Abstract
Background
There is interindividual variation in low-density lipoprotein cholesterol (LDLc) lowering by statins and limited study into the genetic associations of the dose dependant LDLc lowering by statins.
Methods and Results
Five hundred nine patients with hyperlipidemia were randomly assigned atorvastatin 10 mg, simvastatin 20 mg, or pravastatin 10 mg (low-dose phase) followed by 80 mg, 80 mg, and 40 mg (high-dose phase), respectively. Thirty-one genes in statin, cholesterol, and lipoprotein metabolism were sequenced and 489 single nucleotide polymorphisms with minor allele frequencies >2% were tested for associations with percentage LDLc lowering at low doses using multivariable adjusted general linear regression. Significant associations from the analysis at low dose were then repeated at high-dose statins. At low doses, only 1 single nucleotide polymorphism met our experiment-wide significance level, ABCA1 rs12003906. Twenty-six subjects carried the minor allele of rs12003906, which was associated with an attenuated LDLc reduction (LDLc reduction in carriers versus noncarriers −24.1±2.6% versus −32.2±1.5%; P=0.0001). In addition, we replicated the association with the APOE ε3 allele and a reduced LDLc reduction. At high doses, carriers of the minor allele of ABCA1 rs12003906 and the APOE ε3 allele improved their LDLc reduction but continued to have a diminished LDLc reduction compared with noncarriers (−30.5±4.0% versus −42.0±2.4%; P=0.005) and (−38.5±1.9% versus −45.3±2.8%; P=0.009), respectively.
Conclusions
An intronic single nucleotide polymorphism in ABCA1 and the APOE ε3 allele are associated with reduced LDLc lowering by statins and identify individuals who may be resistant to maximal LDLc lowering by statins.
Keywords: cholesterol, genetics, hypercholesterolemia, pharmacogenetics, HMG-CoA
Statins (3-hydroxy-3-methylglutaryl [HMG]-CoA reductase inhibitors) are widely prescribed medications that prevent incident and recurrent coronary artery disease events primarily through the reduction of low-density lipoprotein cholesterol (LDLc).1 The goal of lipid-lowering therapy is to reduce the LDLc cholesterol to <100 mg/dL in patients at high risk and an optional goal of <70 mg/dL for patients at highest risk.2,3 Because of these aggressive recommendations, a substantial proportion (>40%) of patients treated outside of clinical trials remain above their recommended LDLc goal.4,5 Barriers to attaining LDLc goals include failure to initiate therapy, nonadherence, side effects, inappropriate drug/dose selection, and inadequate dose titration. Although several dosing algorithms have been devised to tailor dose selection and to help minimize dose titrations, these are not widely used and can leave >20% of patients above their LDLc goals.6,7
Furthermore, although most patients derive a moderate (30% to 50%) reduction in LDLc cholesterol with statin therapy, there is wide interindividual variation in the dose response to statin therapy.8 Nongenetic factors such as race, age, and smoking status have only mild influence on statin response.9,10 Consequently, there has been considerable investigation into the genetic determinants of the response to statin therapy. Single nucleotide polymorphisms (SNPs) in several genes (eg, SNP1211 and the H7 haplotype12 in HMGCR and the ε3 and ε4 alleles in APOE13) are associated with reduced effectiveness of statin therapy. However, others (eg, CYP7A1) have failed to replicate on subsequent testing. The failure to replicate associations between SNPs and statin response may be attributable to differences in low allele frequency, small effect sizes, small study size, or chance.
Therefore, the current study attempts to overcome some of the shortcomings of prior studies by choosing a relatively large sample size with improved statistical power for detection of less common alleles. We sought to explore whether genetic variants in the statin pharmacokinetic and pharmacodynamic, cholesterol synthesis, and lipoprotein metabolism pathways are determinants of statin-mediated LDLc reduction. We pursued this question in the context of a statin pharmacogenetic study with refined phenotyping, attention to medication adherence, and a comprehensive gene-wide, pathway-based approach to SNP selection. Finally, in addition to identifying SNPs that confer a diminished response to statin therapy, we observed the response to maximal dose statin to determine whether these genetic effects could be modulated.
