Abstract
We investigated distribution of the nucleolar phosphoprotein Nopp140 within mammalian cells, by immunofluorescence confocal microscopy and immunoelectron microscopy. During interphase, three-dimensional image reconstructions of confocal sections revealed that nucleolar labelling appeared as several tiny spheres organized in necklaces. Moreover, after an immunogold labelling procedure, gold particles were detected not only over the dense fibrillar component but also over the fibrillar centers of nucleoli in untreated and actinomycin D-treated cells. Labelling was also consistently present in Cajal bodies. After pulse-chase experiments with BrUTP, colocalization was more prominent after a 10-15 min chase than after a 5 min chase. During mitosis, confocal analysis indicated that Nopp140 organization was lost. The protein dispersed between and around the chromosomes in prophase. From prometaphase to telophase, it was also detected in numerous cytoplasmic nucleolus-derived foci. During telophase, it reappeared in the reforming nucleoli of daughter nuclei. This strongly suggests that Nopp140 could be a component implicated in pre-rRNA processing by the rDNA transcription active sites.
Keywords: nucleolus, Nopp140, confocal microscopy, immunoelectron microscopy, mitosis, interphase, mammalian cells
INTRODUCTION
The nucleolus is a prominent subnuclear compartment assembled around clusters of tandemly repeated rDNA genes, from which pre-RNAs are transcribed and subsequently modified, folded, processed, and assembled into small and large ribosomal subunits (Thiry and Goessens 1996). Three morphologically distinct nucleolar components have been defined in interphase cells: the fibrillar center (FC), the dense fibrillar component (DFC) and the granular component (GC). It is currently thought that each of these three components correlates with a specific step in ribosome biogenesis. During mitosis, in most eukaryotic cells, the nucleoli disappear at the end of prophase and reappear in daughter cells during telophase. Interestingly, several lines of evidence suggest that the mitotic fate of nucleolar components depends on their functional role in ribosome biogenesis (Dundr et al. 1997; Dundr et al. 1998; Dundr and Olson 2000; Hernandez-Verdun and Gautier 1994; Scheer et al. 1993; Thiry and Goessens 1996). Indeed, the elements directly involved in rDNA transcription, such as RNA polymerase I, Upstream Binding Factor (UBF) or DNA topoisomerase I, although inactive from prophase until early telophase, remain mainly associated with the Nucleolus Organizer Region (NOR) throughout the different steps of mitosis (Chan et al. 1991; Guldner et al. 1986; Jordan et al. 1996; Roussel et al. 1996; Scheer and Rose 1984; Sirri et al. 1999; Weisenberger and Scheer 1995). On the contrary, components involved in pre-rRNA processing are concentrated in the perichromosomal sheath or in nucleolus-derived foci (NDFs). They are re-utilized throughout telophase in the formation of new nucleoli through different prenucleolar bodies (PNBs) (Dundr et al. 1997; Dundr et al. 1998; Dundr and Olson 2000; Fomproix and Hernandez-Verdun 1999; Hernandez-Verdun and Gautier 1994; Jimenez-Garcia et al. 1994; Savino et al. 1999; Savino et al. 2001; Verheggen et al. 2000; Weisenberger and Scheer 1995).
The present study focused on the nonribosomal, nucleolar protein, Nopp140 (Meier and Blobel 1990). Originally isolated from rat (Meier and Blobel 1990; Meier and Blobel 1992), this protein was later identified in human (Pai et al. 1995), Xenopus (Cairns and McStay 1995), yeast (Meier 1996), Drosophila (Waggener and DiMario 2002) and Trypanosomes (Kelly et al. 2006). Unlike most other nucleolar proteins, Nopp140 does not carry RNA-binding motifs or glycine/arginine-rich stretches, as deduced from the known primary sequences of different species. On the contrary, its amino- and carboxy-termini are separated by a long, central domain, consisting of ten repeats of acidic serine clusters alternating with lysine-, alanine-, and proline-rich basic stretches. Most serine residues of the acidic repeats are phosphorylated by casein kinase 2, which makes Nopp140 one of the most highly phosphorylated proteins in the cell with ~80 phosphates per molecule (Li et al. 1997; Meier and Blobel 1992; Meier 1996;). Despite these biochemical characteristics, the biological functions of Nopp140 are still unclear. Interaction of Nopp140 with p80-coilin, a constituent of the Cajal body (CB) (Andrade et al. 1991), suggests that Nopp140 functions as a molecular link between the nucleolus and the CBs (Isaac et al. 1998). A mammalian nucleolar protein, NAP57, has also been identified as a Nopp140-associated protein, found in the DFC of the nucleolus and in the CBs (Meier and Blobel 1994). Possible roles for NAP57 were mostly deduced from analysis of its yeast homologue, Cbf5p, characterized as a putative rRNA pseudouridine synthase involved in rRNA synthesis and pre-rRNA processing (Cadwell et al. 1997; Lafontaine et al. 1998). In addition, Nopp140 associates with the two major classes of small nucleolar ribonucleoprotein particles (snoRNPs) (Yang et al. 2000) which mainly catalyse rRNA modification (Lafontaine and Tollervey 1998). Thus, Nopp140 could function as a chaperone for the biogenesis and transport of snoRNPs (Isaac et al. 1998; Yang et al. 2000). However, unlike components implicated in pre-rRNA processing, Nopp140 is not associated with the NDFs and with the PNBs during mitosis (Dundr et al. 1997). On the other hand, Nopp140 was found to interact with the largest subunit of RNA polymerase I, suggesting a role in rDNA transcription (Chen et al. 1999). However, unlike the transcriptional machninery of RNA polymerase I, Nopp140 does not associate with the mitotic NORs (Dundr et al. 1997; Pai et al. 1995; Schmidt-Zachmann et al. 1984; Tsai et al. 2008). Intriguingly, it has been shown previously that Nopp140 functions as an RNA polymerase II transcription coactivator, and interacts with the general transcription factor TFIIB and a specific DNA motif-binding transcription factor (Miau et al. 1997). However, the localization of Nopp140 in the nucleolus and the CBs strongly suggest its involvement in cellular activities carried out within these structures.
In order to shed light on the cellular function of Nopp140, the present study re-investigated the precise location of this protein within different mammalian cells during the cell cycle by immunofluorescence confocal microscopy coupled to three-dimensional image reconstructions and immunogold electron microscopy. The results revealed that Nopp140 was detected in the CBs and in the nucleoli. In the latter case, the labelling of the FC and the surrounding DFC was organised in tiny spheres. These were rapidly associated with ribosomal RNA transcripts as revealed by pulse-chase experiments. During mitosis, Nopp140 was not a NOR-associated protein, but it was dispersed in numerous NDFs. The functional significance these findings is discussed.
