Abstract
Purpose
Previous literature provides some evidence that atopic diseases, IgE levels, and inflammatory gene polymorphisms may be associated with risk of glioblastoma. The purpose of this study was to investigate the affects of certain inflammatory gene single nucleotide polymorphisms (SNPs) on patient survival. Malignant gliomas are the most common type of primary brain tumor in adults, however, few prognostic factors have been identified.
Experimental Design
Using 694 incident adult glioma cases identified between 2001 and 2006 in Harris County, Texas, we examined seven SNPs in the interleukin 4, interleukin 13, and interleukin 4-receptor (IL4R) genes. Cox proportional hazards regression was used to examine the association between the SNPs and overall and long-term survival, controlling for age at diagnosis, time between diagnosis and registration, extent of surgical resection, radiation therapy, and chemotherapy.
Results
We found that among high-grade glioma cases, IL4R rs1805016 (TT vs. GT/GG) was significantly protective against mortality over time (HR: 0.59; 95% CI: 0.40–0.88). The IL4R rs1805016 and rs1805015 TT genotypes were both found to be significantly associated with survival beyond one year among high-grade glioma patients (HR: 0.44; 95% CI: 0.27–0.73 and HR: 0.63; 95% CI: 0.44–0.91, respectively). Furthermore, the IL4R haplotype analysis showed that SNPs in the IL4R gene may be interacting together to affect long-term survival among high-grade glioma cases.
Conclusions
These findings indicate that polymorphisms in inflammation pathway genes may play an important role in glioma survival. Further research on the effects of these polymorphisms on glioma prognosis is warranted.
Keywords: glioma, survival, IL-4 receptor, inflammation
INTRODUCTION
Although malignant gliomas are the most common type of primary brain tumor in adults, there is a lack of definitive information regarding their etiology, and identification of the prognostic factors that influence patient survival remains incomplete. Median survival time for patients with glioblastoma, the most fatal form of brain tumor, is approximately one year, and 90% die within three years after diagnosis.(1) Therefore, it is important to determine the factors that influence survival for this rapidly fatal disease and by doing so, perhaps contribute to the understanding of the complex biological interactions that regulate glioma development and control.
To date, the primary differentiating factor for glioma survival is tumor histology; patients with glioblastoma experience the worst survival regardless of treatment. However, recent reports suggest that glioma survival can also be modified by germline polymorphisms in several genes, including HLA-A, HLA-B, GLTSCR1, ERCC2, and GSTP1 and GSTM1.(2;3) In addition, a polymorphism in the ataxin 2 binding protein gene (A2BP1 rs8057643) was associated with a significant reduction in time to death in a population of 112 newly-diagnosed glioblastoma patients.(3) Wrensch et al. also reported that the ERCC1 C8092A (rs3212986) and GSTT1 deletion polymorphisms were significantly associated with glioma survival.(2) These studies lend support to the hypothesis that genetic factors may be important in glioma prognosis.
Variants in inflammatory genes contribute to individual susceptibility in risk for atopic disorders, which have been linked to protection against various malignancies, including gliomas.(4;5) It is, therefore, relevant to examine such polymorphisms in relation to not only glioma risk but also survival. Interleukin 4 (IL-4) is important, in conjunction with IL-13, in the regulation of allergic inflammation. These two cytokines interact with heterodimers of IL-4 and IL-13 receptors to directly affect inflammation and allergy through activation of the Janus kinase (JAK) and signal transducer and activator of transcription (STAT) pathways. The interactions of several non-synonymous coding single nucleotide polymorphisms (SNPs) in IL4, IL13, and IL4R have been associated with asthma (6–9), infection-related inflammation (10;11), and glioma risk (12).
While some studies have examined the effects of SNPs in inflammation genes on the risk of developing glioma, few have focused on their effects on glioma survival. Previous etiologic studies provide evidence that polymorphisms in the IL4R gene may play important roles in the pathways that regulate glioma development and control, whether by influencing IgE levels and thus impacting the effectiveness of treatment or through a more direct pathway that is currently unknown. Thus, the purpose of the current study was to examine the association between seven common polymorphisms in the IL4, IL13, and IL4R genes and overall, as well as long-term, patient survival.
