Abstract
Nijmegen breakage syndrome (NBS) is characterized by genome instability and cancer predisposition. NBS patients contain a mutation in the NBS1 gene, which encodes the NBS1 component of the DNA double-strand break (DSB) response complex MRE11/RAD50/NBS1. To investigate the NBS phenotype in more detail, we combined the mouse mimic of the most common patient mutation (Nbs1ΔB/ΔB) with a Rad54 null mutation, which diminishes homologous recombination. Double mutant cells were particularly sensitive to treatments that cause single strand breaks (SSBs), presumably because these SSBs can be converted into detrimental DSBs upon passage of a replication fork. The persistent presence of nuclear RAD51 foci and increased levels of chromatid type breaks in metaphase spreads indicated that replication-associated DSBs are repaired inefficiently in the double mutant cells. We conclude that Nbs1 and Rad54 function cooperatively, but in separate pathways to counteract this type of DNA damage and discuss mechanistic implications of these findings.
Keywords: DNA damage response, DNA double-strand break repair, Chromosomal instability, MRE11/RAD50/NBS1 complex, RAD54, Cell cycle checkpoint
1. Introduction
DNA repair is essential for the successful maintenance and propagation of genetic information. Endogenous and exogenous DNA damaging agents are constantly challenging the stability of DNA inside cells. Because a large variety of lesions occur in DNA, it is not surprising that multiple pathways have developed that each repair a subset of these lesions [1]. DNA double-strand breaks (DSBs) form a very genotoxic class of lesions [2,3]. Unrepaired DSBs can lead to cell death or loss of heterozygosity, whereas misrepaired DSBs may result in chromosomal rearrangements that contribute to carcinogenesis. Effective DSB repair is critical for maintaining genome stability. In eukaryotes, two main DSB repair pathways have been identified that differ in their requirements for DNA homology. Non-homologous end-joining (NHEJ) uses little or no sequence homology to rejoin broken ends in a manner that need not be error-free. Homologous recombination (HR) requires extensive regions of DNA sequence homology and repairs DSBs accurately using information on the undamaged sister chromatid.
HR involves a large number of proteins, including RAD51, RAD52 and RAD54 [4]. Upon DNA damage induction of these proteins accumulate into nuclear foci which can be detected by immunofluorescence microscopy [5]. The importance of the HR proteins is underscored by the lethality imposed by disruption of Rad51 [6,7]. However, Rad54−/− mice are viable and therefore provide a suitable model system to study the biological significance of a defect in the mammalian HR pathway [8]. Rad54−/− ES cells are sensitive to ionizing radiation (IR) and the interstrand crosslinking agent Mitomycin C (MMC) whereas the knockout mice are only MMC sensitive [9]. Furthermore, the Rad54 deletion dramatically aggravates the IR sensitivity of NHEJ mutant mice [9,10], showing that HR can function as a backup pathway for NHEJ.
DSBs can occur in all phases of the cell cycle. Progression of the cell cycle is controlled by several checkpoints that prevent cell cycle progression when DNA damage has not been repaired [11,12]. DNA damage can prevent initiation of DNA replication (G1/S checkpoint), slow down S phase progression (intra-S checkpoint) or delay mitosis (G2/M checkpoint). The MRE11/RAD50/NBS1 (MRN) complex is a central player in various aspects of the cellular response to DSBs, including HR, NHEJ and DNA damage checkpoint activation [12–15]. Mutations in NBS1 cause Nijmegen Breakage Syndrome, a human disorder characterized by microcephaly, IR hypersensitivity and predisposition to haematopoietic malignancy. An important function of NBS1 is the maintenance of the intra-S phase checkpoint, which also requires the Ataxia-Telangiectasia mutated (ATM) kinase. Wild type cells inhibit firing of new replication origins to prevent replication forks from running into DNA damage. AT and NBS cells inhibit DNA synthesis less efficiently after DNA damage, which can be observed as radioresistant DNA synthesis (RDS) [16]. The MRN complex activates the ATM kinase, which explains the similarity in cellular phenotypes of both syndromes [14,17]. In addition to the activation of ATM kinase, NBS1 also facilitates ATR-dependent phosphorylation [18]. ATM and ATR phosphorylate CHK2 and CHK1, respectively, leading to activation of the intra-S, G1/S and G2/M DNA damage induced checkpoints.
The role of Nbs1 in mammalian cells has been investigated in more detail using mice, which mimic the mutation that is found in most NBS patients [19]. The Nbs1ΔB/ΔB mice are IR sensitive and cells derived from these mice are sensitive to various DNA damaging agents, show increased levels of chromosomal aberrations after IR treatment and display cell cycle checkpoint defects.
We combined the Rad54− and Nbs1ΔB mutations to get more insight into the genetic interactions of HR and the various functions of NBS1. We found that defective HR aggravates the sensitivity of Nbs1ΔB/ΔB cells to agents that can induce replication-associated DSBs.