MATERIALS AND METHODS
Biological materials
HeLa cells and HEp-2 cells were grown at 37°C under 5% CO2 in Glasgow minimum essential medium (Gibco-BRL, Life Technologies, Gent, Belgium) supplemented with 10% fetal calf serum. Ehrlich ascites tumour cells (ELT) were grown in a medium composed of 40% NCTC 109, 40% Hanks solution, 20% fetal calf serum, 100 U/ml penicillin. Some cultures of ELT cells were treated for 1-2 h with 0.05 or 20μg/ml actinomycin D (Sigma, St Louis, USA).
Immunofluorescence methods
The slides were simultaneously fixed and permeabilised for 4 min at room temperature in 4% formaldehyde and 1% (vol/vol) Triton X-100 in 0.1 M PBS (pH 7.4). After washing in PBS containing 1% BSA (W/V) and normal goat serum (NGS) diluted 1/30, the slides were placed for 30 min at 37°C with anti-Nopp140 polyclonal antibodies (RF12 and RE10; Meier and Blobel, 1992) diluted 1/200 in PBS, containing NGS diluted 1/50 and 0.2% BSA. After rinsing with PBS containing 1% BSA, the slides were incubated for 30 min at 37°C with FITC-conjugated goat anti-rabbit antibody (Sigma, St Louis, USA ) diluted 1/100 in PBS containing 0.2% BSA. After several rinses, the slides were mounted with Citifluor™ AF1 (Agar Scientific, Starsted, U.K.).
For double labelling experiments, the slides were fixed for 5 min at room temperature in 4% formaldehyde in 0.1 M PBS (pH 7.4) containing 1% Triton X-100. They were rinsed first in PBS, then in PBS containing 1% BSA, as well as NGS and normal sheep serum (NSS), both diluted 1/30. They were incubated for 30 min at 37°C with anti-Nopp140 polyclonal antibodies diluted 1/200 in PBS, containing 0.2% BSA and NGS diluted 1/50. The slides were washed with PBS containing 1% BSA and incubated for 30 min at 37°C with an FITC-conjugated goat anti-rabbit antibody diluted 1/100 in PBS containing 0.2% BSA. After rinsing, the slides were incubated for 30 min at 37°C with rabbit anti-CHO nucleolin serum (kindly provided by Dr. F. Amalric) diluted 1/200 in PBS, containing 0.2% BSA and NSS, diluted 1/50. The slides were washed with PBS containing 1% BSA and incubated for 30 min at 37°C with a biotinylated sheep anti-rabbit antibody (Roche Diagnostics) diluted 1/100 in PBS containing 0.2% BSA. This secondary antibody was detected with streptavidin labelled with Texas Red (Amersham Life Science, Little Chalfont, United Kingdom) diluted 1/100 in PBS containing 0.2% BSA for 10 min. After rinsing, the cells were mounted with Citifluor™ AF1 (Agar Scientific, Starsted, U.K.).
Pulse-chase experiments were carried out as previously described by Thiry et al. (2000, 2008).
Confocal microscopy and three-dimensional visualizations
A Biorad 1024ES system (Bio-Rad, Hercules, CA) mounted on an inverted IX70 Olympus optical microscope was used. Acquisitions, made using a planapochromat ×60, 1.4 numerical aperture oil immersion objective, were performed by exciting the FITC with the 488-nm line of a Krypton/Argon laser. The emission light was collected through a band pass filter at 522 ± 16 nm. Phase contrast images were collected simultaneously on a specific detector. For three dimensional investigations, 30 to 50 optical sections were recorded from the top of the cell with a 0.5-μm z-step. Files were then transferred to a Sun Sparc20 workstation (sun Microsystems, mountain View, CA) for processing which was performed using the Analyze software (CNSoftware, Southwater, UK) (Héliot et al. 1997; Klein et al. 1998). Volumes were obtained by re-sampling the number of sections in order to have an identical pixel size in x, y, and z directions. A 3 × 3 × 3-cubic median filter was subsequently applied to remove background voxels that might have appeared in the final volumic visualization pictures. From the reconstructed volumes, a surfacic vizualisation was applied (Cheutin et al. 2002; Klein et al. 1998).
Immunoelectron microscopy
Culture, permeabilization, incorporation and washings were all carried out at room temperature, in Petri dishes. Cells were fixed for 60-90 min in 4% formaldehyde in 0.1 M Sörensen's buffer (pH 7.4). After fixation, the cells were washed in Sorensen's buffer, dehydrated through graded ethanol solutions and then processed for embedding in Lowicryl K4M according to the technique of Roth et al. (1981). For labelling, ultrathin sections were incubated for 30 min in PBS (0.14 M NaCl, 6 mM Na2HPO4, 4 mM KH2 PO4, pH 7.2) containing normal goat serum (NGS) diluted 1/30 and 1% bovine serum albumin (BSA type V; Sigma, St Louis, MI), then rinsed with PBS containing 1% BSA. The sections were then incubated for 4 h with the primary antibodies diluted in PBS, containing NGS 1/50 and 0.2 % BSA. After washing with PBS containing 1% BSA, they were incubated for 60 min with either goat anti-rat IgG or goat anti-rabbit IgG coupled to colloidal gold (10 nm diameter; Amersham Life Science) diluted 1/40 with PBS (pH 8.2) containing 0.2 % BSA. After washing with PBS containing 1% BSA, the ultrathin sections were rinsed in deionized water. Finally, they were mounted on nickel grids and stained with uranyl acetate and lead citrate before examination in a Jeol CX 100 II electron microscope at 60 KV. For immunolabeling, several primary antibodies were used: anti-Nopp140 polyclonal antibodies (diluted 1/200), anti-CHO and human nucleolin serum (diluted 1/200) from rabbit (kindly provided by Dr. F. Amalric), anti-B36 Mab P2G3 (diluted 1/20) (Christenssen et al. 1986; kindly provided by Dr. F. Puvion-Dutilleul) and, anti-p80 coilin polyclonal antibody (diluted 1/100) from rabbit (Carmo-Fonseca et al. 1992; kindly provided by Dr. R. Deltour). Two control experiments were carried out, in which either the primary antibodies were omitted, or the sections were incubated with antibody-free gold particles.
