Abstract
This article considers estimation of constant and time-varying coefficients in nonlinear ordinary differential equation (ODE) models where analytic closed-form solutions are not available. The numerical solution-based nonlinear least squares (NLS) estimator is investigated in this study. A numerical algorithm such as the Runge–Kutta method is used to approximate the ODE solution. The asymptotic properties are established for the proposed estimators considering both numerical error and measurement error. The B-spline is used to approximate the time-varying coefficients, and the corresponding asymptotic theories in this case are investigated under the framework of the sieve approach. Our results show that if the maximum step size of the p-order numerical algorithm goes to zero at a rate faster than n−1/(p∧4), the numerical error is negligible compared to the measurement error. This result provides a theoretical guidance in selection of the step size for numerical evaluations of ODEs. Moreover, we have shown that the numerical solution-based NLS estimator and the sieve NLS estimator are strongly consistent. The sieve estimator of constant parameters is asymptotically normal with the same asymptotic co-variance as that of the case where the true ODE solution is exactly known, while the estimator of the time-varying parameter has the optimal convergence rate under some regularity conditions. The theoretical results are also developed for the case when the step size of the ODE numerical solver does not go to zero fast enough or the numerical error is comparable to the measurement error. We illustrate our approach with both simulation studies and clinical data on HIV viral dynamics.
Key words and phrases: Nonlinear least squares, ordinary differential equation, Runge–Kutta algorithm, sieve approach, spline smoothing, time-varying parameter
1. Introduction
Ordinary differential equations (ODE) are widely used to model dynamic processes in many scientific fields such as engineering, physics, econometrics and biomedical sciences. In particular, new biotechnologies allow scientists to use ODE models to more accurately describe biological processes such as genetic regulatory networks, tumor cell kinetics, epidemics and viral dynamics of infectious diseases [Chen, He and Church (1999), Jansson and Revesz (1975), Michelson and Leith (1997), Daley and Gani (1999), Anderson and May (?AM91), Brookmeyer and Gail (1994), Nowak and May (2000)]. The mathematical modeling approach has made a great impact on these scientific fields over the past decades. For instance, ODE models have been used to quantify HIV viral dynamics which resulted in important scientific findings [Ho et al. (1995), Perelson et al. (1996, 1997)]. Comprehensive reviews of the application of ODE models in HIV dynamics can be found in Perelson and Nelson (1999), Nowak and May (2000), Tan and Wu (2005) and Wu (2005).
Although differential equation models have been widely used in scientific research, very little statistical research has been dedicated to parameter estimation and inference for differential equation models. The existing statistical methods for ODE models include the nonlinear least squares method [Bard (1974), van Domselaar and Hemker (1975), Benson (1979), Li, Osborne and Pravan (2005)], the smoothing-based techniques [Swartz and Bremermann (1975), Varah (1982), Chen and Wu (2008), Liang and Wu (2008), Brunel (2008)], the principal differential analysis (PDA) [Ramsay (1996), Heckman and Ramsay (2000), Poyton et al. (2006), Ramsay et al. (2007), Varziri et al. (?V07)] and the Bayesian approaches [Putter et al. (2002), Huang, Liu and Wu (2006), Donnet and Samson (2007)]. However, very few of these publications rigorously address the theoretical issues and study the asymptotic properties of the proposed estimators when both measurement error and numerical error are significant. In this paper, we intend to investigate statistical estimation methods for both constant and time-varying parameters in ODE models and study the asymptotic properties of the proposed estimators under the framework of the sieve approach.
Denote a general set of ODE models containing only constant parameters as
| (1.1) |
and denote a general set of ODE models with both constant and time-varying parameters as
| (1.2) |
where X(t) = {X1(t),…, XK(t)}T is a K-dimensional state variable vector, β is a d-dimensional vector of unknown constant parameters with true value β0, η(t) is an unknown time-varying parameter with true value η0(t) (here we only consider a single time-varying parameter, the proposed methodology can be extended to include multiple time-varying parameters although it is tedious and cumbersome in notation), F(·) = {F1(·),…, FK(·)}T is a vector of differentiable functions whose forms are known and X(t0) = X0 is the initial value. Equations (1.1) and (1.2) are called state equations. Obviously, equation (1.1) is a special case of (1.2). The function F(t, X, β) in (1.1) or F(t, X, β, η) in (1.2) is assumed to fulfil the Lipschitz assumption to X [with the Lipschitz constant independent of the unknown parameters β and η(·)] ensuring existence and uniqueness of the solutions to (1.1) and (1.2) [see Hairer, Nørsett and Wanner (1993) and Mattheij and Molenaar (2002)]. Let X(t, β) and X(t, β, η(t)) denote the true solutions to (1.1) and (1.2) for given β and η(·), respectively. We usually use notation X(t) to denote X(t, β0) or X(t, β0, η0(t)) in this article. Our objective is to estimate the unknown parameters β and η(·) based on the measurements of the state variables, X(t) or their functions.
If a closed-form solution to (1.1) or (1.2) is available, the standard statistical approaches for nonlinear regression or time-varying coefficient regression models can be used to estimate unknown parameters. In practice, (1.1) and (1.2) usually do not have closed-form solutions for a nonlinear F. In this case, numerical methods such as the Runge–Kutta algorithm [Runge (1895), Kutta (1901)] have to be used to approximate the solution of the ODEs for a given set of parameter values and initial conditions. Consequently, the nonlinear least squares (NLS) principle (minimizing the residual sum of squares of the differences between the experimental observations and numerical solutions) can be used to obtain the estimates of the unknown parameters. The NLS method for (1.1) was first described by mathematicians in 1970s [Bard (1974), van Domselaar and Hemker (1975), Benson (1979)]. The NLS method was also widely used to estimate the unknown parameters in ODE models in the fields of mathematics, computer science and control engineering. In the 1990s, the NLS method was extended to estimate time-varying parameters in (1.2). For example, the NLS method with spline approximation to time-varying parameters has been successfully applied to pharmacokinetic [Li et al. (2002)], physiologic [Thomaseth et al. (1996)] and HIV studies [Adams (2005)].
Though the NLS was the earliest and the most popular method developed for estimating the parameters in ODE models, so far the proposed NLS estimators and their asymptotic properties for ODE models have not been systematically studied, in particular, for time-varying parameter estimates. The influence of the numerical approximation error of ODEs on the asymptotic properties has not been analyzed. All existing studies took the numerical solution as the true solution and did not consider the difference between them. The difficulty is due to the co-existence of both measurement error and numerical error, and the standard theories of the NLS method [Jennerich (1969), Malinvaud (1970), Wu (1981), Delgado (1992)] cannot be directly applied. In this article, we intend to fill this gap.
The rest of the article is organized as follows. In Section 2, we discuss the identifiability problem of ODE models. Then we introduce the numerical solution-based NLS estimators for (1.1) and (1.2), and study their asymptotic properties in Sections 3 and 4, respectively. The asymptotic properties of the proposed estimators, including strong consistency, rate of convergence and asymptotic normalities, are established using the tools of empirical processes [Pollard (1984, 1990), Pakes and Pollard (1989), van der Vaart and Wellner (1996), Ma and Kosorok (2005), Wellner and Zhang (2007)] and the sieve methods [Grenander (1981), Shen and Wong (1994), Huang (1996) and Shen (1997)]. We perform simulation studies to investigate the finite-sample performance of the proposed estimation methods in Section 5. In this section, we also apply the proposed approaches to a set of ODE models for HIV dynamics. We provide a summary and discussion for the proposed methods in Section 6. Finally, the proofs of all the theoretical results are given in the Appendix.
2. Identifiability of ODE models
Identifiability of ODE models is a critical question to answer before parameter estimation. To verify the uniqueness of parameter estimates for given system inputs and outputs, both analytical and numerical techniques have been developed for ODE models since 1950s. Before jumping into technical details, two commonly used definitions of identifiability are given as follows [Bellman and Åström (1970), Cobelli, Lepschy and Jacur (1979), Walter (1987), Ljung and Glad (1994), Audoly et al. (2001), Jeffrey and Xia (2005)].
Definition 1. Globally identifiable: a system structure is said to be globally identifiable if for any two parameter vectors β1 and β2 in the parameter space  , X(t, β1) = X(t, β2) can be satisfied for all t if and only if β1 = β2.
, X(t, β1) = X(t, β2) can be satisfied for all t if and only if β1 = β2.
However, global identifiability is a strong condition to satisfy and usually difficult to verify in practice. Therefore, the definition of at-a-point identifiability was introduced by Ljung and Glad (1994) and Quaiser and Mönnigmann (2009) as follows.
Definition 2. At-a-point identifiable: a system structure is said to be locally (or globally) identifiable at a point β* if for any β within an open neighborhood of β* (or within the entire parameter space), X(t, β) = X(t, β*) can be satisfied for all t if and only if β = β*.
A number of methods have been proposed for identifiability analysis of ODE models, including structural [Bellman and Åström (1970), Ljung and Glad (1994), Xia and Moog (2003)], practical [e.g., Rodriguez-Fernandez, Egea and Banga (2006), Miao et al. (2008)] and sensitivity-based [e.g., Jolliffe (1972), Quaiser and Mönnigmann (2009)] approaches. Due to the limited space, we may not be able to provide an exhaustive list of publications on identifiability of ODE models. In this article, the structural identifiability analysis techniques are of particular interest mainly due to the theoretical completeness.
Various structural identifiability approaches have been proposed, such as power series expansion [Pohjanpalo (1978)], similarity transformation [Vajda et al. (1989), Chappel and Godfrey (1992)] and implicit function theorem method [Xia and Moog (2003), Miao et al. (2008)]. Particularly, Ollivier (1990) and Ljung and Glad (1994) introduced another approach in the framework of differential algebra [Ritt (1950), Kolchin (1973)]. The differential algebra approach is suitable to general nonlinear dynamic systems, and it has been successfully applied to nonlinear differential equation models, including models with time-varying parameters [Audoly et al. (2001)]. In this article, the differential algebra approach is employed to verify the identifiability of ODE models with both constant and time-varying parameters.
For most structural identifiability analysis techniques such as the implicit function theorem method and the differential algebra approach, a key step is the elimination of latent variables via taking derivatives and algebraic operations, which makes such techniques suitable for multivariate ODE models with partially observed state variables. After all unobserved state variables are eliminated, equations involving only given inputs, measured outputs and unknown parameters can be obtained. If we consider the parameters as unknowns, it is easy to verify that the identifiability of unknown parameters is determined by the number of roots of these equations.