2. Materials and methods
2.1. Mouse breeding and cell culture
To investigate the effect of combined RAD54 and NBS1 mutations in mice, we set up crosses to generate Nbs1ΔB/ΔB Rad54−/− double mutant mice. Because Rad54−/− mice are fully fertile and viable [8] we used both Rad54−/− and Rad54+/− mice in the crosses. Since, Nbs1ΔB/ΔB females are subfertile we used Nbs1ΔB/+ females for the crosses [19].
Primary MEFs were obtained from embryos at E13.5 by cell dispersal following removal of organ block tissue. Cells were cultured in DMEM/Ham F10 1:1 medium supplemented with 10% fetal bovine serum and penicillin (100 U/ml)/streptomycin (100 μg/ml) in a humidified 37 °C incubator maintained at 3% O2 and 5% CO2.
ES cells were isolated at E3.5 [9]. ES cells were cultured on gelatin coated dishes in a 1:1 mixture of DMEM and Buffalo Rat Liver (BRL) conditioned medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml)/streptomycin (100 μg/ml), 0.1 mM non-essential amino acids, 50 μM β-mercaptoethanol and 500 U/ml leukemia inhibitory factor.
2.2. Homologous targeting assay
The efficiency of homologous recombination was assessed by homologous gene targeting experiments to the mRad54 locus. Targeting of the locus by mRAD54-GFP was detected by FACS analysis of GFP expression in ES cells containing the targeted locus as described previously [27].
2.3. Sister chromatid exchange (SCE) analysis
SCE analysis was performed according to the standard procedures, with the cells either mock treated or treated with 0.8 μg/ml MMC. 40–60 metaphases per cell line were analyzed for the number of SCEs. For the double mutant cells 6 metaphases were analyzed.
2.4. DNA damage sensitivity assays
Cellular clonogenic survival assays were performed in triplicate as described previously [8]. For UV and IR survival assays, cells were exposed to the specified dose of UV-C light or gamma rays from a 137Cs source at 0.8 Gy/min. For MMC survival assays, cells were incubated in medium containing the specified concentration of MMC for 1 h. In the case of CPT, cells were exposed for 24 h. The cells were then washed with phosphate-buffered saline and replenished with fresh medium. Hydrogen peroxide was added at the indicated concentration; this compound was not removed by changing the media. In the case of the KU58948 PARP inhibitor [22], ES cells were exposed continuously, and after 3 days the medium was replaced by fresh medium. The PARP inhibitor survival is plotted on a double logarithmic scale. For all survival assays, the cells were allowed to grow for 10 days, stained and colonies containing more than 50 cells were counted.
Wild type, Nbs1ΔB/ΔB ES cells, wild type and VH10-NE13 AT-fibroblasts were exposed to 10 μM KU55933 ATM inhibitor [26] and after 30 min exposed to IR. After 24 h, fresh medium without inhibitor was added and survival analysis was performed as described above.
2.5. Immunofluorescence and Western blotting
Immunostaining to detect RAD51 (α-hRad51 nr. 2307, rabbit polyclonal antibody) [34] was performed as described previously [5]. Cells were counted as positive when they showed 5 or more foci. CHK2 phosphorylation was detected by SDS-PAGE separation of whole cell extracts and Western blotting using mouse monoclonal antibodies to CHK2 (BD Transduction Laboratories). For quantification of the signals the ImageJ software tool was used (http://rsb.info.nih.gov/ij/). The area under the curve (AUC) of a specific signal was corrected by the AUC of the loading control.
2.6. Cytogenetic analysis
Frequencies of spontaneous chromosomal aberrations were determined in exponentially growing cell cultures as described previously [35].
2.7. Radioresistant DNA synthesis assay
The RDS assay was performed essentially as described [36] but without the TCA-precipitation step. In short, duplicate 30 mm dishes (four for the unirradiated control) were prelabeled overnight with [14C]-thymidine (Amersham) in HEPES-buffered ES cell medium, then exposed to various doses of γ-rays using a 137Cs source (0.8 Gy/min), or treated with CPT for 1 h, and subsequently labeled with [3H]-thymidine (Amersham) for 2 h. Free thymidine pools were chased by a further 30–45 min incubation in medium containing unlabeled thymidine. Scintillation-counted [3H] to [14C] radioactivity ratios of alkali-lysed cells were taken as a measure of DNA synthesis rates and plotted as percentages of unirradiated cells. The RDS assay with IR was performed 4 times and it was performed twice with CPT treatment.
3. Results
3.1. Generation of Nbs1ΔB/ΔB Rad54−/− cells
NBS1 has been reported to have many different functions in the DNA damage response. We investigated the effect of combining the hypomorphic Nbs1ΔB mutation with a knockout Rad54 mutation. Both single mutants have no overt defects. The double mutant mice are present at the expected Mendelian frequency up to day 18 p.c. However, they were born at sub-Mendelian frequencies (Table 1) and the mice that survived perinatal death had a reduced body weight (by approximately 15%; data not shown). However, no abnormalities were detected in the gastrointestinal tract, lungs and skeleton and the animals developed without obvious defects, although they remained smaller than their wild type and single mutant littermates.