Quantitative evaluations
Since the electron-dense marker used in the immunogold technique is particulate, the density of labelling can be quantified. As demonstrated previously, only antigenic sites exposed at the surface of the sections can interact with the antibodies (Bendayan 1984). Therefore, the labelling density is independent of the section thickness but is directly related to the areas occupied by each of the intracellular compartments. Since differences in observed labelling densities reflect relative differences in the concentration of antigenic sites, only relative comparisons between intensities can be considered. To evaluate the labelling density, the area of each compartment (Sa) was estimated using a morphometrical approach by the point-counting method (Weibel 1969). Then, the number of gold particles (Ni) present over each compartment was counted and the labelling density (Ns) calculated (Ns = Ni/Sa). Values obtained on the resin and the cytoplasm can be considered as background staining.
RESULTS
Localization of Nopp140 during interphase
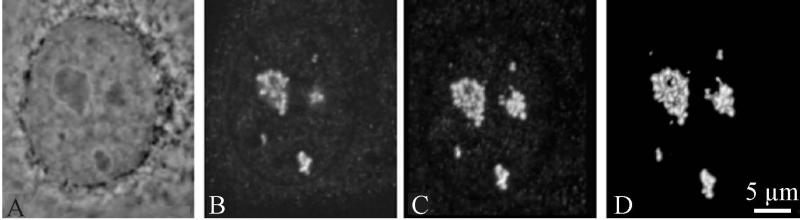
In order to analyse the intracellular location of Nopp140, an indirect immunofluorescence method was applied on HeLa cells, simultaneously fixed with formaldehyde and permeabilized with Triton X100. After probing Nopp140 either with a polyclonal antibody (RF12) more specific for rodents or a polyclonal antibody (RE10) more specific for primates, the labelling was analysed by confocal microscopy. Similar results were obtained in both cases. Comparison between a phase contrast image (Fig. 1a) and a single optical section (Fig. 1b) showed that Nopp140 labelling was found in most parts of the nucleoli. In addition, the labelling was observed in extranucleolar dots, which are reminiscent of Cajal bodies, as observed for cells stained with anti-p80 coilin antibodies (Meier and Blobel 1994). In the control experiments, no label was seen when anti-Nopp140 antibodies were replaced by preimmune serum. The three-dimensional distribution of Nopp140 was obtained by volume reconstruction from the stack (z-series) of confocal optical sections, visualized either as a projection (Fig. 1c), or as a volume with surfacic rendering mode (Fig. 1d). Nopp140 labelling was seen as multiple fluorescent spots that were grouped in clusters (Fig.1, b and c). The fluorescent spots are individual beads, 0.5 μm in diameter, which are organized as a necklace (Fig.1d).
Fig. 1.
Distribution of Nopp140 in a HeLa cell during interphase observed by confocal microscopy. (a): phase contrast. (b): optical section. (c): projection of the z-series. (d): surfacic visualization of the volume. Results show a strong labelling localised in the cell nucleoli and in the nuclear Cajal bodies. As revealed by three-dimensional reconstructions, the nucleolar labelling is organized as spherical spots grouped in clusters (c and d). Bar is 5 μm
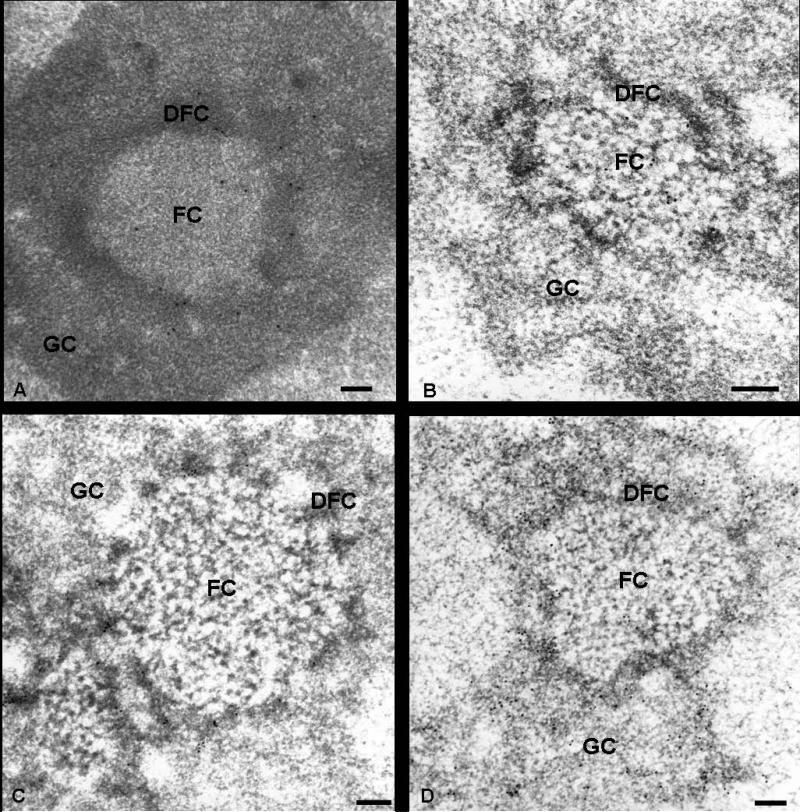
To localize precisely Nopp140 with regard to the nucleolar ultrastructural components, immunoelectron microscopy experiments were performed on ultrathin sections of Lowicryl K4M-embedded HeLa, ELT and HEp-2 cells. The results were similar with all three cell types and both antibodies to Nopp140. Gold particles were clearly evidenced over the DFC and the FC, notably in the peripheral region of this compartment (Fig. 2, a and b). On the contrary, both the GC and the condensed chromatin were completely devoid of labelling. These observations were confirmed after quantification of the labelling density over the different cellular compartments in three different cell types (Table 1). Indeed, the labelling density was very high over the DFCs, but it was still significant over the FCs. By contrast, the labelling density was very low over the GC, and its value was not significant in the nucleoplasm. The distribution of fibrillarin and nucleolin in three cell types was investigated using specific antibodies revealed by gold particles. The electron micrographs showed that anti-B36 antibodies, directed against fibrillarin, preferentially labelled the DFC (Fig. 2c), whereas anti-nucleolin antibodies strongly marked the GC and the DFC (Fig. 2d). However, in both cases, no labelling was observed over the FCs.
Fig. 2.
Immunoelectron microscopy localization of Nopp140, fibrillarin and nucleolin within the nucleolus. Ultrathin sections of HeLa (a) and ELT (b-d) cells were immunolabelled with specific anti-serum RE10 (a) and anti-serum RF12 (b), corresponding to anti-Nopp140 antibodies, and with antibodies directed against fibrillarin (c) and nucleolin (d). (a and b): gold particles are located over the fibrillar centers (FC) and the dense fibrillar component (DFC), while the granular component (GC) is devoid of label. (c): the DFC is labelled by anti-fibrillarin antibodies. The FCs and the GC are not labelled. (d): both the FCs and the GC are strongly labelled by anti-nucleolin antibodies, while the FCs are devoid of labelling. Bars are 0.2 μm
TABLE I.
