cyclase-6 were shown to be targets of miR-96 and miR-182. [DOI] [PubMed] [Google Scholar]
- 22.Michalski N, Michel V, Bahloul A, et al. Molecular characterization of the ankle-link complex in cochlear hair cells and its role in the hair bundle functioning. J Neurosci. 2007;27:6478–6488. doi: 10.1523/JNEUROSCI.0342-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23 *.Hildebrand M, Witmer P, Xu S, et al. miRNA mutations are not a common cause of deafness. Am J Med Genet Part A. 2010;152A:646–652. doi: 10.1002/ajmg.a.33299. This study represents an important strategy for discovery of new deafness genes based on linking them to miRNAs that are expressed in hair cells. Both the miR-183 family, and several of its predicted target genes, were screened in a series of families with inherited deafness. As additional hair-cell-specific miRNAs are identified, more analyses of this sort will be needed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberman MC, Tartaglini E, Fleming JC, Neufeld EJ. Deletion of SLC19A2, the high affinity thiamine transporter, causes selective inner hair cell loss and an auditory neuropathy phenotype. J Assoc Res Otolaryngol. 2006;7:211–217. doi: 10.1007/s10162-006-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26 **.Soukup GA, Fritzsch B, Pierce ML, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. Knockout of miRNA-processing early during development of the mouse inner ear shows that miRNAs are required for normal development. Morphogenesis of the canals and cochlea, the size and distribution of sensory organs, hair cell morphology and axonal innervation patterns were abnormal when the miRNA-processing enzyme, Dicer1, was deleted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soukup GA. Little but loud: Small RNAs have a resounding affect on ear development. Brain Res. 2009;1277:104–114. doi: 10.1016/j.brainres.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trang P, Weidhaas J, Slack F. MicroRNAs as potential cancer therapeutics. Oncogene. 2009;27:S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]