Table 1.
Crosses to generate Nbs1ΔB/ΔB Rad54−/− mice.
| Cross | Expected | Born |
|---|---|---|
| Nbs1ΔB/+ Rad54+/− × Nbs1ΔB/+ Rad54+/− | 1/16 (6.25%) | 1/61 (1.6%) |
| Nbs1ΔB/+ Rad54+/− × Nbs1ΔB/+ Rad54−/− | 1/8 (12.5%) | 6/78 (7.7%) |
| Nbs1ΔB/ΔB Rad54+/− × Nbs1ΔB/+ Rad54+/− | 1/8 (12.5%) | 3/46 (6.5%) |
| Nbs1ΔB/ΔB Rad54+/− × Nbs1ΔB/+ Rad54−/− | 1/4 (25%) | 3/75 (4%) |
| Overall | 11% | 5% |
The expected number of double mutant pups and the numbers of born pups are given for each cross.
We isolated mouse embryonic fibroblasts (MEFs) and embryonic stem (ES) cells. The single and double mutant MEFs and ES cells showed normal cell viability, proliferation and morphology.
3.2. Analysis of sensitivities to DNA damaging agents
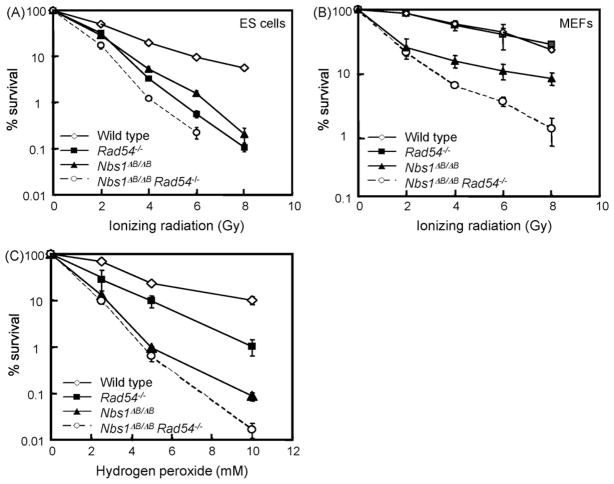
To get more insight into the genetic interactions of RAD54 and NBS1, we exposed the ES cells to various DNA damaging agents. We first tested treatments that can directly create DSBs: IR and hydrogen peroxide (H2O2). The Nbs1ΔB/ΔB and Rad54−/− single mutant ES cells both showed increased levels of IR sensitivity (Fig. 1A). Double mutant ES cells were slightly more IR sensitive than the single mutants, suggesting that NBS1 and RAD54 function in different pathways to counteract the deleterious effects of IR. To test whether the sensitivity found in the double mutant ES cells is cell type specific we also analyzed MEFs. Rad54−/− MEFs were not IR sensitive, whereas Nbs1ΔB/ΔB were [19]. The double mutant MEFs were also hypersensitive to IR, showing that the additive effect of NBS1 and RAD54 deficiency is not ES cell specific (Fig. 1B).
Fig. 1.
Sensitivities of Nbs1ΔB/ΔB Rad54−/− double and single mutants to DSB-inducing agents. (A) ES cell and (B) MEF colony survival assay for IR and (C) ES cells colony survival assay for H2O2. The doses of various genotoxic agents are displayed on the X-axis on a linear scale, while the percentage of surviving colonies is displayed on the Y-axis on a logarithmic scale. Error bars show the standard error of the mean for triplicate experiments.
Rad54−/− and Nbs1ΔB/ΔB ES cells also showed increased sensitivity to H2O2 (Fig. 1C). Interestingly, the Nbs1ΔB/ΔB cells were more sensitive than the Rad54−/− cells, suggesting that NBS1 is especially important to counteract H2O2 induced DNA lesions. H2O2 causes mainly SSBs, which can be converted to DSBs during replication [20]. Apparently, NBS1 is required to counteract the negative effect of these lesions. The double mutant cells were only slightly more sensitive to H2O2 than the Nbs1 single mutant, indicating that NBS1 has a more important role in this type of repair than RAD54.