For illustration purposes, we consider a classical HIV dynamic model with both constant and time-varying parameters [Nowak and May (2000), Huang et al. (?H03), Wu et al. (2005)] as an example
| (2.1) |
where TU is the concentration of uninfected target CD4+ T cells, TI the concentration of infected CD4+ T cells, V(t) the viral load, λ the proliferation rate of uninfected CD4+ T cells, ρ the death rate of uninfected CD4+ T cells, η(t) the time-varying infection rate depending on antiviral drug efficacy, δ the death rate of infected cells, c the clearance rate of free virions, N the number of virions produced by a single infected cell on average. This model will also be used in our numerical studies in Section 5. For notational simplicity, let x1, x2 and x3 denote TU, TI and V, and let y1 = TU + TI = x1 + x2 and y2 = V = x3 denote the measurable outputs, respectively. Then (2.1) can be re-written as
| (2.2) |
where x′1, x′2 and x′3 denote the derivatives of x1, x2 and x3, respectively. We adopt the following ranking for variable elimination [Ljung and Glad (1994)],
| (2.3) |
where β = (λ, ρ, N, δ, c)T is the vector of constant unknown parameters. By taking the higher order derivatives on both sides of (2.2) and using some algebra elimination techniques, we can eliminate x1, x2 and x3 from (2.2) using the ranking (2.3) to obtain
| (2.4) |
| (2.5) |
where and denote the second-order derivative of y1(t) and y2(t), respectively. Therefore, η(t) can be expressed in terms of measurable system outputs and other constant unknown parameters either from (2.4) as
| (2.6) |
or from (2.5) as
| (2.7) |
Thus, η(t) is identifiable if all the constant unknown parameters are identifiable. To verify the identifiability of all unknown parameters θ = (βT, η)T, equations (2.6) and (2.7) can be combined to obtain
| (2.8) |
The equation above only involves measurable system outputs [(TU + TI), V and their derivatives] and constant unknown parameters. We assume that the derivatives of (TU + TI) and V exist and are continuous up to order 2. Although the derivatives of (TU + TI) and V are usually not directly measured in experiments, for theoretical identifiability analysis, they are known once (TU + TI) and V are measured (e.g., via numerical evaluation). Finally, it can be verified that (2.8) is of order 0 and of degree > 1 in θ, so (2.8) satisfies the sufficient conditions given in Ljung and Glad (1994) and β = (λ, ρ, N, δ, c)T is thus at-a-point identifiable at the true parameter point. Therefore, η(t) is also at-a-point identifiable at the true parameter point. For more detailed techniques for identifiability analysis of ODE models, we refer readers to the references listed above.
3. ODE models with constant parameters
Throughout this article, we let ‖a‖ be the Euclidean norm (or L2 norm) of a vector (or a matrix) a; be the supremum norm of an m × n matrix A, where aij is the (i, j)th element of A; A⊗2 = AAT for a matrix A; Cr[a, b] be the class of functions with r-order continuous derivative on the interval [a, b]; ‖f‖∞ = supt |f(t)| be the supremum norm of a function f; and x ∧ y denotes min(x, y). Moreover, for a random vector Z ∼ P, where P is a probability measure, we let ‖f(Z)‖2 = ‖f‖P,2 = (∫f2 dP)1/2 be the L2(P)-norm of a function f.
3.1. Measurement model and estimator
In this section, we consider ODE models with constant parameters, that is, equation (1.1), over the time range of interest I = [t0, T] (−∞ < t0 < T < +∞), where the initial value X0 = X(t0) is assumed to be known in this article. In reality, X(t) cannot be measured exactly and directly; instead, its surrogate Y(t) can be measured. For simplicity, here we assume an additive measurement error model to describe the relationship between X(ti) and the surrogate Y(ti),
| (3.1) |
at random or fixed design time points t1,…,tn, where the measurement errors (ε(t1),…,ε(tn)) are independent with mean zero and a diagonal variance–covariance matrix Σ. Moreover, in the case of random design, assume that the measurement errors are independent of X(t). Equation (3.1) is called the observation or measurement equation.
If (1.1) does not have a closed-form solution, we need to resort to numerical techniques to obtain numerical solutions at discrete time points. In this article, we consider a general one-step numerical method. Let t0 = s0 < s1 < ⋯ < sm−1 = T be grid points on the interval I, hj = sj − sj−1 be the step size and h = max1≤j≤m−1 hj be the maximum step size, and and be the numerical approximations to the true solutions X(sj) and X(sj+1), respectively, which can be typically written as
| (3.2) |
where the specific form of Φ depends on the numerical method. The common numerical methods include the Euler backward method, the trapezoidal rule, the r-stage Runge-Kutta algorithm (r is usually between 2 and 5), and so on. Among these algorithms, the 4-stage Runge-Kutta algorithm [Mattheij and Molenaar (2002), page 53, Hairer, Nørsett and Wanner (1993), page 134] has been well developed and widely used in practice. Therefore, we employ the 4-stage Runge–Kutta algorithm as an example in our numerical studies.
Define , which is called the numerical error or the global discretization error [Hairer, Nørsett and Wanner (1993), page 159, Mattheij and Molenaar (2002), page 57]. If eh = O(hp), p is called the order of the numerical method. It is necessary to establish a relationship between the number of grid points m (or the maximum step size h) and the sample size of measurements n since the asymptotic properties of the proposed estimators are related to both numerical error and measurement error. To our best knowledge, this is the first attempt to establish such as a relationship.
Following Mattheij and Molenaar (2002, page 58), the interpolation technique is commonly used if the measurement points (ti, i = 1, 2,…, n) are not coincident with the grid points (sj, j = 1, 2,…,m − 1) of the numerical method, and the cubic Hermite interpolation is often adopted. Let X̃(t, β) denote the interpolated numerical solution of X(t, β) obtained from the numerical method for given β, and then (3.1) can be approximately rewritten as Y(t) ≈ X̃(t, β0) + ε(t). The simple numerical solution-based NLS estimator β̂n of β0 minimizes
| (3.3) |
Note that if the data are correlated or the measurement variances are heterogeneous, the weighted NLS can be used. The theoretical results can be extended to the weighted NLS. Also note that we can easily obtain the estimator X̂(t) = X̃(t, β̂n) for X(t).
To minimize the NLS objective function (3.3), the standard gradient optimization methods may fail due to the complicated nonlinear ODE model and the NLS objective function may have multiple local minima or may be ill-behaved [Englezos and Kalogerakis (2001)]. Fortunately, various global optimization methods are available to more reliably solve the parameter estimation problem for ODE models, although the global optimization methods are very computationally intensive. Moles, Banga and Keller (2004) compared the performance and computational cost of seven global optimization methods, including the differential evolution method [Storn and Price (1997)]. Their results suggest that the differential evolution method outperforms the other six methods with a reasonable computational cost. Improved performance can be achieved using a hybrid method combining gradient methods and global optimization methods. A hybrid method based on the scatter search and sequential quadratic programming (SQP) has been proposed by Rodriguez-Fernandez, Egea and Banga (2006), who showed that the hybrid scatter search method is much faster than the differential evolution method for a simple HIV ODE model. In addition, Miao et al. (2008) also suggested that global optimization methods should be used for general nonlinear ODE models. Here we combine the differential evolution, the scatter search method and the SQP local optimization technique to implement our NLS minimization.
3.2. Asymptotic properties
In this section, we study the asymptotic properties of the proposed numerical solution-based NLS estimator when both measurement error and numerical error are considered. First we make the following assumptions:
-
A1
β ∈
 , where
, where  is a compact subset of
is a compact subset of  d with a finite diameter Rβ.
d with a finite diameter Rβ. -
A2
Ω = {X(t, β) : t ∈ I, β ∈
 } is a closed and bounded convex subset of
} is a closed and bounded convex subset of  K.
K. -
A3
There exist two constants −∞ < c1 < c2 < +∞ such that c1 ≤ Y(t) ≤ c2 for all t ∈ I.
-
A4
All partial derivatives of F(t, X, β) up to order p with respect to t and X exist and are continuous.
-
A5
The numerical method for solving ODEs is of order p.
-
A6
For any β ∈
 , Et[X(t, β) − X(t, β0)]2 = 0 if and only if β = β0.
, Et[X(t, β) − X(t, β0)]2 = 0 if and only if β = β0. -
A7
The first and second partial derivatives, and , exist and are continuous and uniformly bounded for all t ∈ I and β ∈
 .
. -
A8
For the ODE numerical solution X̃(t, β), the first and second partial derivatives, and , exist and are continuous and uniformly bounded for all t ∈ I and β ∈
 .
. -
A9
Let 0 < c3 < c4 < ∞ be two constants. For random design points, t1,…,tn are i.i.d. The joint density function ϕ(t, y) of (t, Y) satisfies c3 ≤ ϕ(t, y) ≤ c4 for all (t, y) ∈ [t0, T] × [c1, c2].
-
A10
The true parameter β0 is an interior point of
 .
. -
A11
β̃ is an interior point of
 , where β̃ = arg minβ ∈
, where β̃ = arg minβ ∈ E0[Y(t) − X̃(t, β)]T × [Y(t) − X̃(t, β)] and E0 is the expectation with respect to Pβ0, the joint probability distribution of (t, Y(t)) at true value β0.
E0[Y(t) − X̃(t, β)]T × [Y(t) − X̃(t, β)] and E0 is the expectation with respect to Pβ0, the joint probability distribution of (t, Y(t)) at true value β0. -
A12
is positive definite, where Et[g(t)] is expectation of function g(t) with respect to t.
-
A13
is positive definite.
Assumptions A1–A4 are general requirements for existence of numerical solutions of ODE models. Assumption A5 from Mattheij and Molenaar (2002, pages 55 and 56) defines the precision of the numerical algorithm. For example, the Euler backward method, the trapezoidal rule, the 4-stage and 5-stage Runge–Kutta are of order 1, 2, 4 and 5, respectively. Theorem 2.13 in Hairer, Nørsett and Wanner (1993, page 153) provides sufficient and necessary conditions for the numerical method to be of order p. Theorems 3.1 and 3.4 in Hairer, Nørsett and Wanner (1993, pages 157 and 160) give the magnitude of the numerical error of the numerical algorithms. Assumption A6 is required for identifiability and imposed for consistency. From Section 2, we know that the HIV model (2.1) is at-a-point identifiable at the true value β0. This result and assumption A9 are sufficient conditions for assumption A6 to be satisfied. Assumptions A7–A9 are needed for consistency. Assumptions A10–A13 are needed for the proof of asymptotic normality in Theorem 3.2.
Theorem 3.1. Assume that there exists a λ > 0 such that h = O(n−λ), then under assumptions A1–A10, we have β̂n − β0 → 0, almost surely under Pβ0.
Theorem 3.2. (i) For h = O(n−λ) with λ > 1/(p ∧ 4) where p is the order of the numerical method (3.2), under assumptions A1–A10 and A12, we have that .
(ii) For h = O(n−λ) with 0 < λ ≤ 1/(p ∧ 4), under assumptions A1–A9, A11 and A13, we have that with ‖β̃ − β0‖ = O(h(p∧4)/2) = O(n−λ(p∧4)/2) and ‖Ṽ1 − V1‖ = O(h(p∧4)/2) = O(n−λ(p∧4)/2).