Methods
The statin response examined by genetic haplotype markers (STRENGTH) study was a pharmacogenetic study of statin efficacy and safety. In summary, it was a 16-week multicenter, randomized, open label study of 3 commercially available statins that were each tested at starting and maximum labeled doses (as stated in the package inserts) in patients with primary hypercholesterolemia (Figure 1) conducted between 2001 and 2002. The original intent of the STRENGTH trial was to identify common (minor allele frequencies >15%) genetic variants associated with large (15%) within-treatment differences in LDLc lowering as well as gene×treatment interactions. In a post hoc decision to identify rarer (>2%) variants associated with smaller (10%) differences in treatment effect, all 3 treatment arms were pooled for subsequent analyses with adjustment for assigned treatment.
Figure 1.

Study design.
The protocol was approved by a central institutional review board. Subjects provided signed informed consent.
Study Population
Male or female outpatients age 18 to 75 years with a diagnosis of type IIa or IIb hypercholesterolemia who had been on the American Heart Association Step I or Step II diet for at least 6 weeks before the onset of screening were eligible to participate. Subjects could either be treatment naïve or have been previously treated for hypercholesterolemia with any approved medications; however, previously treated subjects must have discontinued antihyperlipidemic medication 5 weeks before drug assignment (8 weeks before screening if clofibrate was in use) to be eligible. Subjects must have demonstrated dietary compliance with the American Heart Association Step I or Step II diet as measured by a food diary at baseline to be eligible for randomization.
Exclusion criteria included an LDLc >240 mg/dL, triglycerides >400 mg/dL, history of myocardial infarction within the past 6 months, unstable angina, angina at rest, or a history of stroke, transient ischemic attacks, or coronary revascularization within the past year.
Medication Assignment and Administration
After the washout period, subjects were randomly assigned to 1 of 3 low-dose treatment groups at Week 0: 10 mg/d atorvastatin, 20 mg/d simvastatin, or 10 mg/d pravastatin, for 8 weeks. At the Week 8 visit, all subjects were then switched to the highest allowable dose, as stated in the package insert of their assigned medication, for the subsequent 8 weeks. The doses for the high-treatment groups were the following: 80 mg/d atorvastatin, 80 mg/d simvastatin, and 40 mg/d pravastatin. All medications were prescribed to be taken once daily in the evening, with or without food, as specified in the respective package insert.
Follow-Up and Laboratory Testing
For entry into the study, 2 fasting, pretreatment lipid profiles were averaged and were required to be within 15% of each other. A 12-hour fasting serum lipid profile was obtained for all subjects at the end of low-dose statin therapy (weeks 6 and 8 averaged) and high-dose therapy (weeks 14 and 16 averaged). In-person visits with a research coordinator were scheduled every other week to assess for adverse events and compliance. Patients were considered to be adherent if >80% of pills were taken by self-report.
Laboratory and Clinical Measurements
All assays were performed at Covance Central Laboratories (Indianapolis, Ind). High-density lipoprotein cholesterol was measured using dextran sulfate and magnesium chloride precipitation (Roche Diagnostics), and total cholesterol and triglycerides by enzyme hydrolysis on Hitachi analyzers. The LDLc level was calculated using the Friedewald equation (LDLc=total cholesterol − high-density lipoprotein − 1/5 triglycerides).
Candidate Gene Selection
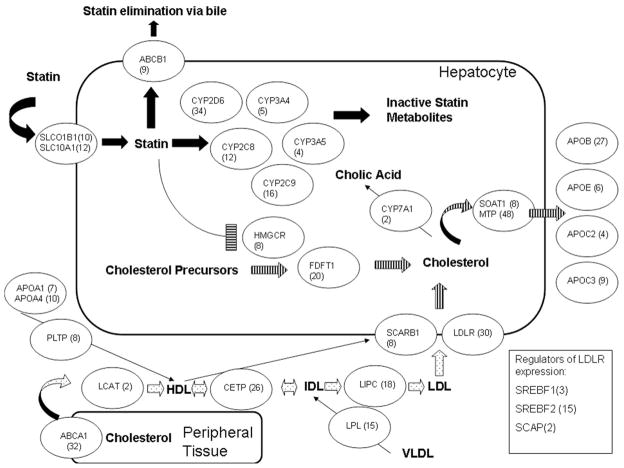
Based on prior studies, genetic variation in 4 different pathways have been implicated in statin-mediated LDLc reduction: (1) pharmacokinetics of statins,13 (2) pharmacodynamics of statins,11 (3) metabolism of cholesterol,15 and (4) metabolism of lipoproteins.13 These studies were limited because only selected SNPs/candidate genes in each pathway were chosen and tested in isolation. As part of the STRENGTH protocol >160 potential genes were sequenced or genotyped. For the purposes of this study, we selected 31 genes for analysis based on prior associations with statin response as compiled by the PharmGKB database (www.pharmgkb.org)16 and recent reviews of statin pharmacogenetics pathways (Figure 2).14
Figure 2.