MATERIALS AND METHODS
Subjects
The study population consisted of incident adult (over age 18) glioma cases identified by hospital physicians in Harris County, Texas between January 2001 and January 2006. Blood samples were collected from the cases before initiation of chemotherapy or radiation therapy, but in most cases this was after initial surgical resection. The original study population (n=761) was restricted to non-Hispanic whites (n=694) for the genetic analyses presented here. The male to female ratio was 1.4:1 with 92% of patients being non-Hispanic white. Other detailed information on the study population has been previously reported.(13) The study was approved by the institutional review boards (IRB) of all participating institutions, and written informed consent was obtained from each participant.
Determination of vital status
Treatment and survival (overall and disease-free) data were collected from medical record review for all cases. This was done in a systematic way to determine the medical treatment course, dates of treatment, and survival information. For patients not followed at M.D. Anderson Cancer Center, the patient or next-of-kin was contacted as allowed by IRB approval to request release of medical records for abstraction, and to update treatment and survival information.
SNP selection and genotyping
SNPs in the IL4, IL13, and IL4 genes were selected for this pathway-based analysis from a panel of pro- and anti-inflammatory genes. Non-synonymous coding SNPs and SNPs previously reported to be associated with atopic disorders or glioma were selected for genotyping using the Sequenom MassARRAY iPLEX™ platform. This process combines the technologies of mass spectrometry and PCR and primer extension to determine each allele. A major advantage of this platform is that it utilizes minimal DNA, only 5–10 ng, per set of multiplexed assays while provideing call rates of greater than 95%. Quality control analysis included genotyping internal positive control samples, no template controls, and replicates for 10% of the samples. Positive, negative, and DNA controls were organized in specific patterns on the genotyping plates to ensure correct plate orientations during processing and to assist in the QC process and data review.
Statistical Analyses
The distribution of population characteristics was examined, overall and by tumor histology, using the chi-square test for categorical variables and student’s t-test for continuous variables. Analyses were stratified by histology because of dramatic differences in survival for high-grade (IV) versus intermediate-/low-grade (III/II) tumors. Survival time was calculated beginning at the date of hospital registration.
Total survival probability over time and survival probability beyond 12 months were visualized, overall and stratified by genotype, using Kaplan-Meier survival curves created with SAS PROC LIFETEST. Log-rank tests were utilized to determine significant differences (α=0.05) in survival curves stratified by genotype. Furthermore, yearly survival probabilities conditional upon surviving the previous year in two IL4R SNPs (IL4R805015 and IL4R805016) and the IL4R haplotype were calculated among high-grade glioma patients using the Kaplan-Meier life table method.
Cox proportional hazards regression, using SAS PROC TPHREG, was utilized to calculate hazard ratios and 95% confidence intervals for each SNP, adjusting for age at diagnosis, chemotherapy, radiation therapy, extent of surgery, and time between hospital registration and diagnosis among the entire cohort and among those surviving beyond one year. Probable haplotypes for the five IL4R SNPs were calculated using SAS PROC HAPLOTYPE utilizing an expectation-maximization algorithm to calculate the maximum-likelihood estimate of the haplotype frequencies.(14) The SNPs were ordered according to numerical position in the gene: rs1805011, rs1805012, rs1805015, rs1801275, and rs1805016. A hazard ratio and 95% confidence interval were computed for the most probable haplotype, compared to all others, with Cox proportional hazards regression. The proportional hazards assumption for each model was tested using log-log plots; there was no evidence that the proportional hazards assumption was violated for any of the models. All statistical analyses were conducted in SAS, Version 9.1 (Cary, NC).
RESULTS
Of the 694 non-Hispanic white cases included in this analysis, 343 (49.4%) had high-grade (IV) glioma and 351 (50.6%) had low (II) or intermediate (III) grade glioma. Table 1 presents the distribution of demographic characteristics and SNP genotypes by histologic type. High-grade glioma cases were older at diagnosis, on average, and were more likely to have received radiation therapy.
Table 1.
