Summary
In this issue of Molecular Cell, Fukao et al. (2009) report that HuD upregulates mRNA translation through direct interaction with eIF4A in the 5' cap-binding complex, revealing a post-transcriptional role for HuD in neuronal development and plasticity.
Hu proteins are a group of RNA-binding proteins (RBPs) that affect almost every post-transcriptional aspect of mRNA metabolism (Hinman and Lou, 2008). In mammals, the Hu family has four members including HuR (HuA), HuB, HuC and HuD. Unlike many other AU-rich-element (ARE)-binding proteins that principally act to destabilize transcripts, Hu proteins stabilize ARE-containing transcripts (DeschÍnes-Furry et al., 2006). Hu proteins also affect translation of target mRNAs by acting mostly as enhancers but occasionally as repressors (Hinman and Lou, 2008); however, the underlying mechanisms of their action remain unclear.
Many studies have linked HuD expression to neuronal development and plasticity. For example, overexpression of HuD in projection neurons and HuD knockout experiments showed that HuD plays an essential role in stimulating morphological differentiation of newly committed and developing neurons through binding to a specific subset of transcripts that encode proteins involved in neuronal development (Akamatsu et al., 2005; Anderson et al., 2001). Although HuD is known to stabilize its mRNA targets and to associate with polysomes or mRNAs actively being translated (Roee et al., 2007; Tiruchinapalli et al., 2008), it is unclear whether HuD’s binding to its target messages may also affect their translation.
HuD consists of three RNA-binding domains (RBDs) with a linker region that separates the second RBD from the third RBD. The linker region contains nucleo-cytoplasmic shuttling signals. RBD1 and RBD2 together account for ARE-binding while RBD3 is responsible for poly(A)-binding (DeschÍnes-Furry et al., 2006). Fukao et al. (2009) identified the linker region and RBD3 (HuD216-385) as necessary for HuD to associate with polysomes in PC12 pheochromocytoma cells, a cellular model frequently used for studying neuronal differentiation. Using purified HuD-containing mRNA-protein complexes (mRNPs), they demonstrated that both the linker region and RBD3 of HuD are necessary and sufficient for its association with actively translating mRNAs. Co-immunoprecipitation and pulldown experiments revealed that HuD directly interacts with eIF4A, a component of the eIF4F complex, assembled at the 5' 7-methylguanosine (m7G) cap of mRNA. eIF4A is an ATP-dependent RNA helicase (Gingras et al., 1999), which plays a key role in translation initiation by unwinding RNA secondary structure in the 5′ UTR, thereby promoting ribosomal scanning (Sonenberg and Hinnebusch, 2009). Fukao et al. (2009) demonstrated that the linker region between amino acids 250 and 302 is necessary for HuD-eIF4A interaction. Moreover, they found that a single point mutation (F278A) in the linker region of HuD abolished its interaction with eIF4A. These results identify HuD as a bona fide eIF4A-interacting partner. The data strongly suggest that HuD is involved in modulating translation initiation.
Using an in vitro translation system, Fukao et al. (2009) showed that both the 5' m7G-cap and 3' poly(A) tail are required for the reporter RNA substrate to display HuD-dependent translation enhancement. Moreover, HuD mutants lacking either poly(A)-binding or eIF4A-binding ability cannot exert a translation-enhancing effect. Fukao et al. (2009) then addressed whether this interaction is physiologically significant for neuronal differentiation. They found that HuD can induce neurite outgrowth in PC12 cells whereas HuD mutants lacking either eIF4A interaction or poly(A)-binding ability cannot induce neurite outgrowth, strongly suggesting that enhancement of cap-dependent translation initiation is a prerequisite for the neurite-inducing activity of HuD. Intriguingly, a HuD216-385 mutant containing just the linker region and RBD3 was capable of inducing neurite outgrowth, even though it lacked the first two RBDs thought to be required for binding to ARE-containing mRNA targets. This observation may imply that HuD exerts a general rather than a transcript-specific enhancing effect on translation. One possibility is that HuD may promote bridging or synergism between the 5' cap-eIF4F and 3' poly(A)-binding protein (PABP) complexes that is important for translation initiation (Sonenberg and Hinnebusch, 2009). Alternatively, HuD may simply enhance eIF4A helicase activity important for spotting the first AUG initiation codon during the scanning process.