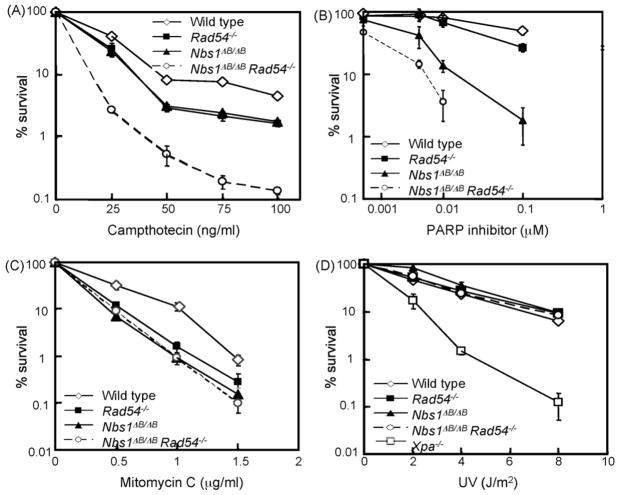
To investigate the effect of S phase specific DSBs in more detail, we used camptothecin (CPT), which acts as an inhibitor of topoisomerase 1 and results in stable covalent DNA–topoisomerase complexes. CPT-induced breaks can be converted to DSBs by advancing DNA polymerases during replication [21]. CPT treatment was given for 24 h to act on all cells in S phase. Clonogenic survival experiments indicated that Nbs1ΔB/ΔB and Rad54−/− ES cells were equally sensitive, whereas the double mutant cells were much more sensitive than the single mutants (Fig. 2A). The double mutant cells showed a much more pronounced hypersensitivity to CPT than to IR treatment, suggesting that double mutant cells are especially deficient in repairing S phase specific DSBs. Hydroxy urea-induced replication fork stalling also resulted in hypersensitivity of double mutant cells (data not shown), confirming that S phase specific DSBs pose a particularly severe problem if both genes are defective.
Fig. 2.
Sensitivities of Nbs1ΔB/ΔB Rad54−/− double and single mutants to various DNA damaging agents. Colony survival assays after treatment of ES cells with (A) CPT, (B) PARP inhibitor (KU58948), (C) MMC and (D) Ultra Violet (UV-C) irradiation in ES cells. The doses of various genotoxic agents are displayed on the X-axis on a linear scale, except for the PARP inhibitor, while the percentage of surviving colonies is displayed on the Y-axis on a logarithmic scale. Error bars show the standard error of the mean for triplicate experiments.
As an alternative approach to increase the number of replication-associated DSBs, we used the PARP inhibitor KU58948, which causes increased levels of SSBs [22,23]. This compound inhibits repair of SSBs in the template DNA. When the replication fork approaches such a lesion, it collapses and replication fork restart involves repair of the broken DNA molecule by HR [2]. The Nbs1 mutant ES cells were much more sensitive to this treatment than Rad54 deficient cells (Fig. 2B), suggesting that replication-associated DSBs may pose a particularly severe problem to cells with diminished function of the MRN complex. Interestingly, the PARP inhibitor caused an additive sensitivity in the double mutant cells, suggesting that Rad54 and Nbs1 function in different pathways that counteract replication-associated DSBs.
Subsequently, ES cells were exposed to the interstrand crosslinking (ICL) agent MMC, because HR is also involved in the repair of these types of lesions. The Nbs1ΔB/ΔB, Rad54−/− and double mutant cells showed similar levels of sensitivity (Fig. 2C), suggesting that both genes function in the same pathway of ICL repair.
Repair of intrastrand crosslinks was assayed by ultraviolet (UV) irradiation, which causes pyrimidine dimers that can be repaired via nucleotide excision repair. The Nbs1, Rad54 and double mutant cells did not show hypersensitivity to UV-light (Fig. 2D).
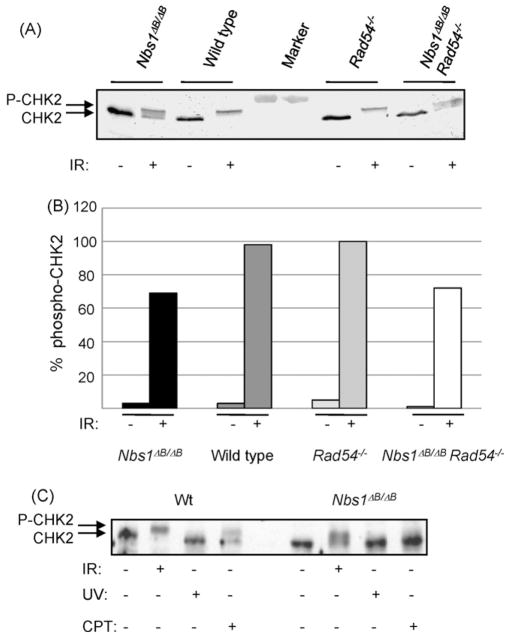
3.3. CHK2 phosphorylation depends on NBS1 after IR and CPT treatment
NBS1 is important for cell cycle checkpoint control through activation of ATM. Therefore, we studied CHK2 phosphorylation by ATM, which is thought to be NBS1-dependent after IR treatment [24,25]. All CHK2 was hyperphosphorylated in IR treated wild type and Rad54−/− cells. However, CHK2 phosphorylation was markedly reduced in the Nbs1ΔB/ΔB cells and double mutant cells (Fig. 3A and B). Residual CHK2 phosphorylation was ATM dependent, as pre-treatment with the ATM inhibitor KU55933 prevented phospho-CHK2 formation (supplementary Fig. SI).
Fig. 3.