Study population characteristics and genotypes by tumor histology
| By Histology |
|||||
|---|---|---|---|---|---|
| All cases (%) | High Grade (%) | Medium/Low Grade | (%) p-value | ||
| (n=694) | (n=343) | (n=351) | |||
| Sex | |||||
| Male | 419 (60) | 210 (61) | 209 (60) | ||
| Female | 275 (40) | 133 (39) | 142 (40) | 0.65 | |
| Chemotherapy | |||||
| Yes | 473 (70) | 233 (72) | 240 (69) | ||
| No | 201 (30) | 92 (28) | 109 (31) | 0.41 | |
| Radiation Therapy | |||||
| Yes | 573 (83) | 306 (89) | 267 (76) | ||
| No | 121 (17) | 37 (11) | 84 (24) | <.0001 | |
| Surgery Extent | |||||
| Gross Total | 305 (44) | 158 (52) | 147 (48) | ||
| Subtotal | 388 (56) | 185 (48) | 203 (52) | 0.28 | |
| Age at diagnosis | |||||
| median (range) | 45.50 (18.00–72.80) | 52.29 (20.10–72.80) | 38.32 (18.00–65.00) | ||
| mean (sd) | 44.62 (11.78) | 50.38 (9.85) | 38.99 (10.76) | <.0001 | |
| IL4 rs243250 | |||||
| CT/TT | 194 (30) | 94 (29) | 100 (31) | ||
| CC | 449 (70) | 227 (71) | 222 (69) | 0.62 | |
| IL13 rs1800925 | |||||
| CT/TT | 240 (37) | 116 (36) | 124 (39) | ||
| CC | 403 (63) | 205 (64) | 198 (61) | 0.53 | |
| IL4R rs1805011 | |||||
| AC/CC | 133 (21) | 68 (21) | 65 (20) | ||
| AA | 511 (79) | 253 (79) | 258 (80) | 0.73 | |
| IL4R rs1805012 | |||||
| CT/CC | 126 (20) | 65 (20) | 61 (19) | ||
| TT | 517 (80) | 257 (80) | 260 (81) | 0.71 | |
| IL4R rs1805015 | |||||
| CT/CC | 188 (29) | 99 (31) | 89 (28) | ||
| TT | 454 (71) | 220 (69) | 234 (72) | 0.33 | |
| IL4R rs1801275 | |||||
| AG/GG | 232 (36) | 121 (38) | 111 (34) | ||
| AA | 412 (64) | 200 (62) | 212 (66) | 0.38 | |
| IL4R rs1805016 | |||||
| GT/GG | 71 (11) | 37 (12) | 34 (11) | ||
| TT | 573 (89) | 284 (88) | 289 (89) | 0.69 | |
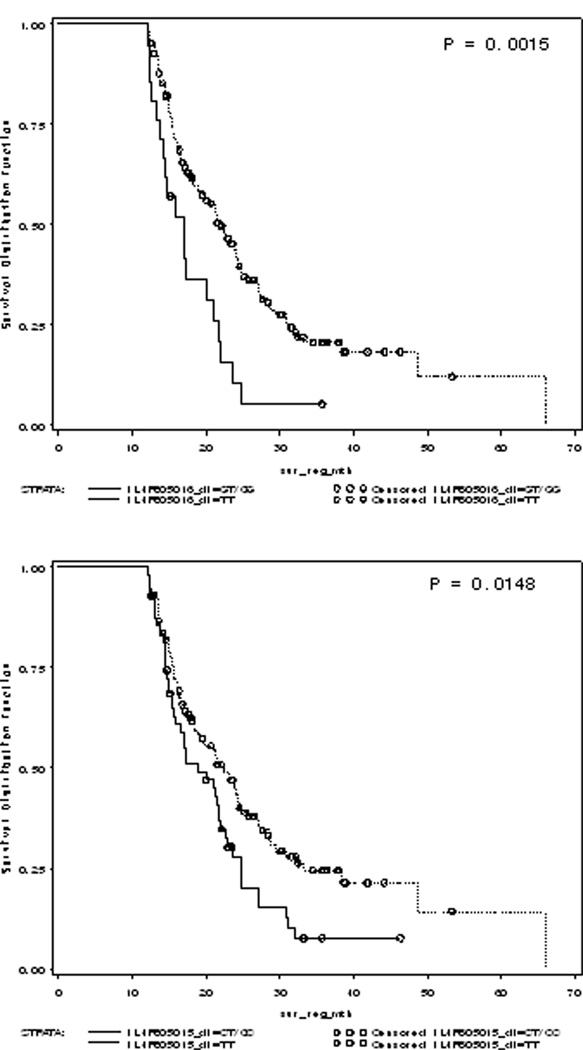
Using Cox regression, we found that the IL4R rs1805016 T allele is significantly protective against mortality over time among high-grade glioma cases (HR: 0.59; 95% CI: 0.40–0.87); although not among the low- and intermediate-grade cases. Furthermore, when we restricted the survival curves to only those high-grade patients who survived beyond 12 months, we saw a significant increase in survival for those carrying the TT genotype of rs1805016 or rs1805015, both in the IL4R gene (Figure 1). This could indicate that events surrounding treatment overwhelm the immune response and once these events have ceased, the genetics of immune function lead to greater differences in survival. None of the other polymorphisms examined were significantly associated with overall or long-term glioma survival for either histological group (Table 2).