The detailed proofs of Theorems 3.1 and 3.2 are provided in the Appendix. The basic idea for the proofs is motivated by Pakes and Pollard (1989) in which a general central limit theorem is proved for a broad class of simulation estimators, that is, the objective function of the estimator is too complicated to evaluate directly, and instead the Monte Carlo simulation is used to approximate the objective function to obtain the estimator. The asymptotic properties of the simulation-based estimator are established using a general central limit theorem under nonstandard conditions given in Huber (1967) and Pollard (1985), which are often called the Huber–Pollard Z-theorem [see Theorem 3.3.1 in van der Vaart and Wellner (1996)]. In this article, we use the same theorem to prove the asymptotic normality of the numerical solution-based NLS estimator for ODEs. Similarly, our objective function Ξ1 (β) in (3.3) cannot be directly evaluated; instead we have to approximate it by solving (1.1) numerically. Thus, similar ideas in Pakes and Pollard (1989) can be borrowed to establish the asymptotic results of our estimator in Theorems 3.1 and 3.2.
Remark 1. Theorems 3.1 and 3.2 can be extended to fixed design points ti ∈ [t0, T] (i = 1,…, n). Assume that there exists a distribution function Q(t) with corresponding density φ(t) such that, with Qn(t), the empirical distribution of (t1,…,tn), supt∈[t0, T] |Qn(t) − Q(t)| = Op(n−1/2) and φ(t) is bounded away from zero and has continuous second-order derivative on [t0, T]. Define Et[g(t)] be the integral for function g(t). Similarly we can prove Theorems 3.1 and 3.2 for the fixed design if we replace assumption A9 by above assumption.
Remark 2. From the proof of Theorem 3.2 in the Appendix, we still have ‖β̃ − β0‖ = O(h(p∧4)/2) and ‖Ṽ1 − V1‖ = O(h(p∧4)/2) for a fixed constant h, which is independent of the sample size n. This suggests that, if the maximum step size h of the numerical algorithm for solving ODEs is a fixed constant, the numerical solution-based NLS estimator is not consistent. Instead the asymptotic bias is in the order of h(p∧4)/2.
Notice that our asymptotic results provide a theoretical foundation for the relationship between the numerical step size and sample size, that control numerical error and measurement error, respectively, for the widely-used NLS estimator based on the numerical solutions of the ODEs. Intuitively, the smaller the numerical step size is, better the estimator is. However, a smaller step size will increase the computational cost and this may become a serious problem when the ODE system is large and the computational cost is high. It is important to study the trade-off between the numerical error and measurement error when the computational cost needs to be taken into consideration. Our theoretical results show that, only when the numerical step size, which controls the numerical error and computational cost, goes to zero with a rate faster than a particular rate n−1/(p∧4), the numerical solution-based NLS estimator converges to the true value of the parameters with the rate of root-n. In addition, the asymptotic variance of the NLS estimator is the one as if the true solution X(t) is exactly known.
The asymptotic variance–covariance matrix needs to be estimated in order to perform statistical inference for unknown parameters β. There are some standard methods that can be used. The first approach is to use the observed pseudo-information matrix based on the NLS objective function (3.3). The observed pseudo-information matrix is defined as . The standard error of β̃n can then be approximated by . In practice, we have noted that the inverse of the observed pseudo-information matrix provides a reasonable approximation to the asymptotic variance-covariance matrix V1. Rodriguez-Fernandez, Egea and Banga (2006) also proposed this approach for parameter inference in ODE models.
The second approach is the weighted bootstrap method [Ma and Kosorok (2005)]. Let Wi, i = 1,…,n, denote n i.i.d. positive random weights with mean one [E(W) = 1] and variance one [Var(W) = 1]. The weights, Wi are independent of {β, t, Y(t)}. For (1.1), the weighted M-estimator satisfies
From Corollary 2 and Theorem 2 in Ma and Kosorok (2005), given {ti, Y(ti)}, and have the same limiting distribution, then the weighted M-estimator can be used for inference on β̂n.
Note that the empirical bootstrap has been used for statistical inference for ODE models [Joshi, Seidel-Morgenstern and Kremling (2006)]. However, the asymptotic properties of the empirical bootstrap estimators are quite difficult to derive. This is why we propose to use the weighted bootstrap method instead of the empirical bootstrap approach.
4. ODE models with both constant and time-varying parameters
4.1. Measurement model and estimator
In this section, we consider (1.2) with both constant and time-varying parameters, where the initial value X0 = X(t0) is assumed to be known. Again, X(t) is not observed directly in practice; instead, we observe its surrogate Y(t) through (3.1).
Let  be the following class of functions,
be the following class of functions,
| (4.1) |
where μ is a nonnegative integer, γ ∈ (0, 1], ϱ = μ + γ > 0.5, and L an unknown constant. The smoothness assumption is often used in nonparametric curve estimation. Usually, either ϱ = 1 (i.e., μ = 0 and γ = 1) or ϱ = 2 (i.e., μ = 1 and γ = 1) should be satisfied in various situations. Denote θ = (βT, η)T. Then the parameter space is denoted by Θ = {θ : β ∈  , η ∈
, η ∈  } =
} =  ×
×  .
.
In this article, we use the method of sieves to approximate η0(t) on the support interval [t0, T] of t. The basic idea of the sieve approach is to approximate an infinite-dimensional parameter space Θ by a series of finite-dimensional parameter spaces Θn, which depend on the sample size n, and then to estimate the parameter on the finite-dimensional spaces Θn instead of Θ. The concept of sieve was first proposed by Grenander (1981). Since then, the sieve method has been a powerful tool in the area of nonparametric and semiparametric statistics [Shen and Wong (1994), Huang (1996), van der Vaart and Wellner (1996), Section 3.4, Shen (1997), Huang and Rossini (1997), Huang (1999), He, Fung and Zhu (2002), Xue, Lam and Li (2004) and Huang, Zhang and Zhou (2007)].
Here we apply the sieve estimation method to (1.2) with a time-varying parameter. First, we approximate η(t) by B-spline functions on the support interval I of t. Let t0 = u0 < u1 < ⋯ < uq = T be a partition of the interval I, where q = O(nv) (0 < v < 0.5) is a positive integer such that max1≤j≤q|uj − uj−1| = O(n−v). Then we have N = q + l normalized B-spline basis functions of order l + 1 ≥ ϱ [see Huang (2003), page 1618] that form a basis for the linear spline space. We denote these basis functions in the forms of a vector π(t) = (B1(t),…,BN(t))T with which η(t) can be approximated by π(t)Tα, where α = (α1,…,αN)T ∈  N is the spline coefficient vector with α0 corresponding to η0 (t). Such approximation is extensively used in nonparametric and semiparametric problems [Stone (1985), Shen and Wong (1994), Shen (1997), Huang (1999) and Huang (2003)]. The readers are referred to Schumaker (1981, page 118) for more details about the construction of the basis functions. Regression spline approximation to a nonparametric function can always be expressed as a linear function of basis functions so that the problem of time-varying coefficients can be transformed into an estimation problem for a number of constant parameters. Thus the estimation methods and computational algorithms developed for (1.1) with constant coefficients in Section 3 can be employed for (1.2) with both constant and time-varying parameters.
N is the spline coefficient vector with α0 corresponding to η0 (t). Such approximation is extensively used in nonparametric and semiparametric problems [Stone (1985), Shen and Wong (1994), Shen (1997), Huang (1999) and Huang (2003)]. The readers are referred to Schumaker (1981, page 118) for more details about the construction of the basis functions. Regression spline approximation to a nonparametric function can always be expressed as a linear function of basis functions so that the problem of time-varying coefficients can be transformed into an estimation problem for a number of constant parameters. Thus the estimation methods and computational algorithms developed for (1.1) with constant coefficients in Section 3 can be employed for (1.2) with both constant and time-varying parameters.
For any θi ∈  ×
×  (i = 1, 2), we define a distance
(i = 1, 2), we define a distance
| (4.2) |
Denote set
where ℓn ≤ n(2l−1)/[2l′(2l+1)] with a constant l′ arbitrarily close to l [see Shen (1997), page 2560], then Θn = {θ : β ∈  , η ∈
, η ∈  n} =
n} =  ×
×  n can be used as a sieve of Θ. In fact, for any θ = (βT, η)T ∈ Θ, by Corollary 6.21 in Schumaker (1981), there exists ηn ∈
n can be used as a sieve of Θ. In fact, for any θ = (βT, η)T ∈ Θ, by Corollary 6.21 in Schumaker (1981), there exists ηn ∈  n such that ‖ηn − η‖∞ = Op(n−vϱ). Denote θn = (βT, ηn)T ∈ Θn, then d(θ, θn) = Op(n−vϱ). Equation (1.2) now becomes
n such that ‖ηn − η‖∞ = Op(n−vϱ). Denote θn = (βT, ηn)T ∈ Θn, then d(θ, θn) = Op(n−vϱ). Equation (1.2) now becomes
For this approximation model, let X̃(t, β, π(t)Tα) be the numerical approximation of X(t, β, η(t)) that can be obtained from the same numerical algorithm as described in Section 3. Equation (3.1) can be approximated by Y(t) ≈ X̃(t, β, π(t)Tα0) + ε(t). The numerical solution-based sieve NLS estimator is defined as
| (4.3) |
When we substitute the sieve NLS estimators θ̂n into the numerical approximation, we can obtain the estimator X̂(t) = X̃(t, β̂n, η̂n(t)).
4.2. Asymptotic properties
The empirical objective function for the sieve NLS method proposed in Section 4.1 is a second-order loss function which is not a likelihood function. We cannot use the standard information calculation of the maximum likelihood estimator (MLE) based on orthogonal projections in semiparametric models [Bickel et al. (1993)], and the asymptotic normality theory for semiparametric MLEs obtained by Huang (1996, Theorem 6.1) does not apply to our case. Fortunately, Ma and Kosorok (2005) and Wellner and Zhang (2007) extended the Huang's asymptotic normality results to more general semiparametric M-estimators by using a so-called pseudo-information calculation. We are able to employ these new asymptotic results to asymptotic properties of the proposed sieve NLS estimator, and the following additional assumptions are needed:
-
B1
The true time-varying parameter η0(·) ∈
 , where
, where  is denoted in (4.1).
is denoted in (4.1). -
B2
All partial derivatives of F up to order p with respect to t, X, and η, respectively, exist and are continuous.
-
B3
For any β ∈
 and η ∈
and η ∈  , Et[X(t, β, η(t)) − X(t, β0, η0(t))]2 = 0 if and only if β = β0 and P{t : η(t) = η0(t)} = 1.
, Et[X(t, β, η(t)) − X(t, β0, η0(t))]2 = 0 if and only if β = β0 and P{t : η(t) = η0(t)} = 1. -
B4
The first and second partial Fréchet-derivatives [van der Vaart and Wellner (1996), page 373] in the norm d defined in (4.2), , , , and exist and are continuous and uniformly bounded for all t ∈ I, β ∈
 and η ∈
and η ∈  .
. -
B5
For the ODE numerical solution X̃(t, β, η), the first and second partial Fréchet-derivatives in the norm d, , , , and exist and are continuous and uniformly bounded for all t ∈ I, β ∈
 and η ∈
and η ∈  .