CHK2 phosphorylation after DSB induction depends on NBS1. Western blot analysis with CHK2 antibodies (A) 1 h after 12 Gy γ-irradiation and (B) quantification of the Western blot analysis. Protein bands were derived from single membranes and band intensities were quantified using ImageJ software tool. The amount of phospho-CHK2 was quantified over the amount of total CHK2 and phoshpho-CHK2. (C) 1 h after 12 Gy γ-irradiation, 24 h after CPT treatment (10 ng/ml) or 1 h after UV-C irradiation (20 J/m2).
CPT also induced CHK2 phosphorylation, suggesting that DSBs activate ATM after this treatment (Fig. 3C). In Nbs1ΔB/ΔB cells, we did not observe any CHK2 phosphorylation after 24 h of CPT treatment (Fig. 3C) and only a trace of the phospho-CHK2 species after 1 h in CPT containing medium (supplementary Fig. SI), showing that signaling from replication-associated DSBs via CHK2 is markedly dependent on NBS1 function. The Rad54−/− cells showed wild type levels of CHK2 phosphorylation after CPT treatment whereas the double mutant cells showed the same pattern as the Nbs1ΔB/ΔB cells (data not shown). We conclude that RAD54 does not influence ATM dependent signaling.
3.4. Intra-S phase checkpoint
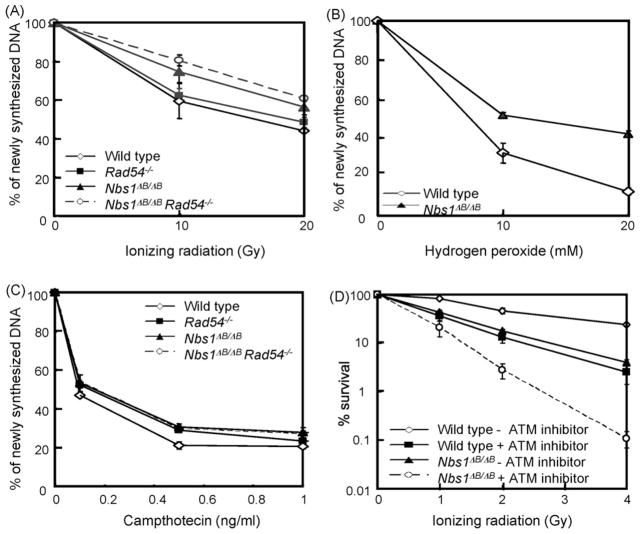
NBS1-dependent phosphorylation events are important to establish the IR-induced intra-S phase checkpoint. We confirmed that wild type and Rad54 mutant cells down regulated DNA synthesis after IR treatment, whereas Nbs1ΔB/ΔB and double mutant cells showed a less pronounced decrease of DNA synthesis under these conditions (Fig. 4A). We observed a similar difference between wild type and Nbs1ΔB/ΔB cells after H2O2 exposure (Fig. 4B). However, 1 h CPT treatment, showed little or no difference in DNA synthesis rate between wild type and Nbs1 mutant cells (Fig. 4C). As the small difference in both curves was not observed in other experiments, we conclude that Nbs1ΔB/ΔB cells do not have a defect in CPT-induced downregulation of DNA synthesis. Furthermore, Rad54−/− cells and Nbs1ΔB/ΔB Rad54−/− cells both displayed the same level of DNA synthesis after CPT treatment, confirming that the intra-S phase checkpoint induced by this compound does not require NBS1. We conclude, that the hypersensitivity of Nbs1ΔB/ΔB Rad54−/− cells cannot be explained by an intra-S phase checkpoint defect.
Fig. 4.
Inhibition of DNA synthesis after DNA damage. The relative level of 3H-Thymidine incorporation was measured after (A) γ-irradiation, (B) H2O2 treatment and (C) 1 h of CPT treatment. (D) ES cells colony survival assay for IR after ATM inhibition. The doses of IR are displayed on the X-axis on a linear scale, while the percentage of surviving colonies is displayed on the Y-axis on a logarithmic scale. Error bars show the standard error of the mean for triplicate experiments.
Additional evidence for NBS1 functions that are independent from ATM signaling in the intra-S phase checkpoint came from epistasis analysis. Nbs1ΔB/ΔB cells and wild type cells were treated with the ATM inhibitor KU55933 and colony survival after IR treatment was determined [26]. Interestingly, both the Nbs1ΔB/ΔB and wild type ES cells became more IR sensitive after treatment with the ATM inhibitor (Fig. 4D). The ATM inhibitor did not cause hypersensitivity of AT-fibroblasts (supplementary Fig. SII), excluding off target effects of the inhibitor. Nbs1ΔB/ΔB cells with ATM inhibitor were more IR sensitive than ATM inhibited wild type cells, indicating that NBS1 has ATM-independent functions, which are different from the intra-S phase checkpoint.