Gold particle densities (number of particles per square micron) over the different structural compartments in HeLa, ELT and HEp-2 cells after immunogold labelling using anti-Nopp140 antibodies (RE10 or RF12). 552, 654, 633 and 228 gold particles were counted, respectively. Student's t-test for cellular compartments vs. resin or cytoplasm
| Mean values ± S.E.M. | ||||
|---|---|---|---|---|
| HeLa cells | ELT cells | HEp-2 cells | ||
| RE10 n = 12 | RF12 n = 19 | RE10 n = 12 | RE10 n = 9 | |
| NUCLEUS | ||||
| Nucleolus | ||||
| Fibrillar centers | 5.36 ± 1.33* | 5.72 ± 2.30* | 6.64 ± 2.53* | 7.36 ± 2.25* |
| Dense fibrillar component | 18.45 ± 5.30* | 20.33 ± 7.18* | 22.59 ± 4.32* | 23.79 ± 5.95* |
| Granular component | 1.63 ± 0.91 | 1.40 ± 0.52 | 1.40 ± 0.51 | 2.52 ± 0.94 |
| Nucleoplasm 1 | 1.01 ± 0.13 | 0.54 ± 0.19 | 0.48 ± 0.14 | 0.36 ± 0.22 |
| Cajal bodies | 18.12 ± 5.45* | 17.11 ± 5.74* | ||
| CYTOPLASM | 0.62 ± 0.49 | 0.55 ± 0.31 | ||
| RESIN | 0.92 ± 0.12 | 0.80 ± 0.27 | ||
P<0.01
condensed chromatin included.
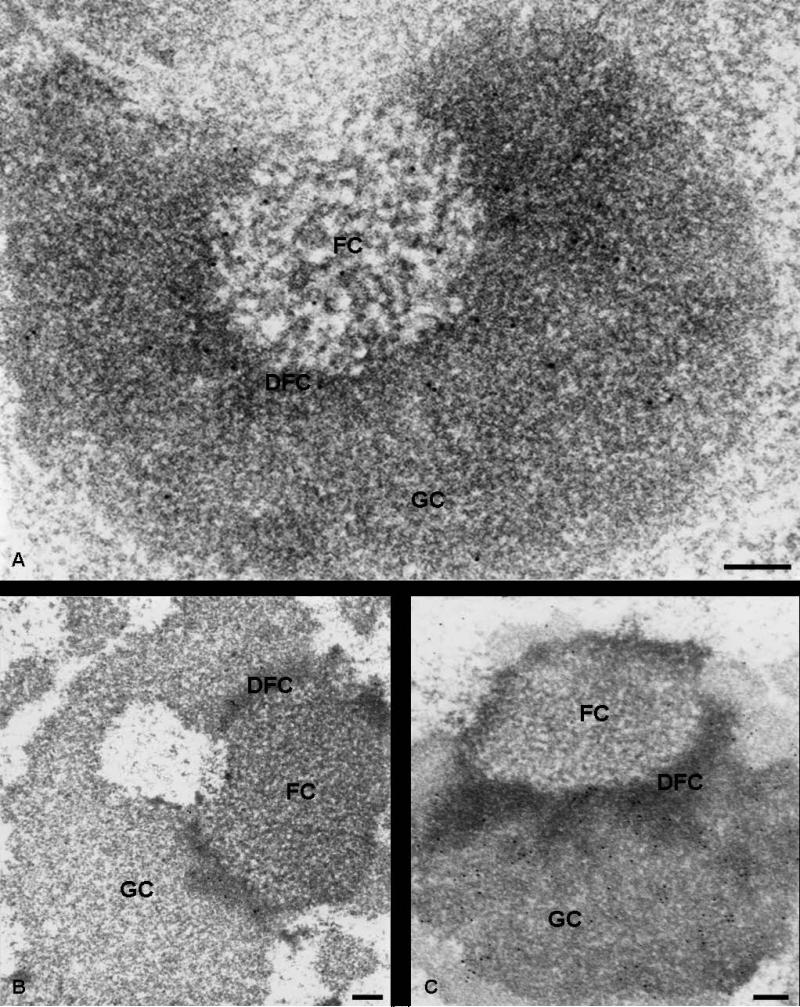
The nucleolar distribution of Nopp140 was also studied in cells treated with actinomycin D. This transcriptional inhibitor induces a complete reorganization of the nucleolar ultrastructural compartments, whose diposition with respect to the condensed chromatin becomes polarized. As observed in previous studies (Thiry and Goessens 1996), the order of succession is always as follows: condensed chromatin-FC-DFC-GC. When cells were submitted to actinomycin D, Nopp140 was located in both fibrillar components (DFC and FC) of segregated nucleoli (Fig. 3a). In contrast, no label was present over the GC. In the same experimental conditions, fibrillarin was detected in the DFC of segregated nucleoli (Fig. 3b), and nucleolin was seen both over the GC and the DFC (Fig. 3c). In both cases, the FCs were totally devoid of gold particles.
Fig. 3.
Immunoelectron microscopy localization of Nopp140 (a), fibrillarin (b) and nucleolin (c) within the nucleolus in cells treated with actinomycin D. Ultrathin sections of ELT cells, treated for 2 h with 0.05 μg/ml actinomycin D, were immunolabelled with anti-Nopp140 (RE10, a), anti-fibrillarin (b) and anti-nucleolin (c) antibodies. (a): in the segregated nucleolus, labelling was clearly evidenced over the FCs and the DFC, while the GC displayed only a few rare gold particles. (b): the DFC of the segregated nucleolus is the only compartment labelled by anti-fibrillarin antibodies. No labelling was observed over the FCs and the GC. (c): both the DFC and the GC were labelled by anti-nucleolin antibodies, while the FC was devoid of particles. Bars are 0.2 μm
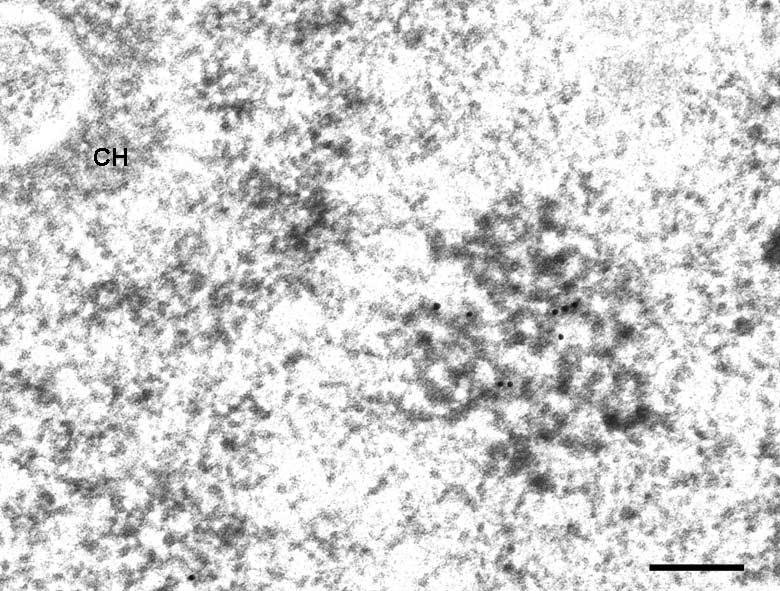
Finally, immunoelectron microscopy experiments were performed on ultrathin sections of HeLa, ELT and HEp-2 cells in order to determine whether the extranucleolar labelling observed with anti-Nopp140 antibodies corresponded to CBs (Fig. 4). An antibody directed against p80 coilin, a specific marker of CBs, unambiguously confirmed that this nuclear compartment was similar to that revealed with the Nopp140 probes (data not shown). After quantification, the labelling density over the CBs was comparable to that obtained over the DFC of the nucleolus (Table 1).