. -
B6
For K ≥ 2, is positive definite, where S1 and S2 are defined in (A.5) and (A.6) in the Appendix, respectively.
-
B7
v satisfies the restrictions 0.25/ϱ < v < 0.5 and v(2 + ϱ) > 0.5, where ϱ is the measure of smoothness of η(t) defined in assumption (B1).
Theorem 4.1. Assume that there exists a λ > 0 such that h = O(n−λ) and under assumptions A1–A4, A9, A10 and B1–B5, we have d(θ̂n, θ0) → 0, almost surely under Pθ0.
Theorem 4.2. Assume that there exists a λ > 1/[2(p ∧ 4)] such that h = O(n−λ) where p is the order of the numerical algorithm (3.2), and under assumptions A1–A4, A9, A10 and B1–B5, we have d(θ̂n, θ0) = Op(n−vϱ + n−(1−v)/2).
From Theorem 4.2, we know that ‖β̂n − β0‖ = Op(n−vϱ + n−(1−v)/2) and ‖η̂n(t) − η0(t)‖2 = Op(n−vϱ + n−(1−v)/2). If v = 1/(1 + 2ϱ), the rate of convergence of η̂n is n−ϱ/(1+2ϱ), which is the same as the optimal rate of the standard nonparametric function estimation [Stone (1982)]. Theorem 4.3 below states that the rate of weak convergence of β̂n achieves n−1/2 under some additional assumptions.
Theorem 4.3. For the maximum step size h = O(n−λ) with λ > 1/(p ∧ 4), under assumptions A1–A4, A9, A10 and B1–B7, and K ≥ 2, we have .
Remark 3. For the case h = O(n−λ) with 1/[2(p ∧ 4)] < λ ≤ 1/(p ∧ 4), similar results to case (ii) in Theorem 3.2 can be obtained.
For K = 1, Theorem 4.3 does not hold, since in this case the special perturbation direction a*(t) given in (A.4) is , which leads to both S1 in (A.5) and S2 in (A.6) to be zero (see the proof of Theorem 4.3 in the Appendix). In this article, we consider one special case that we assume there exists an additive relationship between β and η(·) as follows:
| (4.4) |
which is a special case of (1.2), then the function X(t) has the form of X(t, β + η(t)). In this case, we are able to establish similar asymptotic normality results under the identifiability constraint Etη(t) = 0. Note that Schick (1986) studied a similar problem under a semiparametric regression model and used the same identifiability constraint for the unknown function η(t) to establish the asymptotic normality for the constant parameters. We follow a similar idea and use B-spline approximation for η(t). We center the B-spline estimator of η(t) as follows:
which is subject to the constraints . Under similar assumptions, the strong consistency and the rate of weak convergence of the estimators, similar to those of Theorems 4.1 and 4.2, can be obtained. In particular, the asymptotic normality can be established as follows:
Proposition 1. For (4.4) with K = 1, when the maximum step size h = O(n−λ) with λ > 1/(p ∧ 4), under assumptions A1–A4, A9, A10, B1–B5, B7 and in addition Et [η(t)] = 0, we have , where with ξ = β0 + η0(t).
The proof of this proposition is different from that of Theorem 4.3 and is given in the Appendix.
Remark 4. By combining Theorem 4.3 and Proposition 1, we can see that the proposed sieve NLS estimator is asymptotically normal with a convergence rate of for K ≥ 2 under assumption B6, but we are only able to prove the result for a special ODE model (4.4) for K = 1. This is because the asymptotic covariance V2 defined in B6 is always singular in the case of K = 1, and is only possibly nonsingular in the case of K ≥ 2. Since V2 is always singular for K = 1, we derive the asymptotic distribution for the special ODE model (4.4) using a different approach which results in Proposition 1.
Similar approaches proposed in Section 3 can be used to estimate the asymptotic variance-covariance matrix for (β̂n, η̂n (t)). For the first approach, the observed pseudo-information matrix can be evaluated by replacing η(t) with the spline approximation πT (t)α, that is, to rewrite the objective function Ξ2(θ) in the expression (4.3) as Ξ2(β,α). Then the observed pseudo-information matrix  2(β,α) can be defined as
2(β,α) can be defined as
The standard error of (β̂n, α̂n) is approximately from which the standard error of β̂n can be obtained. We also find that the inverse of the observed pseudo-information matrix provides a reasonable approximation to V2 via our simulation studies in the next section.
Similarly the weighted bootstrap method can also be used. For (1.2), the weighted M-estimators satisfy
Based on Corollary 2 and Theorem 2 in Ma and Kosorok (2005), given {ti, Y(ti)}, and have the same limiting distribution which can be used to justify the weighted bootstrap for inference on β̂n and η̂n(t).
5. Numerical studies
In this section, we consider the HIV dynamic model described in Section 2. Recall that in this system, TU(t), TI (t) and V (t) are state variables and (λ, ρ, δ, N, c, η(t))T are kinetic parameters. By introducing the time-varying infection rate η(t) in this HIV dynamic model, the model can flexibly describe the long-term viral dynamics. In clinical studies, only viral load, V(t) and total CD4+ T cell count, T(t) = TU(t) + TI(t), are closely monitored and measured over time. For easy illustration and computational simplicity, we fix the parameters ρ and δ in our numerical studies, and our objective is to estimate three constant parameters and one time-varying parameter, (λ, N, c, η(t))T based on measurements of viral load and total CD4+ T cell count.
5.1. Monte Carlo simulation study
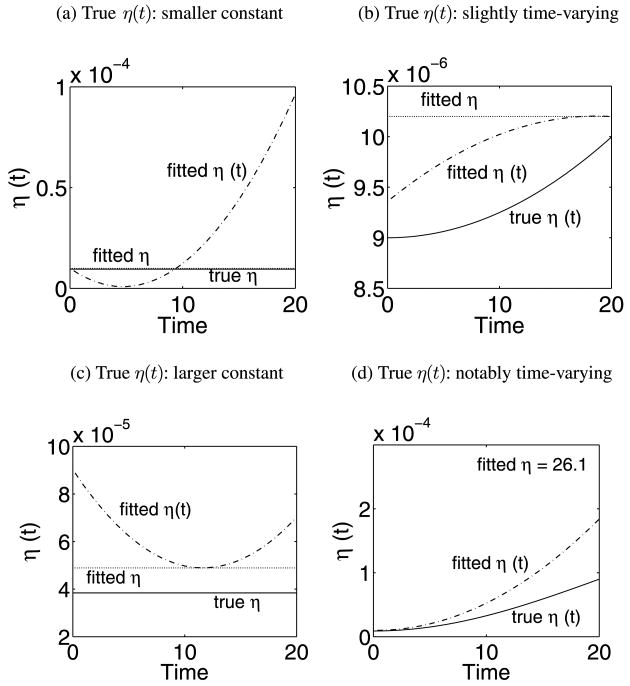
The following parameter values and initial conditions were used to simulate observation data for (2.1): TU(0) = 600, TI (0) = 30, V (0) = 105, λ = 36, ρ = 0.108, N = 1000, δ = 0.5, c = 3. For comparison purpose, we generated the measurement data of V(t) and T(t) for four scenarios in our simulation studies: (i) η(t) = η is a small constant, η = 9.5 × 10−6; (ii) η(t) is time-varying but with a smaller (10%) variation, η(t) = 9 × 10−5 × {1 − 0.9cos(πt/400)}; (iii) η(t) = η is a larger constant, η = 3.84 × 10−5; and (iv) η(t) is time-varying but with a large (10-fold) variation, η(t) = 9 × 10−5 × {1 − 0.9cos(πt/40)}. Note that for cases (i) and (iii), the values of constant η were chosen to be approximately the average of η(t) over the period of time interval for cases (ii) and (iv), respectively.
Let y1 = T = TU + TI denote the total number of infected and uninfected CD4+ cells and y2 = V denote the viral load, the measurement models are given as follows:
where ε1i and ε2i are independent and follow normal distributions with mean zero and variances and , respectively. The HIV dynamic model was numerically solved within the time range [0, 20] to generate the simulated data at each time interval of 0.5 using the 4-stage Runge–Kutta algorithm. Consequently, the corresponding sample size is 40. The 20% measurement errors were added to the numerical results of the ODE model according to the observation equations above. We applied the proposed estimation methods in Sections 3 and 4 to the simulated data for the 4 cases to evaluate the performance of the proposed estimators and the effect of the model misspecification. To stabilize the computational algorithm, we log-transformed the data. We also fixed parameters ρ and δ as their true values.
For evaluating the performance of the estimation methods, we define the average relative estimation error (ARE) as
where θ̂j is the estimate of the parameter vector θ from the j th simulation data set, and M = 500 is the total number of simulation runs.
In Table 1, the AREs of the constant parameters (λ, N, c) are listed. In addition, we also report as the average of the estimated variance by the observed pseudo-information matrix and as the empirical variance based on simulation runs. Based on these results, we can see that, when the change of η(t) is small as a function of time t or η is a small constant, the estimation of parameters is always good by fitting a constant η model as observed by the low ARE values. However, when the change of η(t) is large or η is a large constant, misspecification of η(t) may produce large AREs for all parameter estimates. In particular, when η(t) is time-varying with a large variation, using a constant η model may result in very poor estimates for all constant parameters. The variance estimates based on the pseudo-information agree well with the empirical estimates based on simulations, which shows that the pseudo-information-based variance estimate is reasonably good. The evaluation of the bootstrap variance estimation is prohibited in our simulation study due to high computational cost.
Table 1. Simulation results for constant η and the time-varying η(t) models. The ARE is calculated based on 500 simulation runs for the HIV dynamic model. In addition, is the average of the estimated variance by the observed pseudo-information, and is the empirical variance based on simulations. The sample size is n = 40 and the noise level is about 20%.
| Change of η(t) | True η(t) model | Fitted η(t) model | λ | N | c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARE(%) | ARE(%) | ARE(%) | ||||||||||||
| Small | Constant | Constant | 3.19 | 2.49 | 1.91 | 17.7 | 3.23e+04 | 4.94e+04 | 17.4 | 0.313 | 0.425 | |||
| Time-varying | 6.45 | 9.82 | 8.77 | 22.9 | 7.14e+04 | 8.63e+04 | 20.5 | 0.593 | 0.635 | |||||
| Time-varying | Constant | 3.77 | 2.38 | 2.08 | 17.9 | 3.36e+04 | 4.71e+04 | 19.8 | 0.331 | 0.432 | ||||
| Time-varying | 6.40 | 9.16 | 8.55 | 22.6 | 6.53e+04 | 7.93e+04 | 20.9 | 0.543 | 0.637 | |||||
| Large | Constant | Constant | 6.29 | 8.19 | 12.1 | 72.5 | 1.13e+06 | 8.53e+05 | 67.3 | 9.22 | 6.75 | |||
| Time-varying | 7.34 | 9.19 | 15.6 | 88.8 | 3.25e+06 | 1.13e+06 | 82.5 | 26.2 | 8.96 | |||||
| Time-varying | Constant | 94.2 | 13.7 | 7.02 | 994 | 5.86e+07 | 1.25e+08 | 1899 | 1660 | 3780 | ||||
| Time-varying | 15.6 | 31.5 | 48.2 | 29.5 | 1.67e+05 | 1.67e+05 | 25.1 | 1.81 | 1.37 | |||||
In Figure 1, the average trajectories of estimated η(t) are compared to the true trajectories of η(t) for four different scenarios. From this figure, we observed a similar trend as the constant parameter estimates. The misspecification of η(t) produces estimation error, in particular for the cases with a large variation of η(t) or a large constant η. When the model of η(t) is correctly specified, the estimates based on the proposed methods are reasonably good. In order to evaluate the robustness of the proposed approach, we also performed further simulation studies for a complex function η(t) = 9.0 × 10−6 +9.0 × 10−7 × t ×{1 − 0.5sin(πt/5.8)} under the same simulation settings (i.e., 40 time points, 20% error, 500 simulation runs). The results suggest that the sieve estimator can still capture the essential pattern of the complex η(t) reasonably well (plots not shown).