3.5. Homologous targeting and SCE are not affected in Nbs1ΔB/ΔB cells
Subsequently, we investigated whether a decreased HR capacity could explain the excess residual DSBs in the double mutant cells. We determined the HR levels in ES cells with a homologous gene targeting assay, using a promoterless RAD54-GFP knock-in construct [27]. GFP is only expressed upon correct targeting into the Rad54 locus and not after random integration. Nbs1ΔB/ΔB and wild type ES cells showed a similar targeting efficiency (Fig. 5), indicating that HR was not affected by the Nbs1ΔB mutation. The targeting efficiency in Rad54−/− and double mutant cells was reduced to similar levels (5% and 8%, respectively), suggesting that the repair defect in double mutant cells could not be explained by a general defect in HR. We confirmed this conclusion by measuring sister chromatid exchanges (SCE). As found previously, Rad54 deficiency caused a dramatic decrease in MMC-induced SCE [35]. However, the Nbs1ΔB mutation did not influence SCE induction and the double mutant cells were similar to Rad54 single mutants.
Fig. 5.

Homologous gene targeting does not depend on NBS1. (A) Cells of the indicated genotypes were electroporated with the mRAD54-GFP knock-in construct that results in GFP expression after homologous gene targeting, but not after random integration. GFP expression was analyzed by flow cytometry after 7 days of antibiotic selection. The calculated targeting efficiency is displayed for each experiment. (B) Induction of SCEs by MMC in MEFs. 40–60 metaphases per sample were scored for the number of SCEs per cell and corrected for the amount of chromosomes. Only 6 metaphases were counted in the MMC treated double mutant cells. The error bars indicate the SEM.
3.6. Persistent DSBs in Nbs1ΔB/ΔB Rad54−/− cells
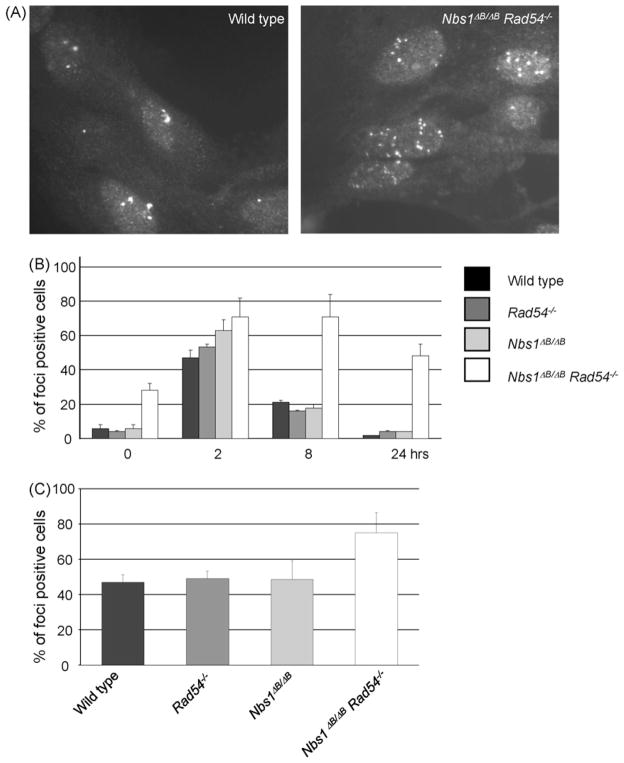
The sensitivity of the double mutant cells after IR and H2O2 treatment could in principle be explained by a combination of a HR defect, caused by RAD54 deletion and an intra-S phase checkpoint defect. However, the hypersensitivity of Nbs1ΔB/ΔB cells compared to wild type cells in the presence of ATM inhibitor suggested that NBS1 has additional, ATM-independent functions in the DNA damage response. Therefore, we investigated the accumulation of RAD51 in subnuclear structures (foci) as a marker for sites of HR. Interestingly, untreated double mutant MEFs showed a 5-fold higher level of RAD51 foci positive cells than the wild type or the single mutant MEFs, indicating that endogenous DNA damage levels were increased in these cells (Fig. 6A and B). RAD51 foci were induced by γ-irradiation in wild type and mutant MEFs. The highest percentage of foci positive cells was observed 2 h after irradiation, which then decreased over the next 24 h to the low level observed before irradiation (Fig. 6B). However, the RAD51 foci in double mutant MEFs persisted for a longer period of time and even after 24 h the number of RAD51 foci had not returned to pre-irradiation levels. Spontaneous RAD51 foci were also increased in double mutant ES cells, indicating that this phenomenon is not cell type specific (Fig. 6C).
Fig. 6.
RAD51 foci persist in Nbs1ΔB/ΔB Rad54−/− MEFs. (A) RAD51 foci formation in untreated MEFs of the indicated genotypes. (B) Quantification of RAD51 focus formation at 0, 2, 8 and 24 h after 2 Gy γ-ray irradiation in MEFs. (C) Spontaneous foci formation in ES cells. At least 75 cells were counted for each genotype in 3 independent experiments. Cells with 5 or more foci were scored as foci positive cells.