Candidate pathways and genes. The 4 different pathways are depicted here: (1) statin pharmacokinetics (solid), (2) statin pharmacodynamics (horizontal stripe), (3) cholesterol metabolism (vertical stripe), and (4) lipoprotein metabolism (dotted). Each of the candidate genes is depicted in ovals with the number of SNPs per gene in parentheses. Not depicted are APOA2 (12) and LRP2 (62).
Sequencing Protocol, SNP Selection, and Power Calculation
For each candidate gene, all exons and associated 100 bp of flanking intronic DNA, 5′UTR, 3′UTR, 1000 bp of 5′ upstream DNA, and 100 bp of 3′ downstream DNA were sequenced as previously described.17 Sequence data were analyzed for the presence of polymorphisms using Polyphred. Polymorphisms were confirmed by sequencing both strands of DNA.17 For the initial analysis, we identified 2481 SNPs and only included those 489 SNPs with a minor allele frequency of >2% for further analysis. The number of SNPs tested per gene is listed in Figure 2. In a post hoc power analysis, this allele frequency cutoff provides >85% power to detect genetic effects that explain >2% of the variance in percent LDLc reduction.
Statistical Methods
General Linear Regression Models for Single SNP Effects
Because of the known association between baseline LDLc and absolute LDLc reduction, we prospectively defined the percent reduction in LDLc cholesterol as the primary end point, as this reduces the influence of baseline LDLc as a confounder. Patients were randomized to 3 different treatment arms; however, because we were interested in those SNPs with effects across statins, all patients were pooled for the subsequent analyses and assigned treatment was included in multivariable models. To understand clinical factors contributing to percent reduction in LDLc cholesterol, we first constructed univariable models with clinical covariates (race, age, sex, and smoking status) and assigned treatment using general linear regression. Predictors (P<0.1) were selected and subsequently entered in the genetic analysis models. For genetic analyses, individual SNPs were tested in additive, recessive, and dominant models using a general linear regression model, adjusting for smoking status, race, assigned treatment, and age. We graphically examined the residuals from the general linear models, and these approximated a normal distribution. Further, we performed sensitivity analyses with the log transformed percent reduction in LDLc, which did not alter the strengths of association with low-dose statins (data not shown). SNPs that were nominally (P<0.05) significant were selected for further analysis by multiple comparisons testing. The least square means method was used for reporting mean differences and standard errors in treatment effect between groups. These analyses were performed in SAS/SAS Genetics (Cary, NC).
Multiple Comparisons Testing
To account for the large number multiple comparisons performed, we calculated an experiment wide significance level by estimating the effective number of independent tests using the method of Li and Ji18 as implemented on http://gump.qimr.edu.au/general/daleN/matSpD/. This methodology takes into consideration linkage disequilibrium patterns among all SNPs and reduces this set to a minimal number of “effective” SNPs. Within the STRENGTH data, the original set of 489 SNPs was reduced to 269 SNPs. The final step is to adjust the test criteria for this number of effective SNPs by the method of Bonferroni (ie, 0.05/269=0.0002), and therefore only those SNPs with a probability value below this level are reported.
Haplotype Analyses
The haplo.stats package through R Statistical Computing was used to identify haplotypes and to provide a measure (haplo.score) of association to disease.19 Haplo.stats expands on the likelihood approach to account for phase ambiguity in case–control studies by using a generalized linear model to test for haplotype association that allows for adjustment of nongenetic covariates.19 This method derives a global score statistic to test the null hypothesis of no association of the trait with the genotype, H0: β = 0. Genetic markers that are immediately adjacent on a chromosome might be statistically independent, whereas those that are hundreds of thousands or more base pairs apart might be highly correlated.20 While controlling for smoking, age, race, and assigned treatment, we used the function haplo.score to derive a matrix of global pairwise probability values for association significance between 2-SNP haplotypes and LDL levels.