Fig. 1. Simulation results for constant η and the time-varying η(t) models. In each figure, the true model of η (solid), the constant η model (dotted) and the time-varying η(t) model (dash-dotted) are plotted and compared.
5.2. Application to AIDS clinical data
To illustrate applicability and feasibility of our proposed methods and theories, we also applied the proposed estimation methods to fit the HIV dynamic model to a clinical data set obtained from an HIV-1 infected patient who was treated with an antiretroviral therapy. Very frequent viral load measurements were collected from this patient after initiating the antiretroviral regimen: 13 measurements during the first day, 14 measurements from day 2 to week 2, and then one measurement at weeks 4, 8, 12, 14, 20, 24, 28, 32, 36, 40, 44, 48, 52, 56, 64, 74 and 76, respectively. In addition, the measurements of total CD4+ T cell counts were also taken at Day 1, weeks 2 and 4, and monthly thereafter. Equation (2.1) was used to estimate HIV kinetic parameters using the viral load and total CD4+ T cell data.
For simplicity of illustration and computation, we fixed the initial conditions of the state variables in (2.1) as TU(0) = 1, TI (0) = 551, V (0) = 6.38 × 104, which were derived from the baseline measurements. We also fixed two parameters, as in the simulation study, ρ = 0.10 and δ = 0.434, which were taken from the estimates in literature. Our objective is to estimate the three constant parameters (λ, N, c) and the time-varying parameter η(t) as in the simulation study. As we proposed in Section 4, we employed B-splines to approximate η(t). We positioned the spline knots at equally-spaced time points (the log-time scale was used since the distribution of observation time points is highly-skewed). We selected the order of splines and the number of spline knots using the model selection criterion AICc given by
where RSS is the residual of the sum of squares obtained from the NLS model fitting, n is the total number of observations and k is the number of unknown parameters [including the coefficients in the spline representation of η(t)]. Note that as a practical guideline, if the number of unknown parameters exceeds n/40 (where n is the sample size), the AICc instead of AIC should be used. For our clinical data, the sample size n is equal to 65, and the number of unknown parameters varies between 6 and 13 for different scenarios, which is much larger than n/40 = 65/40 = 1.6. Thus the AICc is more appropriate for our applications. In general, the AICc converges to the AIC as the sample size gets larger, thus the AICc is often suggested to be employed regardless of the sample size [Burnham and Anderson (2004)]. For our application, we used AICc and compared the models with the splines of order 3 and 4, and the number of knots from 3 to 10. In Table 2, the AICc values for these different models are reported, from which the best model was selected as the spline with order 3 and 5 knots for η(t) approximation.
Table 2. Model selection results for B-spline approximation of the time-varying parameter η(t).
| Model | Spline order | Number of knots | AICc |
|---|---|---|---|
| 1 | 3 | −222.3 | |
| 2 | 4 | −242.8 | |
| 3 | 5 | −252.8 | |
| 4 | 6 | −243.8 | |
| 5 | 3 | 7 | −250.6 |
| 6 | 8 | −246.0 | |
| 7 | 9 | −246.8 | |
| 8 | 10 | −244.4 | |
| 9 | 3 | – | |
| 10 | 4 | −233.2 | |
| 11 | 5 | −230.3 | |
| 12 | 6 | −242.6 | |
| 13 | 4 | 7 | −249.1 |
| 14 | 8 | −245.5 | |
| 15 | 9 | −244.9 | |
| 16 | 10 | −240.5 |
We used the weighted bootstrap method to calculate both the confidence intervals for the constant parameters and the confidence bands for the time-varying parameter. The basic idea of the weighted bootstrap method is provided in Sections 3.2 and 4.2. For the computational implementation, we first generated a positive random weight for each data point in the raw data set from the exponential distribution with mean one and variance one. By repeating this step, a large number of (say, 1000) sets of weights can be generated. Second, for each set of weights, the ODE model is fitted to the data to obtain parameter estimates by minimizing the weighted residual sum of squares (see Sections 3 and 4). Recall that the time-varying parameter in the model has been approximated by B-splines, then both the constant parameters and the constant B-spline coefficients are actually estimated. Once the estimates of the B-spline coefficients are obtained, we construct the B-splines which approximate the time-varying parameter. Thus, we eventually obtain 1000 estimates for each constant parameter and 1000 B-splines for each time-varying parameter. Third, for each constant parameter, we select the 2.5% and 97.5% quantiles of the 1000 estimates to form the 95% confidence intervals for this parameter. For the time-varying parameter, at a single time point, the 1000 B-splines have 1000 values. We also select the 2.5% and 97.5% quantiles of the 1000 values at this time point to eventually form the 95% pointwise confidence bands for the time-varying parameter.
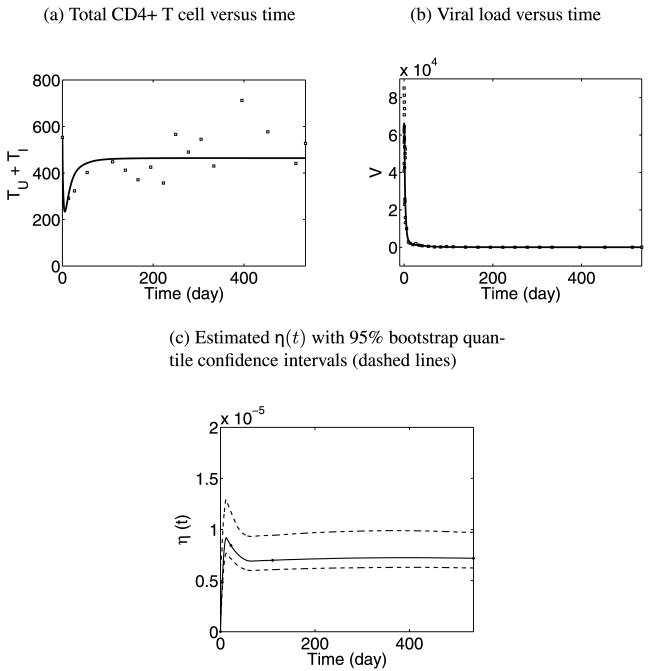
Model fitting results are given in Figure 2 and Table 3. From Figures 2(a) and (b), we can see that the fitting is reasonably good for both CD4+ T cell counts and viral load data. The estimates of constant parameters (λ, N, c) are listed in Table 3, and the 95% bootstrap confidence intervals of the estimates are also provided. The uninfected cell proliferation rate (λ) was estimated as 46.52 cells per day, the average number of virions produced by one infected cell (N) was estimated as 1300 per day and the clearance rate of free virions was 4.35 per day which corresponds to a half-life of 3.8 hours. All these estimates are in the ball-park of similar estimates from other methods [Perelson et al. (1996, 1997)]. In Figure 2(c), the estimated trajectory of the time-varying parameter η(t) (the viral infection rate), is plotted with 95% bootstrap quantile confidence intervals, which shows an initial fluctuation but converges to a constant after 2 to 3 months.
Fig. 2. Model fitting results with η(t) approximated by B-splines of order 3 and 5 knots.
Table 3. The constant parameter estimation results.
| Parameter | Estimate | 95% confidence interval |
|---|---|---|
| λ | 46.52 | [43.20, 51.04] |
| N | 1300.39 | [251.93, 4628.26] |
| c | 4.35 | [0.98, 14.83] |
6. Discussion
In this paper, we have systematically studied numerical solution-based NLS estimators for general nonlinear ODE models which the closed-form solutions are not available. Both constant and time-varying parameters are considered. For the model involved time-varying parameters, we formulated the estimator under the framework of sieve approach. Our main contribution is the establishment of the asymptotic properties for the proposed numerical solution-based NLS estimators (including the sieve NLS estimator for the time-varying parameter) with consideration of both numerical error and measurement error. Our results show that if the maximum step size of the p-order numerical algorithm goes to zero at a rate faster than n−1/(p∧4), the numerical error is negligible compared with the measurement error. This provides guidance in selecting the step size for numerical evaluations of ODEs. Moreover, we have shown that the numerical solution-based NLS estimator and the sieve NLS estimator for the model with a time-varying parameter are strongly consistent. The sieve estimator of constant parameters is asymptotically normal with the same asymptotic co-variance as that of the case where the true solution is exactly known, while the estimator of the time-varying parameter has an optimal convergence rate under some regularity conditions. We also obtained the theoretical results for the case when the step size of the ODE numerical solver does not go to zero fast enough or the numerical error is comparable to the measurement error [see case (ii) of Theorem 3.2 and Remark 3]. To our best knowledge, this is the first time that the sieve method has been extended to the case of ODE models which have no closed-form solutions, and the sieve-based theories were used to establish the asymptotic results and construct confidence intervals (bands) for both constant and time-varying parameters. Note that we only considered a single time-varying parameter in the model, but the methodologies can be extended to multiple time-varying parameters although it is more tedious to implement.
Note that the NLS estimators have good properties under some assumptions and are more accurate compared to other estimates such as those proposed in Ramsay et al. (2007), Chen and Wu (2008) and Liang and Wu (2008). But the price that we have to pay is the high computational cost to obtain the NLS estimates. To reduce the computational burden, we may use the rough estimates from other methods [Ramsay et al. (2007), Chen and Wu (2008), Liang and Wu (2008)] to narrow down the search range for the NLS optimization algorithm. More efficient optimization algorithms may also be employed to speed up the computation. We are also considering to parallel our global optimization algorithms on high-performance computers. Hopefully these efforts can help us to handle a reasonable size of ODE models.
This article only considered the initial value problem (IVP), that is, the initial conditions are assumed to be given. In practice, the initial conditions can be estimated from the data. However, the generalizations of the theoretical results to the cases of estimated initial conditions and other boundary value problems as well as constraints on parameters are not trivial. Also note that, if there is more than one time-varying parameter in the model, similar identifiability techniques in Section 2 may be applied to these parameters one by one, sequentially. Spline approximation to these multiple time-varying parameters can be used for estimation. But the computation and theoretical results are more complicated in this case. However, these generalizations are worth further investigations in future.