Figure 1. Kaplan-Meier survival curves beyond 12 months by genotype for IL4R SNPs among high-grade gliomas.
(A) Patients with the TT genotype for IL4R rs1805016 SNP experienced a median survival 4 months longer than those with the GT/GG genotypes. (B) Patients with the TT genotype for IL4R rs1805015 SNP experienced a median survival of 5 months longer than those with the CT/CC genotypes. The benefit of the TT genotypes seemed to increase as the patients lived longer.
Table 2.
Associations between IL4, IL13, and IL4R SNPs and high and medium/low grade glioma
| High Grade |
Medium/Low Grade |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP rs# | n | n died |
Hazard Ratio* (95% CI) |
n | n died |
Hazard Ratio* (95% CI) |
||
| Overall Survival | ||||||||
| IL4 rs243250 | ||||||||
| CT/TT | 94 | 68 | ref | 100 | 33 | ref | ||
| CC | 227 | 170 | 1.03 (0.76–1.39) | 222 | 78 | 1.10 (0.73–1.66) | ||
| IL13 rs1800925 | ||||||||
| CT/TT | 116 | 83 | ref | 124 | 41 | ref | ||
| CC | 205 | 155 | 1.14 (0.87–1.51) | 198 | 70 | 1.33 (0.90–1.97) | ||
| IL4R rs1805011 | ||||||||
| AC/CC | 68 | 52 | ref | 65 | 28 | ref | ||
| AA | 253 | 186 | 0.97 (0.70–1.34) | 258 | 83 | 0.71 (0.46–1.11) | ||
| IL4R rs1805012 | ||||||||
| CT/CC | 65 | 49 | ref | 61 | 26 | ref | ||
| TT | 257 | 190 | 0.99 (0.71–1.37) | 260 | 83 | 0.66 (0.42–1.04) | ||
| IL4R rs1805015 | ||||||||
| CT/CC | 99 | 78 | ref | 89 | 35 | ref | ||
| TT | 220 | 158 | 0.80 (0.60–1.06) | 234 | 76 | 0.79 (0.53–1.19) | ||
| IL4R rs1801275 | ||||||||
| AG/GG | 121 | 91 | ref | 111 | 43 | ref | ||
| AA | 200 | 147 | 0.90 (0.68–1.18) | 212 | 68 | 0.83 (0.57–1.22) | ||
| IL4R rs1805016 | ||||||||
| GT/GG | 37 | 32 | ref | 34 | 10 | ref | ||
| TT | 284 | 206 | 0.59 (0.40–0.87) | 289 | 101 | 1.42 (0.73–2.75) | ||
| IL4R Haplotype | ||||||||
| A-T-T-A-T† | 220 | 158 | 0.80 (0.61–1.04) | 270 | 90 | 0.87 (0.62–1.21) | ||
| Post 1-year survival | ||||||||
| IL4 rs243250 | ||||||||
| CT/TT | 59 | 43 | ref | 81 | 19 | ref | ||
| CC | 128 | 90 | 0.82 (0.56–1.19) | 180 | 54 | 1.30 (0.77–2.20) | ||
| IL13 rs1800925 | ||||||||
| CT/TT | 72 | 51 | ref | 105 | 29 | ref | ||
| CC | 115 | 82 | 1.00 (0.69–1.43) | 156 | 44 | 1.29 (0..79–2.08) | ||
| IL4R rs1805011 | ||||||||
| AC/CC | 41 | 32 | ref | 51 | 18 | ref | ||
| AA | 146 | 101 | 0.75 (0.49–1.13) | 211 | 55 | 0.70 (0.40–1.23) | ||
| IL4R rs1805012 | ||||||||
| CT/CC | 40 | 31 | ref | 47 | 16 | ref | ||
| TT | 147 | 102 | 0.75 (0.50–1.14) | 214 | 56 | 0.69 (0.39–1.24) | ||
| IL4R rs1805015 | ||||||||