Statement of Responsibility
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
A total of 509 patients were enrolled in the study. The baseline characteristics and medical conditions are listed in Table 1, with no significant differences across the 3 treatment arms. The cohort was primarily white, 53% female, with one or no cardiac risk factors. A minority of the patients had a diagnosis of coronary artery disease or type II diabetes.
Table 1.
Baseline Characteristics and Medical Conditions by Treatment
| Characteristic | Atorvastatin (n=168) | Simvastatin (n=182) | Pravastatin (n=159) | Total (n=509) |
|---|---|---|---|---|
| Gender | ||||
| Male | 74 (44.0) | 99 (54.4) | 67 (42.1) | 240 (47.0) |
| Female | 94 (56.0) | 83 (45.6) | 92 (57.9) | 269 (53.0) |
| Race | ||||
| White | 145 (86.3) | 154 (84.6) | 129 (81.1) | 428 (84.2) |
| Non-white | 23 (13.7) | 28 (15.4) | 30 (18.9) | 81 (15.8) |
| Age, years | 56.3±9.9 | 55.9±10.7 | 57.2±10.5 | 56.2±10.3 |
| Hypertension | 67 (39.9) | 79 (43.4) | 77 (48.4) | 223 (43.4) |
| Type II diabetes | 23 (13.7) | 19 (10.4) | 21 (13.2) | 63 (11.8) |
| Coronary artery disease | 9 (5.4) | 14 (7.7) | 10 (6.3) | 33 (6.0) |
| Arrhythmia | 8 (4.8) | 6 (3.3) | 9 (5.7) | 23 (4.3) |
| Smoker | 20 (18) | 37 (20) | 33 (21) | 90 (19) |
| Baseline lipid values, mg/dL | ||||
| Total cholesterol | 257±34 | 258±31 | 256±30 | 257±31 |
| High-density lipoprotein cholesterol | 50±13 | 47±11 | 49±12 | 49±12 |
| Low-density lipoprotein cholesterol | 171±26 | 174±25 | 173±24 | 173±25 |
| Triglycerides | 177±68 | 180±66 | 173±69 | 177±68 |
Data are presented as n (%) or mean±SD
At the end of the low-dose phase, the average LDLc reduction was 30±15%, and after the high-dose phase, the LDLc reduction averaged 40±16%. In the univariable analysis of nongenetic covariates with low-dose statin older age, nonsmokers, black ethnicity, and assigned statins were associated with the percent reduction in LDLc with low-dose statins (Table 2). Gender was not associated with percent LDLc reduction (P=0.9).
Table 2.
Significant Univariable Associations With Low-Dose Statin Percent LDLc Lowering
| Variable | Mean Effect±SE | P Value | |
|---|---|---|---|
| Age | +1±0.5% per decade | 0.04 | |
| Assigned statin | |||
| Atorvastatin 20 mg | −39±1% | ||
| Simvastatin 20 mg | −35±1% | <0.0001 | |
| Pravastatin 10 mg | −21±1% | ||
| Percent LDLc Lowering With Variable | Percent LDLc Lowering Without Variable | ||
| Smoking | −33±1% | −29±1% | 0.01 |
| Black ethnicity* | −26±3% | −32±1% | 0.05 |
Comparison of blacks vs whites.