Persistent RAD51 foci are indicative of persistent DNA damage, which predicts that Nbs1ΔB/ΔB Rad54−/− cells should display a high incidence of chromosomal aberrations. Therefore, we made metaphase spreads of wild type and mutant MEFs and ES cells (Table 2). Interestingly, double mutant cells exhibited a much higher level of spontaneous chromosomal instability than wild type or single mutant cells. The majority of these aberrations affected only one chromatid, which is consistent with the idea that the most deleterious type of damage for the double mutant cells is a SSB that is converted to a DSB in S phase, which frequently remains unjoined and finally can lead to cell death.
Table 2.
Nbs1ΔB/ΔB Rad54 −/− cells acquire spontaneous chromosomal aberrations.
| Cell type | Genotype | Metaphases analyzed | aberrant metaphases (%) | Chromatid type aberrations (%) | Chromosome type aberrations (%) |
|---|---|---|---|---|---|
| ES cells | Wild type | 58 | 3 | 1.5 | 1.5 |
| Rad54−/− | 60 | 3 | 1.5 | 1.5 | |
| Nbs1ΔB/ΔB | 60 | 7 | 2 | 5 | |
| Nbs1ΔB/ΔB | 60 | 13 | 8 | 5 | |
| Rad54−/− | |||||
| pMEFs | Wild type | 22 | 0 | 0 | 0 |
| Rad54−/− | 20 | 5 | 0 | 5 | |
| Nbs1ΔB/ΔB | 20 | 0 | 0 | 0 | |
| Nbs1ΔB/ΔB | 38 | 29.5 | 22 | 7.5 | |
| Rad54−/− | |||||
| tMEFs | Wild type | 56 | 2 | 0 | 2 |
| Rad54−/− | 52 | 0 | 0 | 0 | |
| Nbs1ΔB/ΔB | 70 | 1 | 0 | 1 | |
| Nbs1ΔB/ΔB | 55 | 60 | 51 | 20 | |
| Rad54−/− |
Chromatid type aberrations: aberrations restricted to a single chromatid, including gaps, breaks and radial structures. Chromosome type aberrations: aberrations involving both sister chromatids, including gaps and breaks. ES cells: embryonic stem cells, pMEFs: primary MEFs (MEF cultures up to passage 8), tMEFS: transformed MEFs (MEF cultures from passage 8 on).
4. Discussion
The MRN complex has multiple functions in the maintenance of chromosomal stability. We investigated how the Nbs1ΔB mutation, which mimics the mutation found in NBS patients, affects genome stability and how this phenotype is influenced by a defect in HR (Rad54−/−). We found that the double mutant cells are hypersensitive to many DNA damaging agents and show a high level of chromosomal instability, even in an unchallenged situation.
We considered several possible reasons for the synergistic effects in Nbs1ΔB/ΔB Rad54−/− cells. Nbs1 mutations have been found to compromise the intra-S checkpoint, both in patient cells and in mouse cells. This has been interpreted as a defect in signaling to the replication machinery that should inhibit initiation of new replicons during S phase. We therefore reasoned that the hypersensitivity to various agents might be caused by such a signaling defect. However, this could not explain the hypersensitivity of double mutant cells to CPT: although CHK2 phosphorylation was decreased in Nbs1ΔB/ΔB cells this did not result in a measurable intra-S phase checkpoint defect. Furthermore, Nbs1ΔB/ΔB cells could be sensitized by ATM inhibitor to a much higher level than wild type cells, showing that NBS1 has ATM-independent functions. Therefore, we also considered other possible explanations for the observed hypersensitivities. From our analysis, it is clear that agents that induce S phase specific DSBs (CPT and PARP inhibitor) cause the highest level of hypersensitivity in the double mutant cells. This leaves three possible explanations: (1) NBS1 could signal in an ATM-independent fashion, for example from DSBs to the nearby replication fork, which might be necessary for proper restart of replication, especially when HR is compromised, or (2) NBS1 could prevent formation of DSBs from SSBs during S phase or (3) NBS1 might be necessary for the DSB repair reaction itself.
The first two possibilities would both result in increased formation of DSBs. This signaling could take place after DSB formation by a traversing replication fork, or one could imagine that a signal is generated from a SSB that would prevent DSB formation by halting the replication fork until the damage has been repaired. It is difficult to discriminate these two possibilities. In both scenarios one would expect an increase in persistent DSBs and chromatid type chromosomal aberrations in the double mutant cells. The classic signaling pathway that involves NBS1 is via ATM kinase activation, resulting in an impaired CHK2 phosphorylation. However, MRN dependent activation of the ATR kinase activity after replication fork stalling or UV irradiation has also been reported [18], suggesting that non-DSB type lesions may also require the MRN complex for efficient detection and/or signaling. Interestingly, depletion of the MRN complex from Xenopus egg extracts caused many DSBs during replication [28]. A similar phenomenon was found after caffeine treatment, which inhibits both ATM and ATR kinases, or after depletion of these two protein kinases, suggesting that DSB formation or a lack of repair can result from an inability to activate the ATM and ATR kinases. Both possibilities are not mutually exclusive and may contribute to the chromosomal instability observed in Nbs1ΔB/ΔB and double mutant cells.