Fig. 4.
Immunoelectron microscopy localization of Nopp140 in extranucleolar regions of cell nuclei. The micrograph displays a Cajal body of a HEp-2 cell in which Nopp140 is detected. CH: condensed chromatin. Bar is 0.05 μm
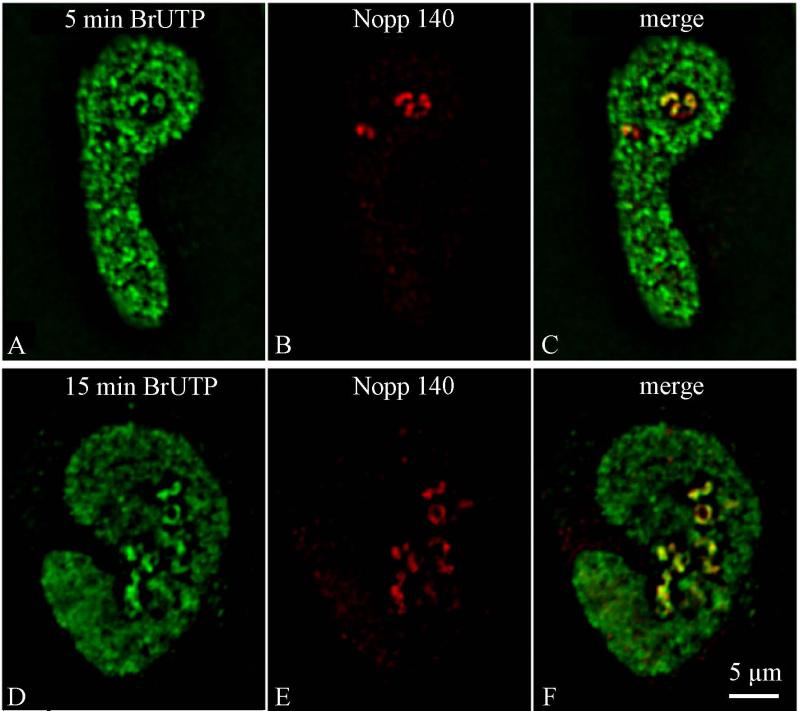
Having localized Nopp140, we attempted to determine whether this location corresponds to a particular stage in the formation of pre-rRNAs within the nucleolus. To perform this study, the spatial distribution of pre-rRNAs (Fig. 5, green) obtained in HeLa cells lipofected with BrUTP for increasing times of chase was compared to that of Nopp140 (Fig. 5, red) on single optical sections. After short chase periods, the relative surface of colocalization of the two signals in the nucleoli was low. BrUTP labelling was detected as round spots at the center of nucleoli, whereas the Nopp140 labelling occupied a wider zone (Fig. 5, a-c). Colocalization of Nopp140 and BrUTP was assessed with MetaMorph software. A scatter-plot was used to select the pixels which displayed high levels of both green and red, thus corresponding to colocalizing species. As quantified from Fig. 5 (a and b), 65.3% of Nopp140 labelling overlapped with the BrUTP signal within the nucleoli after 5 min incorporation. Fifteen min after transfection (Fig. 5, d and e), quantification of the two images revealed that 89.3% of Nopp140 colocalized with the nucleolar transcripts. Indeed, BrRNAs and Nopp140 displayed almost identical patterns in the nucleoli, as seen on the overlay (Fig. 5f).
Fig. 5.