In the initial screen with low-dose statins, we tested the effects of the 489 SNPs in the 31 candidate genes in combination with age, smoking status, assigned statin, and ethnicity and found that one SNP in ABCA1 (rs12003906, in Hardy–Weinberg equilibrium, minor allele frequency in STRENGTH=0.03) met our experiment wide significance level and was associated with an attenuated LDLc reduction (percent LDLc reduction in carriers versus noncarriers −24.1±2.6% versus −32.2±1.5%; P=0.0001). None of the carriers of the minor alleles of SNPs in HMGCR (including SNP1211) nor carriers of the ε4 allele in APOE had a significantly different percent LDLc lowering compared with noncarriers. However, as previously reported, we observed that at low statin doses carriers of the major allele in APOE rs7412 (ε3 allele, allele frequency in STRENGTH=0.03) had a reduced LDLc lowering compared with carriers of the minor allele (ε2 allele; percent LDLc reduction in carriers versus noncarriers −30.4±1.5% versus −36.4±2.4%; P=0.005). However, this finding did not meet our experiment-wide significance level. In a univariable analyses, ABCA1 rs12003906 explained 4% (R2=0.037) and APOE rs7412 explained 1% (R2=0.011) of the variation in percent LDLc reduction. In a multivariable model that included both ABCA1 rs12003906 and APOE rs7412 SNPs as well as age, assigned statin, gender, smoking status, and ethnicity, both SNPs remained significant (P<0.01) predictors of the percent LDLc reduction.
To quantify the effects of maximal dose titration in carriers identified during the low-dose phase of the study, we then observed the effect of the highest prescribed dose for each of the low-dose associations (Table 3). Both carriers of the ABCA1 SNP rs12003906 and the APOE ε3 alleles improved their LDLc reduction with maximal doses. However, at maximal statin doses, carriers continued to have a diminished LDLc lowering compared with noncarriers.
Table 3.
Effect SNPs Identified From Low-Dose Statin Analysis on High Dose Stain Response
| Gene | SNP rsID | % LDLc Reduction±SE for Marker Positive* | % LDLc Reduction±SE Marker Negative* | P Value |
|---|---|---|---|---|
| ABCA1 | rs12003906 | −30.5±4.0 | −42.0±2.4 | 0.005 |
| APOE | rs7412 | −38.5±1.9 | −45.3±2.8 | 0.009 |
Compared with baseline, adjusted for race, gender, age, smoking, and treatment group.
Because medication adherence and baseline LDLc cholesterol could confound these results, additional analyses were performed to assess their effects. Self-reported compliance was routinely assessed in the study cohort at regular intervals. With low- and high-dose statins, 31 (6%) and 97 (19%) patients, respectively, were not adherent with their assigned medication. There was no association between adherence and statin type (P>0.3). To explore whether nonadherence and baseline LDLc cholesterol influenced our findings, additional analyses including adjustment for these variables did not appreciably alter the findings at low- or high-dose statins (data not shown). To explore whether haplotypes within ABCA1 influenced LDL lowering, we examined all pairwise haplotypes for their association with low-dose percent LDLc lowering. We found 1 haplotype pair that included rs12003906 but was no more significant than rs12003906 alone (0.0003 versus 0.0001). Finally, there was no evidence that unbalanced randomization occurred across ABCA1 rs12003906 (P=0.2) or APOE ε3 (P=0.6) genotypes.
Discussion
The STRENGTH study is a comprehensive study of the pharmacogenetics of dose-dependent LDLc lowering to statins. We extensively examined the effects of SNPs related to statin pharmacokinetics, statin pharmacodynamics, cholesterol, and lipoprotein metabolism, and the LDLc-lowering response with low doses of statins. This study was the first to take the additional step of assessing the dose response to various SNPs associated with reduced LDLc lowering. The main findings of this study are that (1) patients who carry the minor allele ABCA1 rs12003906 have a reduced LDLc lowering with low doses of statins, and (2) maximal statin doses improve, though may not overcome, the difference in LDLc reduction seen in carriers of the minor allele of ABCA1 rs12003906 and the ε3 allele of APOE.
Despite the proven efficacy of statins in the treatment and prevention of coronary artery disease, there is a significant proportion of patients who do not achieve their target LDLc goal.4,5 Among the possible reasons why patients do not reach their target LDLc goal are (1) the interindividual variability in the response to statin medications,8 and (2) the dose-related toxicity and side effects that may lead to nonadherance.21 Prospective identification of patients who have a diminished response to statins, particularly with higher doses, may improve the risk-to-benefit ratio of statin therapy.