| CT/CC | 57 | 46 | ref | 69 | 20 | ref | ||
| TT | 129 | 86 | 0.63 (0.44–0.91) | 193 | 53 | 0.93 (0.55–1.57) | ||
| IL4R rs1801275 | ||||||||
| AG/GG | 67 | 50 | ref | 88 | 25 | ref | ||
| AA | 120 | 83 | 0.83 (0.58–1.19) | 174 | 48 | 0.97 (0.60–1.59) | ||
| IL4R rs1805016 | ||||||||
| GT/GG | 21 | 19 | ref | 28 | 5 | ref | ||
| TT | 166 | 114 | 0.44 (0.27–0.73) | 234 | 68 | 1.93 (0.77–4.84) | ||
| IL4R Haplotype | ||||||||
| A-T-T-A-T† | 129 | 86 | 0.68 (0.48–0.96) | 226 | 64 | 0.91 (0.61–1.37) | ||
Adjusted for age at diagnosis, time between diagnosis and registration, chemotherapy, extent of surgery, and radiation therapy
SNPs are ordered rs1805011, rs1805012, rs1805015, rs1801275, and rs1805016. Referent group includes individuals without haplotype genotype.
When examining combinations of SNPs for the IL4R gene, the most probable haplotype (A-T-T-A-T) had an estimated frequency of 79 percent. Compared to all other haplotypes, it was associated with a 20% decrease in mortality hazard of borderline statistical significance (HR=0.80; 95% CI= 0.61–1.04) among those with high-grade gliomas, but not medium/low-grade tumors (Table 2). This IL4R haplotype was also significantly associated with long-term survival among high-grade glioma patients (HR=0.68; 95% CI=0.48–0.96).
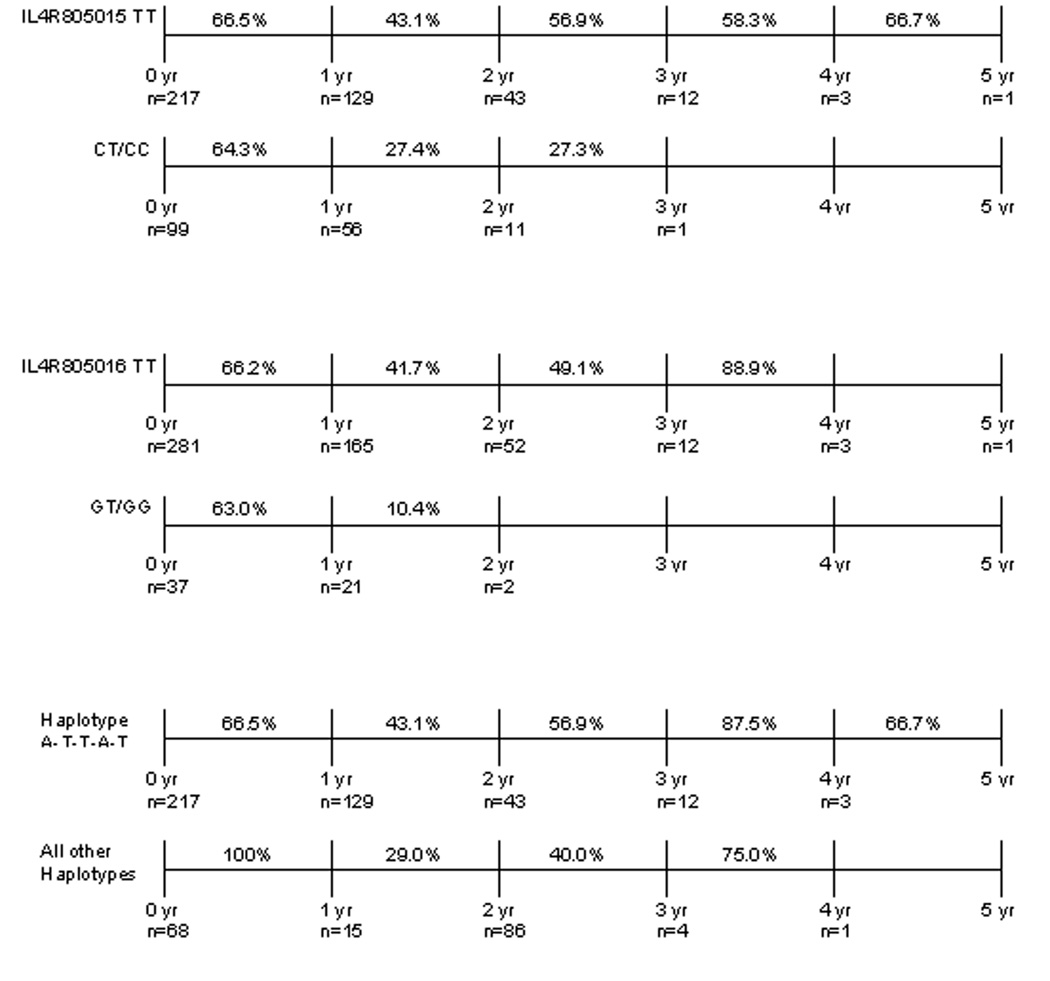
Yearly survival estimates conditional on surviving the previous year were calculated for two of the IL4R SNPs and for the IL4R haplotype that showed significant effects in the Cox models among high-grade gliomas. The results shown in Figure 2 are consistent with those seen in Figure 1. High-grade patients with the IL4R805015 CT/CC genotype had poorer year-to-year survival, the longest surviving just past three years, compared to those patients with the TT genotype. A very similar trend is seen with the IL4R805016 SNP. Among high-grade patients, those with the TT genotype had about 41.7% survival between year one and year two of follow-up, whereas those with the GT/GG genotype only had 10.4% survival between those years. Finally, the impact of having the IL4R haplotype on year-to-year survival was favorable comparable to not having the haplotype and showed a moderate protective effect.
Figure 2. Conditional yearly survival estimates for IL4R SNPs and Haplotype among high-grade gliomas.