Acknowledgments
The authors thank Drs. Hua Liang, Xing Qiu and Jianhua Huang for helpful discussions, and Ms. Jeanne Holden-Wiltse for assistance in editing the manuscript. We also highly appreciate the two referees and the Associate Editors for their insightful comments and useful suggestions that have helped us to greatly improve this manuscript.
Appendix: Proofs
Lemma 1. Under conditions A1–A5, supt∈I ‖X̃(t, β) − X(t, β)‖∞ = O(hp∧4) for any given β ∈  in (1.1).
in (1.1).
Proof. By Theorem 3.4 in Hairer, Nørsett and Wanner (1993, page 160), under conditions A1–A5, for the pth order numerical algorithm (3.2) for (1.1), its global discretization error satisfies
When t is not coincident with the grid points of the numerical algorithm, the cubic Hermite interpolation [de Boor (1978), page 51] will be used to obtain the solution at time t. In this case,
Then it follows that
In general, h is less than 1, O(h4) + O(hp) = O(hp∧4), which completes the proof.
Moreover, Lemma 1 can be extended to the ODE model (1.2) with both constant and time-varying parameters, since for this model, it can be verified that the result of Theorem 3.1 in Hairer, Nørsett and Wanner (1993, page 157) is still valid for any given β ∈  and η ∈
and η ∈  under condition B2 (it can be derived using the Taylor expansion and the Chain rule), which leads to the same conclusion as Theorem 3.4 in Hairer, Nørsett and Wanner (1993, page 160). For Theorems 3.1 and 3.2, the proofs for the univariate and multivariate cases are the same. For presentation and notation simplicity, we only outline the proof for the univariate case below.
under condition B2 (it can be derived using the Taylor expansion and the Chain rule), which leads to the same conclusion as Theorem 3.4 in Hairer, Nørsett and Wanner (1993, page 160). For Theorems 3.1 and 3.2, the proofs for the univariate and multivariate cases are the same. For presentation and notation simplicity, we only outline the proof for the univariate case below.
Proof of Theorem 3.1. Denote , and M(β) = [Y(t) − X(t, β)]2.
First, we claim that E0[M(β)] reaches its unique minimum at β = β0. In fact,
where the third equality holds because the intersection term equals zero according to the following calculation:
because of E0[ε(t)] = 0. Moreover, Et[X (t, β) − X (t, β0)]2 = 0 if and only if β = β0 from assumption A6. Thus the above claim holds. Under assumption A10, it follows that the first-order derivative of E0[M(β)] at β0 equals to zero and the second-order derivative of E0[M(β)] at β0 is positive definite. By assumptions A7 and A9, the second-order derivative of E0[M(β)] in a small neighborhood of β0 is bounded away from 0 and ∞. Then the second-order Taylor expansion of E0[M(β)] gives that there exists a constant 0 < C < ∞ such that
Thus it is sufficient to prove E0[M(β0)] − E0[M(β̂n)] → 0, a.s.
Let N 1 (ε,  ,
,  ) be the covering number of the class
) be the covering number of the class  in the probability measure
in the probability measure  , as given in Pollard (1984, page 25). From Lemma 4.1 in Pollard (1990), we have that
. Let
, as given in Pollard (1984, page 25). From Lemma 4.1 in Pollard (1990), we have that
. Let  n be the set {Mn(β): β ∈
n be the set {Mn(β): β ∈  }. With the Taylor expansion, for any β1, β2 ∈
}. With the Taylor expansion, for any β1, β2 ∈  , we can easily obtain
, we can easily obtain
where C is some constant. Then for any probability measure Q, we have
Then by Theorem II.37 in Pollard (1984), supβ |Mn(β) − E0M(β)| → 0, a.s., under Pβ0. Then we have Mn(β̂n) − E0[M(β̂n)] → 0 and Mn(β0) − E0[M(β0)] → 0, a.s.
Next, by Lemma 1,
| (A.1) |
Then
and
Hence M̃n(β̂n) − E0[M(β0)] → 0, a.s. Thus
Since β0 is the unique minimum point for E0[M(β)], β̂n is almost surely consistent with respect to Pβ0.
Proof of Theorem 3.2. For the proof of part (i), it suffices to verify conditions of Theorem 2 in Pollard (1985). Denote , and . Obviously, Ĝn(β̂n) = 0 and from E0[Y(t)∣t] = X(t, β0).
First, we verify the following result: . For fixed t, according to the multivariate inequality of Kolmogorov type for L2-norms of derivatives [Babenko, Kofanov and Pichugov (1996), page 9], we have for two constants C and C′, where the second inequality holds because of the uniform boundedness of both and under conditions A7 and A8. Based on supt∈I ‖X̃(t, β) − X(t, β)‖∞ = O(n−λ(p∧4)) from Lemma 1, it follows that . Considering that Y(ti) − X(ti, β0) and are bounded, we have
When λ > 1/(p ∧ 4), O(n−λ(p∧4)/2+1/2) = o(1). So for the above expression, we have
Based on the general central limit theorem, with
Second, let δn ↓ 0. For ‖β − β0‖ ≤ δn, we want to show that
In fact, from the first step above, for any β ∈  , we have that
. Then
, we have that
. Then
From Lemma 4.1 in Pollard (1990), we have that
. Let Λn be the set {Gn(β): β ∈  } for any X ∈
} for any X ∈  . Using a Taylor series expansion, for any β1, β2 ∈
. Using a Taylor series expansion, for any β1, β2 ∈  , we can easily obtain
, we can easily obtain
where C is some constant. Then for any probability measure Q, we have
and thus
Since , Λn is a P-Donsker class by Theorem 2.5.2 in van der Vaart and Wellner (1996). Hence .
Third, with some simple calculations, we have , then and . Denote . Then by using the Taylor series expansion again, the function G(β) is Fréchet-differentiable at β0 with nonsingular derivative H2.
Thus all conditions of Theorem 2 in Pollard (1985) are satisfied, then Theorem 3.2(i) holds with .
For the proof of case (ii) of Theorem 3.2, it is easy to verify the conditions of Theorem 2 in Pollard (1985) for the asymptotic normality. Now we just need to show β̃ = β0 + O(h(p∧4)/2) and Ṽ1 = V 1 + O(h(p∧4)/2). Denote M̃(β) = [Y(t) − X̃(t, β)]2 and . Since E0 [M̃(β)] reaches its minimum at β = β̃, then the first-order derivative of E0[M̃(β)] at β̃ equals to 0, that is, G̃(β̃) = 0. Then similar to the proof of case (i) above, we have
It follows that G̃(β̃) = G(β̃) + O(h(p∧4)/2), then G(β̃) = O (h(p∧4)/2) from G̃(β̃) = 0. The Taylor series expansion yields that there exist constants 0 < c1, c2 < ∞ such that
Thus ‖β̃ − β0‖ = O(h(p∧4)/2) from G(β0) = 0. Similarly we can show that ‖Ṽ1 − V1‖ = O(h(p∧4)/2).
Some definitions and notation are necessary in order to prove Theorems 4.1–4.3. Denote , and . We define a semidistance ρ on Θ as
where Ṁ1 is the score function of M for β, and Ṁ2 is the score operator of M for η, both evaluated at the true parameter value θ0. Similarly to the proof in Huang and Rossini (1997, page 966), when V2(θ0), defined in assumption B6, is positive definite, and Ṁ1 and Ṁ2 are bounded away from +∞ and −∞, if ρ (θ̂n, θ0) = Op(rn) then d (θ̂n, θ0) = Op(rn); and if ρ (θ̂n, θ0) → 0 almost surely under Pθ0, then d(θ̂n, θ0) → 0 almost surely under Pθ0.
Proof of Theorem 4.1. Similarly to the proof of Theorem 3.1, we have that E0[M(θ0)] reaches its unique minimum at θ = θ0. It follows that
where C is some constant. Thus if E0[M(θ0)] − E0[M(θ̂n)] → 0, almost surely under Pθ0, then d (θ̂n, θ0) → 0, almost surely under Pθ0.
Let
be the set {η ∈  n, ‖η − ηn0‖2 ≤ δ} and
be its bracketing number with respect to L∞ [see Definition 2.1.6, van der Vaart and Wellner (1996)], where ηn0 is the map point of η0 in the sieve
n, ‖η − ηn0‖2 ≤ δ} and
be its bracketing number with respect to L∞ [see Definition 2.1.6, van der Vaart and Wellner (1996)], where ηn0 is the map point of η0 in the sieve  n. By the calculation of Shen and Wong (1994, page 597), for any ε ≤ δ, we have
n. By the calculation of Shen and Wong (1994, page 597), for any ε ≤ δ, we have
where N = q + l is the number of B-splines basis functions. Let  n be the set {Mn(θ) : ‖β − β0‖ ≤ δ, η ∈
n be the set {Mn(θ) : ‖β − β0‖ ≤ δ, η ∈  n, ‖η − ηn0‖2 ≤ δ}. For any θ1, θ2 ∈ Θn, we can easily obtain
n, ‖η − ηn0‖2 ≤ δ}. For any θ1, θ2 ∈ Θn, we can easily obtain
using Taylor's expansion. Hence
Note that, since N2(ε, L∞,  n) depends on n in the above expression, we cannot directly use Theorem II.37 in Pollard (1984) to obtain sup
n) depends on n in the above expression, we cannot directly use Theorem II.37 in Pollard (1984) to obtain sup n |Mn(θ) − E0[M(θ)]| → 0, a.s., under Pθ0. Fortunately, we can still get this result based on (A.2) in Xue, Lam and Li (2004). Thus we have Mn(θ̂n) − E0[M(θ̂n)] → 0 and Mn(θn0) − E0[M(θn0)] → 0, a.s., where θn0 is the map point of θ0 in the sieve Θn.
n |Mn(θ) − E0[M(θ)]| → 0, a.s., under Pθ0. Fortunately, we can still get this result based on (A.2) in Xue, Lam and Li (2004). Thus we have Mn(θ̂n) − E0[M(θ̂n)] → 0 and Mn(θn0) − E0[M(θn0)] → 0, a.s., where θn0 is the map point of θ0 in the sieve Θn.
From the extension of Lemma 1 for any given β ∈  and η(t) ∈
and η(t) ∈  in (1.2), similarly to (A.1), we have
in (1.2), similarly to (A.1), we have
| (A.2) |
Then the remaining steps are similar to those in the proof of Theorem 3.1.
Proof of Theorem 4.2. We apply Theorem 3.4.1 in van der Vaart and Wellner (1996) to obtain the rate of convergence.
For θn0 in the proof of Theorem 4.1, define θn0 ↦ ρ1(θ, θn0) beamapfrom Θn to [0, ∞) as . Choose δn = ρ(θ0, θn0). For δn < δ < ∞, denote Ω = {θ:θ ∈ Θn, δ/2 < ρ(θ, θn0) ≤ δ}. From the definition of ρ1, we have .