There have only been a few larger studies of statin pharmacogenetics to date, each enrolling >1500 patients. In the first, 43 SNPs were selected from 16 candidate genes that were chosen based on a literature-based search for genes implicated in the statin response.13 Although this study was limited because it only focused on SNPs previously identified in the literature and there was no accounting for multiple comparisons, it identified carriers of the ε3 allele in APOE as having a reduced LDLc response. In STRENGTH, we found an association of similar size and direction in carriers of the ε3 allele; however, this association did not meet our experiment-wide multiple comparisons testing cutoff. The second study tested 148 SNPs guided by common mutations and linkage disequilibrium (LD) patterns in 10 candidate genes related to lipoprotein metabolism and accounted for multiple comparisons testing.11 Although it only tested a limited number of genes, it identified a SNP in HMGCR (SNP 12) associated with a diminished LDLc response. The third and most recent evaluation only focused on HMGCR in the response to a single dose of simvastatin.12 Unlike the present evaluation, few of the prior studies examined the effects of nongenetic effects or assessed the dose response of their findings. Therefore, the STRENGTH study builds on these 3 prior studies by examining the statin dose response in conjunction with direct sequencing of an unbiased, pathway-based approach to candidate SNP selection in a cohort of patients larger than most statin studies.
If replicated, the SNPs identified in this study would be unique in that they do not relate to the pharmacokinetics (ie, metabolism) or pharmacodynamics (ie, HMGCR) of statins. Instead, they relate to lipoprotein metabolism (ABCA1 and APOE). Our failure to identify significant variants in pharmacokinetic pathways may be because pravastatin has minimal Cytochrome P450 (CYP) metabolism, whereas atorvastatin and simvastatin are CYP3A (3A4 and A5) metabolized and genetic variation in these genes has only small effects on their pharmacokinetics.14 The 3 large scale (>900 patients) attempts to study SNPs in HMGCR and statin response yielded mixed results, with 2 studies finding a reduced LDLc reduction in heterozygotes and the other finding no significant effect.11–13 Therefore, the lack of association with HMGCR SNPs in our study may be attributable to underpowering, population admixture, or lack of a true effect. That SNPs outside of HMGCR inhibition are implicated in LDLc lowering suggests that there may be novel pharmaceutical targets for LDLc reduction.
In addition to identifying SNPs associated with LDLc lowering, we also identified older age and nonsmokers as having an enhanced LDLc lowering response to statins. These findings are consistent with the direction and magnitude of effect seen in 2 prior studies.9,10 The individual response to medications is a complex biological trait that is determined by both genetic and nongenetic factors. For example, although the dose response to warfarin is characterized by a wide interindividual variability that is determined by genetic (eg, CYP2C9 and VKORC1 genotypes) factors, the addition of nongenetic factors (eg, age, body mass index, and smoking) factors improves the ability to predict the dose requirement.22,23 Although the individual effects of age and smoking are mild and the mechanism is unclear, the effect is consistent across studies and may add to the predictive ability of a pharmacogenetics based approach to statin prescription.
Although there have been several prior studies that identified genetic determinants of LDLc lowering with statins, none have evaluated the dose response to these predictors. The novel sequential design of the STRENGTH study allowed us to assess the effects of maximal statin dose with SNPs of interest in the same patient population. At the highest doses of statins, those patients who carried the minor alleles of the ABCA1 SNP, rs12003906, and the APOE SNP rs7412 improved their LDLc lowering, but continued to have a significantly reduced response. These findings suggest that dose titration will be of limited benefit in these patients and may increase the risk of adverse events. In STRENGTH, we also observed that the relative potencies of statins was preserved (atorvastatin>simvastatin>pravastatin) across genotypes. Therefore, a possible strategy for the patient with genetically mediated statin resistance would be to switch to a more potent statin to achieve adequate LDLc goals before escalating dose. Although the risk of serious rhabdomyolysis or hepatotoxicity is rare, the risk of drug discontinuation attributable to musculoskeletal complaints is common and increases with increased doses of statins.24 This pattern was evident in the current investigation with a near tripling of the nonadherence rate with high-dose statin. However, the appropriate strategy to overcome genetically mediated statin resistance without increasing the risk of adverse events is not readily apparent from the present study. If validated, additional approaches should be specifically targeted to these individuals in prospectively designed clinical trials.