Patients with the TT genotype for either SNP or with the A-T-T-A-T haplotype experienced better survival beyond one year when compared to other genotypes or haplotypes. This is consistent with and supports both the Cox regression models and the Kaplan-Meier curves for these SNPs and the haplotype. These genotypic differences were not experienced by patients with low-grade or anaplastic tumors.
DISCUSSION
Age and tumor grade are key prognostic factors in glioma survival.(15) While some germline genetic factors have been suspected of playing an important role in prognosis, none have been firmly established. Previous investigations into SNPs in inflammatory genes have mostly focused on their effects on glioma risk, not disease prognosis. However, the current study found that two non-synonymous SNPs in the IL4R gene, rs1805015 and rs1805016, are significantly associated with long-term survival among high-grade glioma patients. In addition, the most common haplotype of the IL4R SNPs (A-T-T-A-T) showed a moderate protective effect overall and a statistically significant long-term protective affect, among high-grade glioma cases.
Median survival time for glioblastoma patients is usually considered to be about one year. Indeed, in our study group, overall median survival was 14.8 months for high-grade glioma patients compared to just over 6 years for those in the low-/intermediate-grade group. However, among high-grade patients surviving past the median survival time, the effects of SNPs in the inflammatory genes appear to be more pronounced. It is clear that the range of survival times for those without the TT genotype at the IL4R805015 and IL4R805016 SNPs was much shorter among these high-grade glioma patients. The TT genotype for IL4R SNP rs1805016 was significantly associated with both overall and long-term survival, and the TT genotype of the IL4R SNPs rs1805015 was also significantly associated with survival past 12 months among high-grade glioma patients. It is possible that during the first year after diagnosis, the effects of the disease and treatment mask the modulatory effects of these SNPs on survival. Our findings also lend support to this hypothesis by showing that during the first year of follow-up, the survival probabilities between the patients with the different genotypes of the IL4R805015 and IL4R805016 SNPs were very similar. However, the one-to-two year survival probabilities between the genotypes diverge, indicating that the TT genotype for both SNPs may confer a protective effect after the first year of follow-up, and thus the effects of the inflammatory SNPs themselves would only be detectable among long-term survivors.