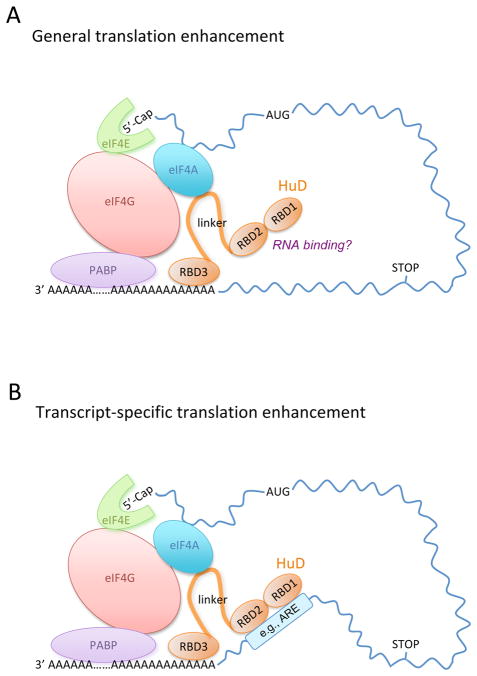
Some key questions arise from this study. For example, does HuD indiscriminately upregulate translation of all poly(A)+ mRNAs (Figure 1A), as implied by this study, or does it enhance translation of only a specific set of mRNAs (Figure 1B)? Other studies showing that HuD selectively associates with mRNAs containing AU-rich and/or U-rich sequences in neuronal cells (Bolognani et al., 2009; DeschÍnes-Furry et al., 2006) favor the hypothesis that HuD only enhances translation of a specific set of mRNAs. These studies demonstrated that target mRNA stabilization by HuD is crucial for neuronal development and plasticity. In contrast, Fukao et al. (2009) demonstrated in vitro that the translation-enhancing effect of HuD is not due to an increase in reporter mRNA stability. It will be important to determine in vivo whether the translation enhancing and mRNA stabilizing effects of HuD are coupled. It will also be interesting to figure out how the HuD216-385 mutant, which is less active than wild-type HuD in the translation assay, can be as active as wild-type HuD in inducing neurite outgrowth in PC12 cells. Another question concerns the poly(A)-binding ability of RBD3 of HuD. This interaction is necessary for both translation enhancement and neurite outgrowth. Since mRNA 3' poly(A) tails are coated with high affinity PABPs in vivo, accessibility of the RBD3 domain of HuD to poly(A) tails of its mRNA targets is a critical mechanistic issue. Answers to these questions will help us better understand the biological functions exerted by HuD and other Hu proteins.
Figure 1. Models for HuD-mediated translation enhancement.
(A) General translation enhancement. This model proposes that HuD functions as an autonomous translation activator through its interactions with eIF4A and the poly(A) tail.
(B) Transcript-specific translation enhancement. This model proposes that HuD first associates with mRNA targets through binding to AU-rich or U-rich sequences (e.g., ARE) in the 3' UTRs to exert its translation enhancing effect.
References
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. Proc Natl Acad Sci USA. 2005;102:4625–30. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Experimental Neurology. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Nucl Acids Res. 2009 October 21; doi: 10.1093/nar/gkp863. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeschÍnes-Furry J, Perrone-Bizzozero NI, Bernard JJ. BioEssays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.11.013. this issue. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Annual Review of Biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Hinman M, Lou H. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roee A, Leah B, Stav S, Irith G. J Neurosci Res. 2007;85:173–183. [Google Scholar]
- Sonenberg N, Hinnebusch AG. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Ehlers MD, Keene JD. RNA Biology. 2008;5:157–168. doi: 10.4161/rna.5.3.6782. [DOI] [PubMed] [Google Scholar]