Despite the unique findings and characteristics of this study, several limitations require consideration. The most important is the lack of an independent replication cohort to validate these findings. Although not in an independent cohort, the findings at high doses highlight the importance of these SNPs by demonstrating the continued difference in LDLc lowering in carriers versus noncarriers of the ABCA1 and APOE minor alleles. Second, we did not use LD patterns to guide SNP selection. Using LD to help guide SNP selection would increase our ability to identify significant SNPs,25 but comes at the expense of missing the rare SNP effects found in this study. Third, although we accounted for race in the genetic analyses, this type of adjustment has limitations and population stratification may still confound these results, because ABCA1 rs12003906 was most significantly associated with low-dose statin response in blacks (P=0.006) and the APOE ε3 allele was only significant (P=0.04) in whites. Because of small sample sizes, though, we could not adequately report race-stratified analyses. Fourth, although we accounted for multiple comparisons testing in the low-dose analyses, we did not account for the multiple genetic models (dominant, recessive, and additive). Therefore, our “experiment-wide” significant level could be lowered to consider all tests performed in this study. Fifth, these SNPs explained a small (<4%) of the variation in LDLc lowering, suggesting that other genetic variants outside of the pathways identified in the present study (such as the PCSK9 and the NPC1L1 gene) may be more important in determining statin efficacy. Last, many of the SNPs identified in this study were intronic SNPs, and we did not identify a functional exonic SNP in significant LD with any of the ABCA1 SNPs (data not shown), therefore the functional effect of these SNPs is unknown and will require further investigation.
In summary, the STRENGTH study is the first to identify a SNP in ABCA1 associated with a reduced LDLc lowering effect and to assess the dose response to statin mediated LDLc lowering with respect to specific genotypes. The dose response to these SNPs suggests that simple dose escalation will not overcome the differences between carriers and noncarriers. Because toxicity and adherence are related more to statin dose and less to LDLc reduction, carriers of these SNPs may benefit more from high potency statins or combination therapy to reach aggressive LDLc goals. If replicated, these findings could begin to provide a foundation for tailored therapy for patients with cardiovascular disease.
CLINICAL PERSPECTIVE.
There is wide variability in the low-density lipoprotein cholesterol (LDLc) response to statins. Outside of clinical trials >20% of patients do not achieve their target LDLc goal. In this pharmacogenetic statin study, we prospectively administered low doses of statins (atorvastatin, simvastatin, and pravastatin) for 8 weeks followed by high doses of statins for 8 weeks to 509 patients with hyperlipidemia. Thirty-one genes in statin, cholesterol, and lipoprotein metabolism were sequenced, and 489 polymorphisms were tested for associations with percentage LDLc lowering at low doses. We found that carriers of a polymorphism in ABCA1 and another in APOE were associated with an attenuated LDLc response to starting doses of statin. In addition, we observed that the highest prescribable statin doses could improve LDLc lowering but could not overcome the difference between carriers and noncarriers. However, at each dose range, the more potent statins provided more LDL reduction than the lower potency statin regardless of genotype. Our observations suggest that genetically mediated statin resistance may not receive the full benefit of dose titration. Upfront knowledge of genetically defined statin resistance may guide clinicians to use a low dose of a more potent statin than a less potent one. Or, if the patient is already on a statin, switching to a more potent drug rather than dose escalation. This strategy may reduce the risk of adverse events because these are more associated with statin dose rather than statin potency.
Footnotes
Disclosures
The STRENGTH trial was conducted by Genaissance Pharmaceuticals, which is now a part of Clinical Data Inc. Drs Reed and Salisbury are employees and stock holders of Clinical Data Inc. All other authors report no disclosures.