A few studies have reported on the effects of inflammatory pathways on glioma etiology, and several inflammatory gene SNPs have been shown to be important in glioma risk. For example, Schwartzbaum et al. examined SNPs in IL4R, IL13, and ADAM33 in a population-based case-control study of 111 glioblastoma cases from Swedish regional cancer registries and 422 randomly-selected controls.(16) The IL4R SNPs T478C (C allele) (rs1805015) and A551G (A allele) (rs1801275) were significantly associated with increased glioma risk (OR= 1.64 (95% CI 1.05–2.55); OR=1.61 (95% CI 1.05–2.47), respectively). Another recent report by Wiemels et al. examined SNPs in the IL4, IL4R, and IL13 genes among 456 glioma cases and 541 controls.(17) They found that the IL13 Arg110Gln (rs20541) and C-1112T (rs1800925) SNPs were significantly associated with higher IgE levels in the controls (p <0 .05 for both). Furthermore, the T allele of the IL13 C-1112T polymorphism was protective against being a case (p=0.05). While they did not find a significant association between single polymorphisms in the IL4 and IL4R genes, they did find an IL4R haplotype, different from our current findings, that was associated with an increase in glioma risk, although of borderline statistical significance (OR=1.5; 95% CI=1.0–2.3). Furthermore, they found that another rare IL4 haplotype was inversely associated with glioma risk (OR=0.23; 95% CI=0.07–0.83). These etiologic studies provide some clues about how polymorphisms in the IL4R gene are involved in the pathways that regulate glioma development and, and in conjunction with our current findings, about glioma control and prognosis.
Having a history of immune hyperactivity, such as allergies, asthma, other atopic diseases, is associated with decreased glioma risk.(4;16) Cytokine-responsive genes include those that code for IgE, as well as the alpha component of the IL-4 receptor. The IL-4 receptor is expressed in several tissues, including brain, which reflects the fact that this receptor has a wide range of functions. While the specific mechanisms by which the IL4R polymorphisms examined here may affect patient survival are unknown, there are several endpoints of the pathway, potentially impacted by these genetic variants, which are relevant to the carcinogenic process. For example, activation of the IL-4 pathway may lead to increased cell proliferation, cell growth, or apoptosis depending on which signal transduction pathway becomes initiated. Furthermore, certain IL4R SNPs, such as rs1805010, have already been shown to have a functional effect on IgE level by up-regulating the receptor’s response to IL-4, which in turn results in activation of the Stat6 pathway.(18) While the functional affects of many of the SNPs examined here are largely unkown, if they lead to an over- or under-expression of IgE, the resulting change in the inflammatory response could have an impact on treatment efficacy, therefore potentially impacting survival. Therefore, future studies of the functionality of these SNPs are warranted to fully understand their effects on brain tumor control.
This study adds to the small, yet growing, body of literature examining the role of genetic prognostic factors for malignant gliomas. The number of glioma patients included in this analysis allowed us to examine genetic effects by histologic subtype. Given the dramatic differences in prognosis and treatment by histologic type, this is important when examining survival for these patients. In the future, we hope to examine the effects of these SNPs on treatment outcome, including adverse events, as well as survival. This study was limited to non-Hispanic white patients, a consequence of limited access to minority patients, due partly to the fact that glioma incidence among these populations is lower than the incidence among non-Hispanic whites. While we found significant genetic effects on survival for two SNPs in the IL4R gene, further studies in other populations are needed to validate and support our findings.
Acknowledgments
This study was supported by grants from the National Cancer Institute (CA070917; PI: ML Bondy). Fellowship support for E. Amirian was provided through the Cancer Prevention Research Training Program (CA056452; PI: RM Chamberlain).