Let Ξn be the set {Mn(θ) – M(θn0) : θ ∈ Θn} and J̃(δ, L2(P), Ξn) be the L2(P)-norm bracketing integral of the sieve Θn. From the proof of Theorem 4.1, we have
Let
Obviously, ϕn(δ)/δ1+τ is a decreasing function in δ for 0 < τ < 1. Then by Lemma 3.4.2 in van der Vaart and Wellner (1996), we have
For λ > 1/[2(p ∧ 4)], from (A.2), it follows that
Then we have that
Then the conditions of Theorem 3.4.1 in van der Vaart and Wellner (1996) are satisfied for the δn, ρ1 and ϕn(δ) above. Therefore we have , where rn satisfies . It follows that rn = N−1/2n1/2 = n(1−v)/2. Thus ρ1(θ̂n, θn0) = Op(n−(1−v)/2).
Now, we define a distance ρ2 as
Let ς be a positive constant. Similarly to the proof of Theorem 3.2 in Huang (1999), it is easy to follow that for any θ with ρ2(θ, θn0) ≤ ς, there exist constants 0 < c1, c2 < ∞ such that
Therefore, for a constant c2 > 0,
Because d(θn0, θ0) ≤ ρ2(θ0, θ0) = Op(n−vϱ), we have d(θ̂n, θ0) = Op(n−vϱ + n−(1−v)/2).
Proof of Theorem 4.3. We prove this theorem using Theorem 6.1 in Wellner and Zhang (2007). It suffices to validate conditions A1–A6 of Theorem 6.1 in Wellner and Zhang (2007). From the proof of Theorems 4.1 and 4.2 above, it is easy to see that condition A1 regarding consistency and rate of convergence and condition A2 for Theorem 6.1 in Wellner and Zhang (2007) hold.
For condition A3, we need to calculate the pseudo-information matrix. For any fixed η ∈  , let
, let  0 = {ηω(·) : ω in a neighborhood of 0 ∈
0 = {ηω(·) : ω in a neighborhood of 0 ∈  } be a smooth curve in
} be a smooth curve in  running through η0 at ω = 0, that is, ηω=0(t) = η0(t). Denote
and the space generated by such a(t) as
running through η0 at ω = 0, that is, ηω=0(t) = η0(t). Denote
and the space generated by such a(t) as  . The score functions of β and η are
. The score functions of β and η are
We also set
and
where a1(t), a2(t) ∈  . Following the idea from the proofs of the asymptotic results for semiparametric M-estimator in Ma and Kosorok (2005) and Wellner and Zhang (2007), we assume that the special perturbation direction
with
for 1 ≤ i ≤ d, satisfies E0{Ṁ12[a] − Ṁ22[a*, a]} = 0 for any a ∈
. Following the idea from the proofs of the asymptotic results for semiparametric M-estimator in Ma and Kosorok (2005) and Wellner and Zhang (2007), we assume that the special perturbation direction
with
for 1 ≤ i ≤ d, satisfies E0{Ṁ12[a] − Ṁ22[a*, a]} = 0 for any a ∈  . Some calculations yield
. Some calculations yield
It follows that
| (A.3) |
Since E0[Y(t)∣t] = X(t), a*(t) in (A.3) can be simplified as
| (A.4) |
For K ≥ 2, both
| (A.5) |
and
| (A.6) |
are nonsingular. Let . Thus condition A3 of finite variance for Theorem 6.1 in Wellner and Zhang (2007) is satisfied.
Condition A4 for Theorem 6.1 in Wellner and Zhang (2007) follows similar arguments as condition (i) of the proof of Theorem 4 in Xue, Lam and Li (2004). Condition A5 of stochastic equicontinuity is similar to verify conditions C3 of the proof of Theorem 4 in Xue, Lam and Li (2004). Condition A6 of smoothness of the model can be easily verified using a straightforward Taylor expansion where n−c1 is just the rate of convergence in Theorem 4.2 and faster than n−1/4, and c2 = 2, which completes the proof.
Proof of Proposition 1. Let  be the set of a real valued functions g on [a, b] which are absolutely continuous and satisfy
and Et g(t) = 0. Similarly to the proof of Theorem 4.3, for any fixed η ∈
be the set of a real valued functions g on [a, b] which are absolutely continuous and satisfy
and Et g(t) = 0. Similarly to the proof of Theorem 4.3, for any fixed η ∈  , let
, let  0 = {ηω(·): ω in a neighborhood of 0 ∈
0 = {ηω(·): ω in a neighborhood of 0 ∈  } be a smooth curve in
} be a smooth curve in  running through η0 at ω = 0, that is, ηω=0(t) = η0(t). Denote
and restrict a(t) ∈
running through η0 at ω = 0, that is, ηω=0(t) = η0(t). Denote
and restrict a(t) ∈  . Denote the space generated by such a(t) as
. Denote the space generated by such a(t) as  . The score functions of β and η are
. The score functions of β and η are
with ξ = β0 + η0(t). Let Ṗ be the linear span of Ṁ2[a]. Since E0{Ṁ1Ṁ2[a]} = 0 for any a(t) ∈  , it follows that Ṁ1 is orthogonal to Ṗ. Thus the efficient score function of β is just Ṁ1. Then the pseudo-information is
. The rest of the proof is similar to that of Theorem 4.3, where the efficient score function and the pseudo-information are updated as discussed before, and the least favorable direction can be selected by any a ∈
, it follows that Ṁ1 is orthogonal to Ṗ. Thus the efficient score function of β is just Ṁ1. Then the pseudo-information is
. The rest of the proof is similar to that of Theorem 4.3, where the efficient score function and the pseudo-information are updated as discussed before, and the least favorable direction can be selected by any a ∈  .
.
Footnotes
AMS 2000 subject classifications. Primary 60G02, 60G20; secondary 60G08, 62P10.
Contributor Information
Hongqi Xue, Email: Hongqi_Xue@urmc.rochester.edu.
Hongyu Miao, Email: Hongyu_Miao@urmc.rochester.edu.
Hulin Wu, Email: hwu@bst.rochester.edu.
References
- Adams BM. Ph D thesis. North Carolina State Univ; 2005. Non-parametric parameter estimation and clinical data fitting with a model of HIV infection. MR2623319. [Google Scholar]
- Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford Univ. Press; Oxford: 1996. [Google Scholar]
- Audoly S, Bellu G, D'Angio L, Saccomani MP, Cobelli C. Global identifiability of nonlinear models of biological systems. IEEE Trans Biomed Eng. 2001;48:55–65. doi: 10.1109/10.900248. [DOI] [PubMed] [Google Scholar]
- Babenko VF, Kofanov VA, Pichugov SA. Multivariate inequalities of Kolmogorov type and their applications. In: Nuraberger G, Schmidt JW, Walz G, editors. Multivariate Approximation and Splines. Birkhauser; Basel: 1996. pp. 1–12. MR1484990. [Google Scholar]
- Bard Y. Nonlinear Parameter Estimation. Academic Press; New York: 1974. MR0326870. [Google Scholar]
- Bellman R, Åström KJ. On structural identifiability. Math Biosci. 1970;7:329–339. [Google Scholar]
- Benson M. Parameter fitting in dynamic models. Ecol Mod. 1979;6:97–115. [Google Scholar]
- Bickel PJ, Klaassen CAJ, Ritov Y, Wellner JA. Efficient and Adaptive Estimation for Semiparametric Models. Johns Hopkins Univ Press; Baltimore: 1993. MR1245941. [Google Scholar]
- Brookmeyer R, Gail MH. AIDS Epidemiology: A Quantitative Approach Monographs in Epidemiology and Biostatistics. Vol. 23. Oxford Univ Press; New York: 1994. [Google Scholar]
- Brunel N. Parameter estimation of ODE's via nonparametric estimators. Electron J Statist. 2008;2:1242–1267. MR2471285. [Google Scholar]
- Burnham KP, Anderson DR. Multimodel inference: Understanding AIC and BIC in model selection. Sociol Methods Res. 2004;33:261. MR2086350. [Google Scholar]
- Chappel MJ, Godfrey KR. Structural identifiability of the parameters of a nonlinear batch reactor model. Math Biosci. 1992;108:245–251. doi: 10.1016/0025-5564(92)90058-5. MR1154720. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu H. Efficient local estimation for time-varying coefficients in deterministic dynamic models with applications to HIV-1 dynamics. J Amer Statist Assoc. 2008;103:369–384. MR2420240. [Google Scholar]
- Chen T, He HL, Church GM. Modeling gene expression with differential equations. Pac Symp Biocomput. 1999:29–40. [PubMed] [Google Scholar]
- Cobelli C, Lepschy A, Jacur R. Identifiability of compartmental systems and related structural proerties. Math Biosci. 1979;44:1–18. [Google Scholar]
- Daley DJ, Gani J. Epidemic Modeling. Cambridge Univ Press; Cambridge: 1999. [Google Scholar]
- de Boor C. A Practical Guide to Splines. Springer; New York: 1978. MR0507062. [Google Scholar]
- Delgado MA. Semiparametric generalized least squares in the multivariate nonlinear regression model. Econometric Theory. 1992;8:203–222. MR1179510. [Google Scholar]
- Donnet S, Samson A. Estimation of parameters in incomplete data models defined by dynamical systems. J Statist Plann Inference. 2007;137:2815–2831. MR2323793. [Google Scholar]
- Englezos P, Kalogerakis N. Applied Parameter Estimation for Chemical Engineers. Dekker; New York: 2001. [Google Scholar]
- Grenander U. Abstract Inference. Wiley; New York: 1981. MR0599175. [Google Scholar]
- Hairer E, Nørsett SP, Wanner G. Solving Ordinary Differential Equations I: Nonstiff Problems. 2nd. Springer; Berlin: 1993. MR1227985. [Google Scholar]
- He X, Fung WK, Zhu ZY. Estimation in a semiparametric model for longitudinal data with unspecified dependence structure. Biometrika. 2002;89:579–590. MR1929164. [Google Scholar]
- Heckman NE, Ramsay JO. Penalized regression with model-based penalties. Canad J Statist. 2000;28:241–258. MR1792049. [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Huang J. Efficient estimation for the proportinal hazards model with interval censoring. Ann Statist. 1996;24:540–568. MR1394975. [Google Scholar]
- Huang J. Efficient estimation of the partly linear additive Cox model. Ann Statist. 1999;27:1536–1563. MR1742499. [Google Scholar]
- Huang J, Rossini AJ. Sieve estimation for the proportional-odds failure-time regression model with interval censoring. J Amer Statist Assoc. 1997;92:960–967. MR1482126. [Google Scholar]
- Huang JZ. Local asymptotics for polynomial spline regression. Ann Statist. 2003;31:1600–1635. MR2012827. [Google Scholar]
- Huang JZ, Zhang L, Zhou L. Efficient estimation in marginal partially linear models for longitudinal/clustered data using plines. Scand J Statist. 2007;34:451–477. MR2368793. [Google Scholar]
- Huang Y, Liu D, Wu H. Hierarchical Bayesian methods for estimation of parameters in a longitudinal HIV dynamic system. Biometrics. 2006;62:413–423. doi: 10.1111/j.1541-0420.2005.00447.x. MR2227489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber PJ. The behavior of maximum likelihood estimates under nonstandard conditions. Fifth Berkeley Symposium on Mathematical Statistics and Probability; Berkeley: Univ California; 1967. pp. 221–233. MR0216620. [Google Scholar]