Time-dependent migration of BrUTP-rRNAs in comparison with Nopp140-positive sites within the nucleolus. Double immunofluorescence detection of BrUTP-labelled rRNAs (green, a and d) and Nopp140 (red, b and e) in HeLa cells cultured in medium without BrUTP for 5 min (a-c) and 15 min (d-f) after lipofection with BrUTP-FuGene-6 complexes and observed on single optical sections. Overlay of green and red signals (c and f). Bar is 5 μm
Fate of Nopp140 throughout mitosis
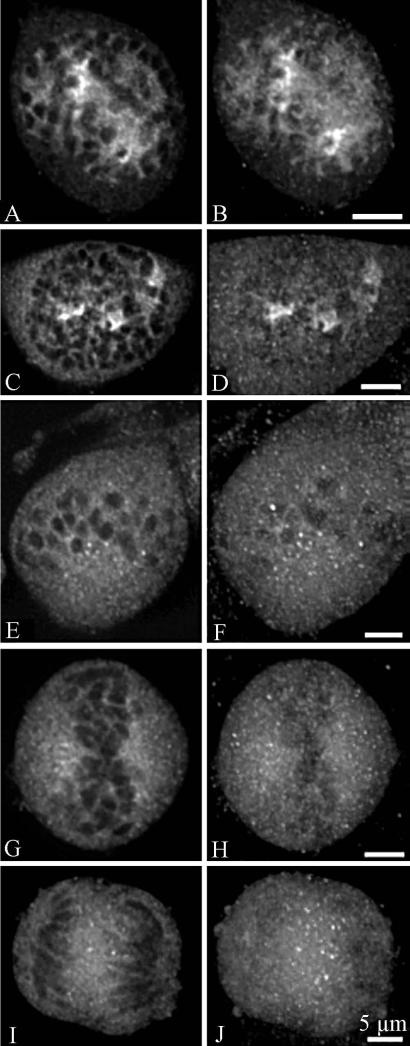
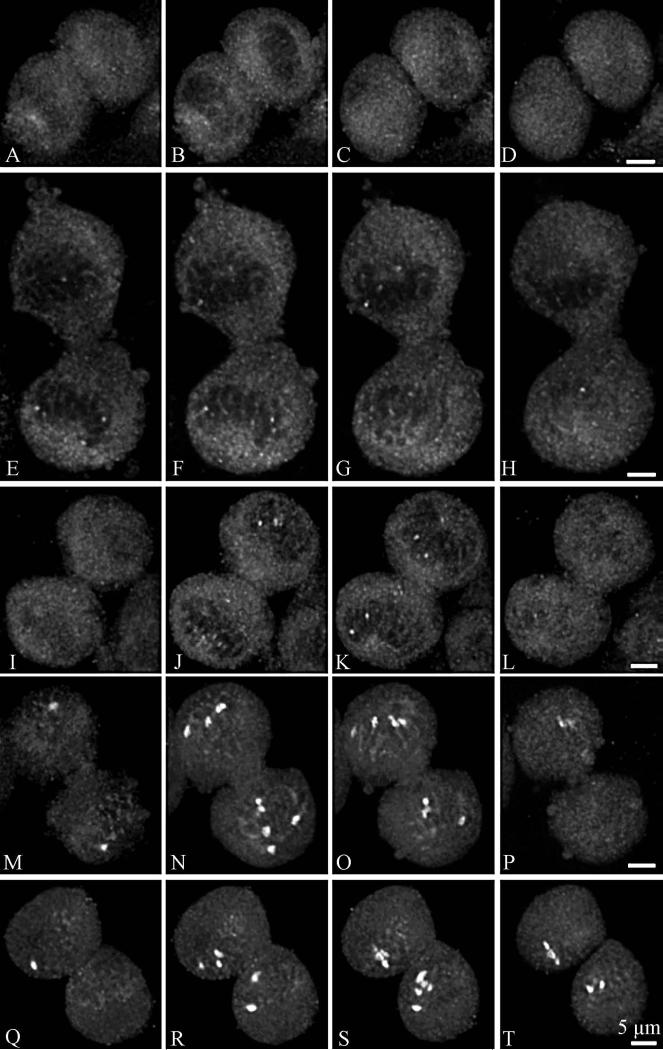
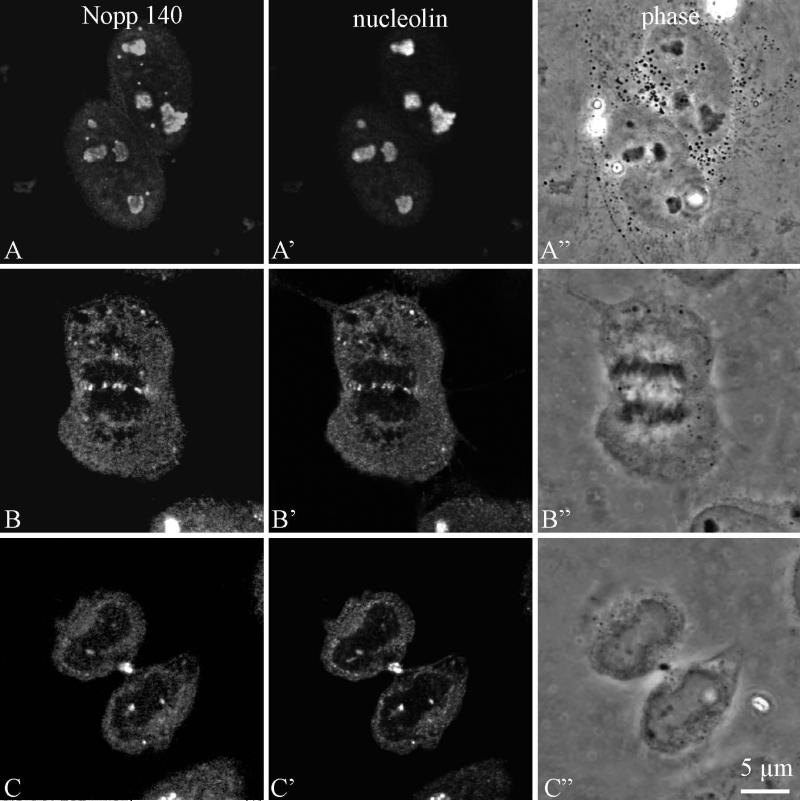
Nopp140 labelling was analysed in HeLa cells by confocal microscopy during the various steps of mitosis, after an indirect immunofluorescence labelling procedure. During prophase, Nopp140 was associated with disintegrated nucleoli (Fig. 6, a-f). The labelling was less structured than during interphase and consisted of several large spots and many fine fluorescent foci. It progressively dispersed in the nucleoplasm between the chromosomes (Fig. 6c) and after the breakdown of the nuclear envelope became cytoplasmic (Fig. 6, e and f). In metaphase (Fig. 6, g and h), the labelling was totally disorganized. However, it seemed preferentially located in the perichromosomal regions on both sides of the metaphase plate (Fig. 6g). Although most labelling was diffuse, brighter spots were also evidenced. Clearly, NORs were not stained by anti-Nopp140 antibodies, in contrast to what was observed with antibodies directed against RNA polymerase I (Héliot et al. 1997), UBF (Klein et al. 1998) or pp135 (Weisenberger and Scheer 1995). In anaphase (Fig. 6, i and j), the labelling was homogeneously distributed throughout the cytoplasm, but brighter spots were also detected between the separating chromosomes (Fig. 6i). During telophase (Fig. 7, a-l), Nopp140 labelling was clearly cytoplasmic, as it was mainly seen around the reforming daughter nuclei (Fig. 7, b-f). However, a few faint nuclear spots were seen in mid telophase (Fig. 7, e-h). In later stages of telophase, the intensity of these spots progressively increased (Fig. 7, i-l). The presence of Nopp140 in the cytoplasm of cells during telophase was clearly evidenced by using a double labellings with nucleolin, a marker of NDFs (Fig. 8).
Fig. 6.
Confocal microscopy analyses of Nopp140 distribution in HeLa cells from prophase to anaphase. Single optical sections (a, c, e, g, i) and projections of the corresponding z-series (b, d, f, h, j) show Nopp140 distribution during the first steps of mitosis. (a-d): early prophase. (e-f): late prophase. (g-h): metaphase. (i-j): anaphase. Bars are 5 μm
Fig. 7.
Confocal microscopy analyses of Nopp140 distribution in HeLa cells during telophase. (a-d): selection of four optical sections of z-series displaying a cell in early telophase. (e-h and i-l): selection of four optical sections of z-series displaying a cell in mid-telophase. (m-p): selection of four optical sections of z-series displaying a cell in late telophase. (q-t): selection of four optical sections of z-series displaying a cell at the beginning of G1. Bars are 5 μm
Fig. 8.