Reference List
- 1.American Cancer Society. Cancer Facts and Figures. American Cancer Society; 2007. [Google Scholar]
- 2.Wrensch M, Wiencke JK, Wiemels J, Miike R, Patoka J, Moghadassi M, McMillan A, Kelsey KT, Aldape K, Lamborn KR, Parsa AT, Sison JD, Prados MD. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006:4531–4541. doi: 10.1158/0008-5472.CAN-05-4032. [DOI] [PubMed] [Google Scholar]
- 3.Wrensch M, McMillan A, Wiencke J, Wiemels J, Kelsey K, Patoka J, Jones H, Carlton V, Miike R, Sison J, Moghadassi M, Prados M. Nonsynonymous coding single-nucleotide polymorphisms spanning the genome in relation to glioblastoma survival and age at diagnosis. Clin. Cancer Res. 2007:197–205. doi: 10.1158/1078-0432.CCR-06-1199. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Diepgen TL. Is atopy a protective or a risk factor for cancer? A review of epidemiological studies. Allergy. 2005:1098–1111. doi: 10.1111/j.1398-9995.2005.00813.x. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Ericksen MB, Levin LS, Khurana Hershey GK. Functional effect of the R110Q IL13 genetic variant alone and in combination with IL4RA genetic variants. J. Allergy Clin. Immunol. 2004:553–560. doi: 10.1016/j.jaci.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 7.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am. J. Hum. Genet. 2002:230–236. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moissidis I, Chinoy B, Yanamandra K, Napper D, Thurmon T, Bocchini J, Jr, Bahna SL. Association of IL-13, RANTES, and leukotriene C4 synthase gene promoter polymorphisms with asthma and/or atopy in African Americans. Genet. Med. 2005:406–410. doi: 10.1097/01.gim.0000170994.24960.48. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Xing ZM, Lu C, Ma YX, Yu DL, Yan Z, Wang SW, Yu LS. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum. Genet. 2003:387–390. doi: 10.1007/s00439-003-1001-x. [DOI] [PubMed] [Google Scholar]
- 10.Kouriba B, Chevillard C, Bream JH, Argiro L, Dessein H, Arnaud V, Sangare L, Dabo A, Beavogui AH, Arama C, Traore HA, Doumbo O, Dessein A. Analysis of the 5q31-q33 locus shows an association between IL13-1055C/T IL-13-591A/G polymorphisms and Schistosoma haematobium infections. J. Immunol. 2005:6274–6281. doi: 10.4049/jimmunol.174.10.6274. [DOI] [PubMed] [Google Scholar]
- 11.Vladich FD, Brazille SM, Stern D, Peck ML, Ghittoni R, Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J. Clin. Invest. 2005:747–754. doi: 10.1172/JCI22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartzbaum J, Ahlbom A, Malmer B, Lonn S, Brookes AJ, Doss H, Debinski W, Henriksson R, Feychting M. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005:6459–6465. doi: 10.1158/0008-5472.CAN-04-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okcu MF, Selvan M, Wang LE, Stout L, Erana R, Airewele G, Adatto P, Hess K, Ali-Osman F, Groves M, Yung AW, Levin VA, Wei Q, Bondy M. Glutathione S-transferase polymorphisms and survival in primary malignant glioma. Clin. Cancer Res. 2004:2618–2625. doi: 10.1158/1078-0432.ccr-03-0053. [DOI] [PubMed] [Google Scholar]
- 14.SAS Genetics Brochure. SAS/Genetics Software Brochure. 2007. Jul 1, 2007. [Google Scholar]
- 15.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. (Berl) 2005:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 16.Schwartzbaum J, Ahlbom A, Malmer B, Lonn S, Brookes AJ, Doss H, Debinski W, Henriksson R, Feychting M. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005:6459–6465. doi: 10.1158/0008-5472.CAN-04-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiemels JL, Wiencke JK, Kelsey KT, Moghadassi M, Rice T, Urayama KY, Miike R, Wrensch M. Allergy-related polymorphisms influence glioma status and serum IgE levels. Cancer Epidemiol. Biomarkers Prev. 2007:1229–1235. doi: 10.1158/1055-9965.EPI-07-0041. [DOI] [PubMed] [Google Scholar]
- 18.Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin JM. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat. Genet. 1998:119–120. doi: 10.1038/472. [DOI] [PubMed] [Google Scholar]