As a third possibility we considered that Nbs1 may be required for DSB repair by HR and/or NHEJ. Previously, combined defect in HR and NHEJ was found to result in synergistic effects in mice and cells [9,10,29]. We excluded a role for NBS1 in homologous targeting, suggesting that the core HR functions are still intact. However, we cannot exclude that NBS1 is involved in a subpathway of HR that is important for replication-associated DSB repair. Although the increased IR sensitivity in double mutant MEFs might be explained by NBS1-related NHEJ defects, this does not explain the high level of hypersensitivity for agents that mainly cause DSBs associated with S phase progression. Therefore, we do not expect that the involvement of NBS1 in the direct mechanisms of HR or NHEJ is a likely explanation for the phenotype of the double mutant cells. However, the MRN complex may have a function in keeping DNA ends in close proximity, which facilitates the actual joining reaction [30]. The MRN complex forms foci after various DSB-inducing treatments, independently of other DSB repair proteins [31]. The MRN complexes interact via a Zinc-hook at the tip of their RAD50 subunit and can bridge two DNA ends [32,33]. This function might help to keep the DSB together until the HR (or NHEJ) repair machinery has properly aligned the DNA ends for the actual joining reaction. Since the NBS1 mutation does not support MRN foci formation, the microenvironment of DNA ends may have changed in such a way, that the ends loose juxtaposition much more readily and therefore remain unrepaired. Obviously, this phenotype would be enhanced when HR is compromised.
In conclusion, the enhanced sensitivities of double mutant cells to various DNA damaging agents point to a function for NBS1 that is separate from the actual repair reaction. We propose that DNA damage signaling and a changed microenvironment of the DSB may contribute to this hypersensitivity. Investigation of the effect of combining Rad54 deficiency with other factors that either fail to accomplish proper signaling (such as ATR point mutations) or IR-induced foci formation (such as γ-H2AX) may help to discriminate these possibilities.
Supplementary Material
Acknowledgments
We would like to thank Drs. G. Smith and M. O’Connor (Kudos Pharmaceuticals) for their kind gifts of ATM and PARP inhibitors. We want to thank Dr. N.G.J. Jaspers for stimulating discussions, Dr. H.B. Beverloo for advise on analysis of chromosomal aberrations and M. van Brakel for technical assistance. This research was supported by the Netherlands Cancer Foundation (NKB/KWF grants EUR 1999-1966 and EMCR 2002-2734), the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO), and the European Community (projects RISC-RAD (FI6R-CT-2003-508842), DNA Repair (LSHG-CT-2005-512113)).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.dnarep.2009.09.002.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Contributor Information
Jeroen Essers, Email: j.essers@erasmusmc.nl.
Dik C. van Gent, Email: d.vangent@erasmusmc.nl.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 9.Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, Alt FW, Jasin M. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 12.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 13.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 14.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 15.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 16.Painter RB. Radioresistant DNA synthesis: an intrinsic feature of ataxia telangiectasia. Mutat Res. 1981;84:183–190. doi: 10.1016/0027-5107(81)90061-0. [DOI] [PubMed] [Google Scholar]
- 17.Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 18.Stiff T, Reis C, Alderton GK, Woodbine L, O’Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- 20.Dahm-Daphi J, Sass C, Alberti W. Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int J Radiat Biol. 2000;76:67–75. doi: 10.1080/095530000139023. [DOI] [PubMed] [Google Scholar]
- 21.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 23.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O’Connor J, Tutt MAN, Zdzienicka MZ, Smith GC, Ashworth A. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 24.Buscemi G, Carlessi L, Zannini L, Lisanti S, Fontanella E, Canevari S, Delia D. DNA damage-induced cell cycle regulation and function of novel Chk2 phosphoresidues. Mol Cell Biol. 2006;26:7832–7845. doi: 10.1128/MCB.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buscemi G, Savio C, Zannini L, Micciche F, Masnada D, Nakanishi M, Tauchi H, Komatsu K, Mizutani S, Khanna K, Chen P, Concannon P, Chessa L, Delia D. Chk2 activation dependence on Nbs1 after DNA damage. Mol Cell Biol. 2001;21:5214–5222. doi: 10.1128/MCB.21.15.5214-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 27.Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, Beverloo B, Hendriks RW, de Klein A, Kanaar R, Hoeijmakers JH, Maas A. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 31.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 32.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 33.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 34.Essers J, Hendriks RW, Wesoly J, Beerens CE, Smit B, Hoeijmakers JH, Wyman C, Dronkert ML, Kanaar R. Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair (Amst) 2002;1:779–793. doi: 10.1016/s1568-7864(02)00110-6. [DOI] [PubMed] [Google Scholar]
- 35.Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaspers NG, Zdzienicka MZ. Inhibition of DNA synthesis by ionizing radiation: a marker for an S-phase checkpoint. Methods Mol Biol. 2006;314:51–59. doi: 10.1385/1-59259-973-7:051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.