- Jansson B, Revesz L. Analysis of the growth of tumor cell populations. Math Biosci. 1975;19:131–154. [Google Scholar]
- Jeffrey AM, Xia X. Identifiability of HIV/AIDS model. In: Tan WY, Wu H, editors. Deterministic and Stochastic Models of AIDS Epidemics and HIV Infections with Intervention. World Scientific; 2005. [Google Scholar]
- Jennerich RI. Asymptotic properties of non-linear least squares estimators. Ann Math Statist. 1969;40:633–643. MR0238419. [Google Scholar]
- Jolliffe IT. Discarding variables in a principal component analyssis. I: Artificial data. J Roy Statist Soc Ser C Appl Statist. 1972;21:160–172. MR0311034. [Google Scholar]
- Joshi M, Seidel-Morgenstern A, Kremling A. Exploiting the boostrap method for quantifying parameter confidence intervals in dynamic systems. Metabolic Engineering. 2006;8:447–455. doi: 10.1016/j.ymben.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Kolchin E. Differential Algebra and Algebraic Groups. Academic Press; New York: 1973. MR0568864. [Google Scholar]
- Kutta W. Beitrag zur näherungsweisen itegration totaler differentialgleichungen. Zeitschr Math Phys. 1901;46:435–453. [Google Scholar]
- Li L, Brown MB, Lee KH, Gupta S. Estimation and inference for a splineenhanced pupulation pharmacokinetic model. Biometrics. 2002;58:601–611. doi: 10.1111/j.0006-341x.2002.00601.x. MR1933534. [DOI] [PubMed] [Google Scholar]
- Li Z, Osborne MR, Pravan T. Parameter estimation of ordinary differential equations. IMA J Numer Anal. 2005;25:264–285. MR2126204. [Google Scholar]
- Liang H, Wu H. Parameter estimation for differential equation models using a framework of measurement error in regression models. J Amer Statist Assoc. 2008;103:1570–1583. doi: 10.1198/016214508000000797. MR2504205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung L, Glad T. On global identifiability for arbitrary model parametrizations. Automatica. 1994;30:265–276. MR1261705. [Google Scholar]
- Ma S, Kosorok MR. Robust semiparametric M-estimation and the weighted bootstrap. J Multivariate Anal. 2005;96:190–217. MR2202406. [Google Scholar]
- Malinvaud E. The consistancy of nonlinear regressions. Ann Math Statist. 1970;41:956–969. MR0261754. [Google Scholar]
- Mattheij R, Molenaar J. Ordinary Differential Equations in Theory and Practice. SIAM; Philadelphia: 2002. MR1946758. [Google Scholar]
- Miao H, Dykes C, Demeter LM, Cavenaugh J, Park SY, Perelson AS, Wu H. Modeling and estimation of kinetic parameters and replicative fitness of HIV-1 from flow-cytometry-based growth competition experiments. Bull Math Biol. 2008;70:1749–1771. doi: 10.1007/s11538-008-9323-4. MR2430325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson S, Leith JT. Tumor heterogeneity and growth control. In: Adam JA, Bellomo N, editors. Tumor Heterogeneity and Growth Control. Boston: Birkhauser; 1997. pp. 295–326. [Google Scholar]
- Moles CG, Banga JR, Keller K. Solving nonconvex climate control problems: Pitfalls and algorithm performances. Appl Soft Comput. 2004;5:35–44. [Google Scholar]
- Nowak MA, May RM. Virus Dynamics: Mathematical Principles of Immunology and Virology. Oxford Univ. Press; Oxford: 2000. MR2009143. [Google Scholar]
- Ollivier F. Ph D thesis. École Polytechnique; Paris, France: 1990. Le problème de l'identifiabilité globale: Étude thé orique, méthodes effectives et bornes de complexité. [Google Scholar]
- Pakes A, Pollard D. Simulation and the asymptotics of optimization estimators. Econometrica. 1989;57:1027–1057. MR1014540. [Google Scholar]
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- Perelson AS, Nelson PW. Mathematical analysis of HIV-1 dynamics in vivo. SIAM Rev. 1999;41:3–44. MR1669741. [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Pohjanpalo H. System identifiability based on the power series expansion of the solution. Math Biosci. 1978;41:21–33. MR0507373. [Google Scholar]
- Pollard D. Convergence of Stochastic Processes. Springer; New York: 1984. MR0762984. [Google Scholar]
- Pollard D. New ways to prove central limit theorems. Econometric Theory. 1985;1:295–314. [Google Scholar]
- Pollard D. Empirical Processes Theory and Applications. IMS; Hayward, CA: 1990. MR1089429. [Google Scholar]
- Poyton AA, Varziri MS, McAuley KB, McLellen PJ, Ramsay JO. Parameter estimation in continuous-time dynamic models using principal differential analysis. Computers and Chemical Engineering. 2006;30:698–708. [Google Scholar]
- Putter H, Heisterkamp SH, Lange JM, de Wolf F. A Bayesian approach to parameter estimation in HIV dynamical models. Stat Med. 2002;21:2199–2214. doi: 10.1002/sim.1211. [DOI] [PubMed] [Google Scholar]
- Quaiser T, Mönnigmann M. Systematic identifiability testing for unambiguous mechanistic modeling—application to JAK-STAT, MAP kinase, and NF-κB signaling pathway models. BMC Sys Bio. 2009;3:50. doi: 10.1186/1752-0509-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JO. Principal Differential Analysis: Data Reduction by Differential Operators. J Roy Statist Soc Ser B. 1996;58:495–508. MR1394362. [Google Scholar]
- Ramsay JO, Hooker G, Campbell D, Cao J. Parameter estimation for differential equations: A generalized smoothing approach (with discussion) J R Stat Soc Ser B Stat Methodol. 2007;69:741–796. MR2368570. [Google Scholar]
- Ritt JF. Differential Algebra. Amer Math Soc; Providence, RI: 1950. MR0035763. [Google Scholar]
- Rodriguez-Fernandez M, Egea JA, Banga JR. Novel metaheuristic for parameter estimation in nonlinear dynamic biological systems. BMC Bioinformatics. 2006;7:1–18. doi: 10.1186/1471-2105-7-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge C. Ueber die numerische Auflösung von Differentialgleichungen. Math Ann. 1895;46:167–178. MR1510879. [Google Scholar]
- Schick A. On asymptotically efficient estimation in semiparametric models. Ann Statist. 1986;14:1139–1151. MR0856811. [Google Scholar]
- Schumaker LL. Spline Functions. Wiley; New York: 1981. MR0606200. [Google Scholar]
- Shen X. On methods of sieves and penalization. Ann Statist. 1997;25:2555–2591. MR1604416. [Google Scholar]
- Shen X, Wong WH. Convergence rate of sieve estimates. Ann Statist. 1994;22:580–615. MR1292531. [Google Scholar]
- Stone CJ. Optimal global rates of convergence for nonparametric regression. Ann Statist. 1982;10:1040–1053. MR0673642. [Google Scholar]
- Stone CJ. Additive regression and other nonparametric models. Ann Statist. 1985;13:689–705. MR0790566. [Google Scholar]
- Storn R, Price K. Differential evolution—a simple and efficient heuristic for global optimization over continuous spaces. J Global Optim. 1997;11:341–359. MR1479553. [Google Scholar]
- Swartz J, Bremermann H. Discussion of parameter estimation in biological modeling: Algorithms for estimation and evaluation of the estimates. J Math Biol. 1975;1:241–275. doi: 10.1007/BF01273746. [DOI] [PubMed] [Google Scholar]
- Tan WY, Wu H. Deterministic and Stochastic Models of AIDS Epidemics and HIV Infections With Intervention. World Scientific; Singapore: 2005. MR2169300. [Google Scholar]
- Thomaseth K, Alexandra KW, Bernhard L, et al. Integrated mathematical model to assess β-cell activity during the oral glucose test. Amer J Phisiol. 1996;270:E522–531. doi: 10.1152/ajpendo.1996.270.3.E522. [DOI] [PubMed] [Google Scholar]
- Vajda S, Rabitz H, Walter E, Lecourtier Y. Qualitative and quantitative identifiability analysis of nonlinear chemical kinetiv-models. Chem Eng Commun. 1989;83:191–219. [Google Scholar]
- van der Vaart AW, Wellner JA. Weak Convergence and Empirical Processes. Springer; New York: 1996. MR1385671. [Google Scholar]
- van Domselaar B, Hemker PW. Report NW18/75. Mathematical Centrum; Amsterdam: 1975. Nonlinear parameter estimation in initial value problmes. [Google Scholar]
- Varah JM. A spline least squares method for numerical parameter estimation in differential equations. SIAM J Sci Comput. 1982;3:28–46. MR0651865. [Google Scholar]
- Varziri MS, Poyton AA, McAuley KB, McLellen PJ, Ramsay JO. Selecting optimal weighting factors in iPDA for parameter estimation in continuous-time dynamic models. Comp Chem Eng. 2008;32:3011–3022. [Google Scholar]
- Walter E. Identifiability of Parameteric Models. Pergamon Press; Oxford: 1987. [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Wellner JA, Zhang Y. Two likelihood-based semiparametric estimation methods for panel count data with covariates. Ann Statist. 2007;35:2106–2142. MR2363965. [Google Scholar]
- Wu CF. Asymptotic theory of nonlinear least squares estiamtion. Ann Statist. 1981;9:501–513. MR0615427. [Google Scholar]
- Wu H. Statistical methods for HIV dynamic studies in AIDS clinical trials. Stat Methods Med Res. 2005;14:1–22. doi: 10.1191/0962280205sm390oa. MR2135921. [DOI] [PubMed] [Google Scholar]
- Wu H, Huang Y, Acosta EP, et al. Modeling long-term HIV dynamics and antiretroviral response: Effects of drug potency, pharmacokinetics, adherence, and drug resistance. JAIDS. 2005;39:272–283. doi: 10.1097/01.qai.0000165907.04710.da. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhu H, Miao H, Perelson AS. Identifiability and statistical estimation of dynamic parameters in HIV/AIDS dynamic models. Bull Math Biol. 2008;70:785–799. doi: 10.1007/s11538-007-9279-9. MR2393024. [DOI] [PubMed] [Google Scholar]
- Xia X. Estimation of HIV/AIDS parameters. Automatica J IFAC. 2003;39:1983–1988. MR2142834. [Google Scholar]
- Xia X, Moog CH. Identifiability of nonlinear systems with applications to HIV/AIDS models. IEEE Trans Automat Control. 2003;48:330–336. MR1957979. [Google Scholar]
- Xue H, Lam KF, Li G. Sieve maximum likelihood estimator for semiparametric regression models with current status data. J Amer Statist Assoc. 2004;99:346–356. MR2062821. [Google Scholar]