References
- 1.Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes R. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 3.Executive Summary of the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 4.Yan AT, Yan RT, Tan M, Hackam DG, Leblanc KL, Kertland H, Tsang JL, Jaffer S, Kates ML, Leiter LA, Fitchett DH, Langer A, Goodman SG. Contemporary management of dyslipidemia in high-risk patients: targets still not met. Am J Med. 2006;119:676–683. doi: 10.1016/j.amjmed.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Davidson MH, Maki KC, Pearson TA, Pasternak RC, Deedwania PC, McKenney JM, Fonarow GC, Maron DJ, Ansell BJ, Clark LT, Ballantyne CM. Results of the National Cholesterol Education (NCEP) Program Evaluation ProjecT Utilizing Novel E-Technology (NEPTUNE) II survey and implications for treatment under the recent NCEP Writing Group recommendations. Am J Cardiol. 2005;96:556–563. doi: 10.1016/j.amjcard.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Martineau P, Gaw A, de Teresa E, Farsang C, Gensini GF, Leiter LA, Langer A. Effect of individualizing starting doses of a statin according to baseline LDL-cholesterol levels on achieving cholesterol targets: the Achieve Cholesterol Targets Fast with Atorvastatin Stratified Titration (ACTFAST) study. Atherosclerosis. 2006 doi: 10.1016/j.atherosclerosis.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 7.McKenney JM, Davidson MH, Saponaro J, Thompson PD, Bays HE. Use of a treatment algorithm to achieve NCEP ATP III goals with atorvastatin. J Cardiovasc Pharmacol. 2005;46:594–599. doi: 10.1097/01.fjc.0000180901.70607.72. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz G, Drobnik W. Pharmacogenomics and pharmacogenetics of cholesterol-lowering therapy. Clin Chem Lab Med. 2003;41:581–589. doi: 10.1515/CCLM.2003.088. [DOI] [PubMed] [Google Scholar]
- 9.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the cholesterol and pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 10.Shear CL, Franklin FA, Stinnett S, Hurley DP, Bradford RH, Chremos AN, Nash DT, Langendorfer A. Expanded clinical evaluation of lovastatin (EXCEL) study results. Effect of patient characteristics on lovastatin-induced changes in plasma concentrations of lipids and lipoproteins. Circulation. 1992;85:1293–1303. doi: 10.1161/01.cir.85.4.1293. [DOI] [PubMed] [Google Scholar]
- 11.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 12.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JF, Man M, Johnson KJ, Wood LS, Lira ME, Lloyd DB, Banerjee P, Milos PM, Myrand SP, Paulauskis J, Milad MA, Sasiela WJ. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 2005;5:352–358. doi: 10.1038/sj.tpj.6500328. [DOI] [PubMed] [Google Scholar]
- 14.Mangravite LM, Thorn CF, Krauss RM. Clinical implications of pharmacogenomics of statin treatment. Pharmacogenomics J. 2006;6:360–374. doi: 10.1038/sj.tpj.6500384. [DOI] [PubMed] [Google Scholar]
- 15.Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. A promoter polymorphism in cholesterol 7alpha-hydroxylase interacts with apolipoprotein E genotype in the LDL-lowering response to atorvastatin. Atherosclerosis. 2005;180:407–415. doi: 10.1016/j.atherosclerosis.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, Rubin DL, Shafa F, Stuart JM, Altman RB. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 17.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 19.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- 21.Pasternak RC, Smith SC, Jr, Bairey-Merz CN, Grundy SM, Cleeman JI, Lenfant C. ACC/AHA/NHLBI Clinical Advisory on the Use and Safety of Statins. Circulation. 2002;106:1024–1028. doi: 10.1161/01.cir.0000032466.44170.44. [DOI] [PubMed] [Google Scholar]
- 22.Millican EA, Lenzini PA, Milligan PE, Grosso L, Eby C, Deych E, Grice G, Clohisy JC, Barrack RL, Burnett RSJ, Voora D, Gatchel S, Tiemeier A, Gage BF. Genetic-based dosing in orthopedic patients beginning warfarin therapy. Blood. 2007;110:1511–1515. doi: 10.1182/blood-2007-01-069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91:87–94. doi: 10.1160/TH03-06-0379. [DOI] [PubMed] [Google Scholar]
- 24.Silva M, Matthews ML, Jarvis C, Nolan NM, Belliveau P, Malloy M, Gandhi P. Meta-analysis of drug-induced adverse events associated with intensive-dose statin therapy. Clin Ther. 2007;29:253–260. doi: 10.1016/j.clinthera.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Jones TS, Yang W, Evans WE, Relling MV. Using HapMap tools in pharmacogenomic discovery: the thiopurine methyltransferase polymorphism. Clin Pharmacol Ther. 2007;81:729–734. doi: 10.1038/sj.clpt.6100135. [DOI] [PubMed] [Google Scholar]