Confocal microscopy analyses (one optical section) of Nopp140 and nucleolin distribution in HeLa cells during interphase (a-c), anaphase (d-f), end of telophase (g-i). Bar is 5 μm
DISCUSSION
This study revealed that the nonribosomal protein Nopp140 is located in nucleoli and CBs of mammalian cells during interphase, and is associated with the NDFs during mitosis. Its behaviour during mitosis is similar to that of proteins involved in pre-rRNA processing. Indeed, we showed that Nopp140, like components implicated in pre-rRNA processing, including U3 snoRNA, fibrillarin, nucleolin, and proteins B23 and p52, accumulates in perichromosomal regions and in numerous NDFs between prometaphase and late telophase (DiMario 2004; Dundr et al. 1997; Dundr et al. 1998; Hernandez-Verdun and Gautier, 1994). However, using double immunofluorescence labelling of specific NDF markers, Dundr et al. (1997) concluded that Nopp140 was not associated with the NDFs from anaphase to telophase in different types of mammalian cells. This discrepancy probably stems in the antibodies directed to Nopp140. In previous immunofluorescence experiments, p130, the human homolog of Nopp140, appeared to be located in granular structures, resembling the PNBs at telophase (Pai et al. 1995). Similar observations were obtained with the Xenopus homolog of the rat nucleolar protein Nopp140, xNopp180 (Schmidt-Zachmann et al. 1984). These data already suggest that Nopp140 could belong to components involved in pre-rRNA processing. More recently, it has been demonstrated that Nopp140 interacts with both classes of snoRNPs, required for rRNA modification and processing, and that it functions as a chaperone for their biogenesis and intranuclear transport (Isaac et al. 1998; Yang et al. 2000). In agreement with our ultrastructural localizations of Nopp140, snoRNAs have been detected preferentially in the DFC of nucleoli. By in situ hybridisation at the electron microscope level, U3 snoRNA has been visualised essentially in the DFC (Puvion-Dutilleul et al. 1991). This RNA catalyses the initial processing of the 5’ external transcribed spacer and subsequent processing events around the 18S region (Hughes et al. 1996; Kass et al. 1990; Savino and Gerbi 1990). Other snoRNAs, such as MRP and U8, which catalyse respectively the processing within internal transcribed spacer I, and at the 5.8S and 28S borders (Chu et al. 1994; Peculis and Steitz 1993; Schmitt and Clayton 1993), have also been located in the DFC or in a subregion thereof (Jacobson et al. 1995; Matera et al. 1994; Reimer et al. 1988).
Our three-dimensional image reconstruction of confocal optical sections through interphase cells clearly indicates that the Nopp140 labelling appears as beads organized in necklaces in the nucleolar volume. Moreover, we showed that Nopp140 is present in the DFC of nucleoli, as previously reported (Meier and Blobel 1992), but is also seen in the FCs. This discrepancy probably arises from differences in fixation procedures. Indeed, we noticed that the presence of 0.1% glutaraldehyde in the fixation buffer abolishes the labelling in both fibrillar components of nucleoli. In the first assay for locating Nopp140 at the ultrastructural level, 0.05% glutaraldehyde was employed in the fixation solution, and a clear labelling was only found in the DFC. Interestingly, this nucleolar distribution coincides rapidly with the three-dimensional and ultrastructural location of ribosomal transcripts as observed in intact cells lipofected with BrUTP for short lengths of time (the present study and Thiry et al. 2000, 2008). In previous double immunofluorescence labelling experiments, it has been further demonstrated that Nopp140 colocalized with RNA polymerase I in the nucleolus (Baran et al. 2001; Chen et al. 1999, Tsai et al. 2008). However, contrary to proteins involved in rDNA transcription such as RNA polymerase I, UBF, DNA topoisomerase I, TATA-binding protein (TBP), TBP-associated proteins and transcription terminator factor TTF-1 (Chan et al. 1991; Guldner et al. 1986; Jordan et al. 1996; Roussel et al. 1996; Scheer and Rose 1984; Sirri et al. 1999; Weisenberger and Scheer 1995), our results also show that Nopp140 is not associated with the NORs throughout mitosis. These results indicate that Nopp140 is not an element of RNA polymerase I transcriptional machinery, even though it is located in the vicinity of the rDNA sites during transcription. Interestingly, it has been shown that RNA polymerase I transcription was arrested in nucleoli depleted of snoRNPs, raising the possibility of a feedback mechanism between rRNA modification and transcription (Yang et al. 2000). All these data suggest that Nopp140 should be a component implicated in pre-rRNA processing at the rDNA transcription active sites.
Contrary to previous immunogold labelling assays, we observed never curvilinear tracks that extended for microns across the nucleoplasm from the DFC of the nucleolus to the nuclear pore complexes. However, such tracks have never been visualised in immunofluorescence preparations and our ultrastructural results are entirely consistent with the previous immunofluorescence observations (Meier and Blobel 1992, 1994). Indeed, the only nuclear structures to be labelled outside the nucleolus are the CBs. In the present study, we also showed the labelling distribution at the ultrastructural level. The presence of Nopp140 in CBs supports the view that these nuclear structures are involved in nucleolar functions (Isaac et al. 1998; Meier and Blobel 1994; Verheggen et al. 2001). Their content in different snRNAs suggests that CBs may play a general role in the cell such as import, assembly and storage of components implicated in different steps of both nucleolar and extranucleolar RNA metabolism.
ACKNOWLEDGEMENTS
The authors wish to thank Dr. F. Amalric, Dr. C. Faucher and Dr. R. Deltour for their generous gift of antibodies. They also acknowledge the skillful technical provided by F. Skivée and D. Bourguignon. This work received financial support from the “Fonds de la Recherche Scientifique Médicale” (grant n°3. 3.4540.06) to M.T., from the “Ligue Régionale de la Marne”, from the “Fondation pour la Recherche Médicale” (grant n°40001837–01) to D.P, and from the National Institute of Health (HL079566) to U.T.M. FL and NT are PhD grant holders of the F.N.R.S.
REFERENCES
- Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran V, Brochard V, Renard JP, Flechon JE. Nopp 140 involvement in nucleologenesis of mouse preimplantation embryos. Mol Reprod Dev. 2001;59:277–284. doi: 10.1002/mrd.1032. [DOI] [PubMed] [Google Scholar]
- Bendayan M. Protein A-gold electron microscopic immunocytochemistry: methods, applications and limitations. J Electr Microsc Technique. 1984;1:243–270. [Google Scholar]
- Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns C, McStay B. Identification and cDNA cloning of a Xenopus nucleolar phosphoprotein, xNopp180, that is the homolog of the rat nucleolar protein Nopp140. J Cell Sci. 1995;108:3339–3347. doi: 10.1242/jcs.108.10.3339. [DOI] [PubMed] [Google Scholar]
- Chan EK, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med. 1991;174:1239–1244. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Pai CY, Huang JY, Yeh NH. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol Cell Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ME, Moloo J, Swischuk JL, Schelling ME. Characterization of the nucleolar protein, B-36, using monoclonal antibodies. Exp Cell Res. 1986;166:77–93. doi: 10.1016/0014-4827(86)90509-4. [DOI] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario P. Cell and molecular biology of nucleolar assembly and disassembly. Int Rev Cytol. 2004;239:99–178. doi: 10.1016/S0074-7696(04)39003-0. [DOI] [PubMed] [Google Scholar]
- Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjold ML, Olson MO. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Dundr M, Olson MO. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol Biol Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N, Hernandez-Verdun D. Effects of anti-PM-Scl 100 (Rrp6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp Cell Res. 1999;251:452–464. doi: 10.1006/excr.1999.4578. [DOI] [PubMed] [Google Scholar]
- Guldner HH, Szostecki C, Vosberg HP, Lakomek HJ, Penner E, Bautz FA. Scl 70 autoantibodies from scleroderma patients recognize a 95 kDa protein identified as DNA topoisomerase I. Chromosoma. 1986;94:132–138. doi: 10.1007/BF00286991. [DOI] [PubMed] [Google Scholar]
- Heliot L, Kaplan H, Lucas L, Klein C, Beorchia A, Doco-Fenzy M, Menager M, Thiry M, O'Donohue MF, Ploton D. Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol Biol Cell. 1997;8:2199–2216. doi: 10.1091/mbc.8.11.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Gautier T. The chromosome periphery during mitosis. Bioessays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Segura-Valdez M de L, Ochs R, Rothblum LI, Hanwan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kelly S, Singleton W, Wickstead B, Ersfeld K, Gull K. Characterization and differential nuclear localization of Nopp140 and a novel Nopp140-like protein in trypanosomes. Eukaryotic Cell. 2006;5:876–879. doi: 10.1128/EC.5.5.876-879.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Cheutin T, O'Donohue MF, Rothblum L, Kaplan H, Beorchia A, Lucas L, Heliot L, Ploton D. The three-dimensional study of chromosomes and upstream binding factor- immunolabeled nucleolar organizer regions demonstrates their nonrandom spatial arrangement during mitosis. Mol Biol Cell. 1998;9:3147–3159. doi: 10.1091/mbc.9.11.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Meier UT, Dobrowolska G, Krebs EG. Specific interaction between casein kinase 2 and the nucleolar protein Nopp140. J Biol Chem. 1997;272:3773–3779. doi: 10.1074/jbc.272.6.3773. [DOI] [PubMed] [Google Scholar]
- Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Meier UT, Blobel G. A nuclear localization signal binding protein in the nucleolus. J Cell Biol. 1990;111:2235–2245. doi: 10.1083/jcb.111.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miau LH, Chang CJ, Tsai WH, Lee SC. Identification and characterization of a nucleolar phosphoprotein, Nopp140, as a transcription factor. Mol Cell Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Chen HK, Sheu HL, Yeh NH. Cell-cycle-dependent alterations of a highly phosphorylated nucleolar protein p130 are associated with nucleologenesis. J Cell Sci. 1995;108:1911–1920. doi: 10.1242/jcs.108.5.1911. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Mazan S, Nicoloso M, Christensen ME, Bachellerie JP. Localization of U3 RNA molecules in nucleoli of HeLa and mouse 3T3 cells by high resolution in situ hybridization. Eur J Cell Biol. 1991;56:178–186. [PubMed] [Google Scholar]
- Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Roth J, Bendayan M, Carlemalm E, Villiger W, Garavito M. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J Histochem Cytochem. 1981;29:663–671. doi: 10.1177/29.5.6166664. [DOI] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino TM, Bastos R, Jansen E, Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Savino TM, Gebrane-Younes J, De Mey J, Sibarita JB, Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Rose K. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Thiry M, Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hugle B, Scheer U, Franke WW. Identification and localization of a novel nucleolar protein of high molecular weight by a monoclonal antibody. Exp Cell Res. 1984;153:327–346. doi: 10.1016/0014-4827(84)90604-9. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Clayton D. Nuclear RNaseMRP in required for correct processing of pre-5.8S in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- Thiry M, Cheutin T, O'Donohue MF, Kaplan H, Ploton D. Dynamics and three-dimensional localization of ribosomal RNA within the nucleolus. RNA. 2000;6:1750–1761. doi: 10.1017/s1355838200001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Goessens G. The Nucleolus during the Cell Cycle. RG Landes Company, Chapman and Hall; New York: 1996. [Google Scholar]
- Thiry M, Lamaye F, Thelen N, Chatron-Colliet A, Lalun N, Bobichon H, Ploton D. A protocol for studying the kinetics of RNA within cultured cells: application to ribosomal RNA. Nat Protoc. 2008;3:1997–2004. doi: 10.1038/nprot.2008.198. [DOI] [PubMed] [Google Scholar]
- Tsai Y-T, Lin C-I, Chen H-K, Lee K-M, Hsu C-Y, Yang S-J, Yeh N-H. Chromatin tethering effects of hNopp140 are involved in the spatial organization of the nucleolus and the rRNA gene transcription. J Biomed Sci. 2008;15:471–486. doi: 10.1007/s11373-007-9226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Le Panse S, Almouzni G, Hernandez-Verdun D. Maintenance of Nucleolar Machineries and pre-rRNAs in Remnant Nucleolus of Erythrocyte Nuclei and Remodeling in Xenopus Egg Extracts. Exp Cell Res. 2001;269:23–34. doi: 10.1006/excr.2001.5304. [DOI] [PubMed] [Google Scholar]
- Verheggen C, Mouaikel J, Thiry M, Blanchard JM, Tollervey D, Bordonne R, Lafontaine DL, Bertrand E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggener JM, DiMario PJ. Two Splice Variants of Nopp140 in Drosophila melanogaster. Mol Biol Cell. 2002;13:362–381. doi: 10.1091/mbc.01-04-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E. Stereological principles for morphometry in electron microscopic cytology. Int Rev Cyt. 1969;26:235–302. doi: 10.1016/s0074-7696(08)61637-x. [DOI] [PubMed] [Google Scholar]
- Weisenberger D, Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]