Abstract
We examined the analysis of nucleotides and nucleotide sugars by chromatography on porous graphitic carbon with mass spectrometric detection, a method that evades contamination of the MS instrument with ion pairing reagent. At first, adenosine triphosphate (ATP) and other triphosphate nucleotides exhibited very poor chromatographic behavior on new columns and could hardly be eluted from columns previously cleaned with trifluoroacetic acid. Satisfactory performance of both new and older columns could, however, be achieved by treatment with reducing agent and, unexpectedly, hydrochloric acid. Over 40 nucleotides could be detected in cell extracts including many isobaric compounds such as ATP, deoxyguanosine diphosphate (dGTP), and phospho-adenosine-5′-phosphosulfate or 3′,5′-cyclic adenosine 5'-monophosphate (AMP) and its much more abundant isomer 2′,3′-cylic AMP. A fast sample preparation procedure based on solid-phase extraction on carbon allowed detection of very short-lived analytes such as cytidine 5'-monophosphate (CMP)-2-keto-deoxy-octulosonic acid. In animal cells and plant tissues, about 35 nucleotide sugars were detected, among them rarely considered metabolites such as uridine 5'-diphosphate (UDP)-l-arabinopyranose, UDP-l-arabinofuranose, guanosine 5'-diphosphate (GDP)-l-galactofuranose, UDP-l-rhamnose, and adenosine diphosphate (ADP)-sugars. Surprisingly, UDP-arabinopyranose was also found in Chinese hamster ovary (CHO) cells. Due to the unique structural selectivity of graphitic carbon, the method described herein distinguishes more nucleotides and nucleotide sugars than previously reported approaches.
Formation of glycosidic linkages and, hence, the synthesis of protein- and lipid-linked oligosaccharides (including even glycogen) requires the availability of the relevant nucleotide sugars. These are initially synthesized from monosaccharides and nucleoside triphosphates with subsequent sugar interconversions.(1) Thus, nucleotides and nucleotide sugars are close neighbors on the metabolic flux map. The physicochemical properties of nucleotides and nucleotide sugars are similar, and therefore, analytical methods usually measure both classes of metabolites in parallel. Mass spectrometric detection is increasingly gaining importance and supplanting the traditional UV detection due to its superior selectivity. Some researchers obtained good results with capillary electrophoretic methods with MS(2) or UV detection,(3) but in general, chromatographic methods are more widely applied. Ion exchange chromatography, though frequently used for nucleotide sugar and nucleotide analysis,4−6 appears refractory to electrospray ionization due to the necessary high salt concentrations. Application of a pH instead of a salt gradient may, however, allows mass spectrometric detection.7,8 Hydrophilic interaction chromatography bears great promise for the analysis of hydrophilic metabolites including nucleotides,9−11 but separation of isobaric nucleotide sugars is hardly realized on, e.g., zwitterionic stationary phases.(11) A widely used approach for charged compounds is ion-pair (IP) reversed-phase (RP) chromatography, which, in conjunction with UV detection, has been successfully applied to nucleotide sugars.12−14 Volatile ion-pairing reagents are applied in IP-HPLC-MS combinations for nucleotide analysis,15−21 but only to a limited extent for nucleotide sugars.(18) Because of the “sticky” nature of the ion-pairing reagents, occasional use of this method on a multipurpose instrument is undesirable.
Porous graphitic carbon (PGC) has recently been applied to nucleotides.22−24 No ion-pairing reagent is required to achieve proper retention of nucleotides on this stationary phase even though diethylamine was added in a recent study focusing on nucleotide triphosphates.(25) In our experience, electrical grounding and a certain ionic strength have proven essential for the elution of nucleotide triphosphates.(26) Carbon columns have been found to be affected by redox reagents such as hydrogen peroxide or sulfite,27,28 which lead to retention time shifts of oligosaccharides.(29) It has not yet been studied how these subtleties of the carbon surface chemistry affect nucleotides.
The carbohydrate anabolism of green plants receives attention in diverse areas such as plant cell wall research30,31 or production of pharmaceutical glycoproteins in plants.(32) The nucleotide sugar repertoire of plants comprises activitated l-rhamnose and l-arabinose. Despite the enormous significance of arabinogalactan-proteins for plant cell wall structure and growth,(33) the biosynthesis of arabinans still remains largely unknown. Only recently, uridine 5'-diphosphate (UDP)-arabinofuranose has been identified as the likely sugar donor for arabinosyltranferase(s).(34) UDP-l-Araf is formed from UDP-l-Arap by the action of UPD-arabinose mutase.(35) As the equilibrium of this conversion lies on the side of the pyranose and as plant cells contain large amounts of the isobaric UDP-xylose, the determination of UDP-Araf poses a considerable analytical challenge.
Plant cell walls contain some l-galactopyranose residues in rhamnogalacturonan II of cell wall pectin and occasionally in xyloglucans.36,37 These stem from guanosine 5'-diphosphate (GDP)-l-Gal, which together with GDP-l-gulose is also a vital component of the plant-exclusive “Smirnoff-Wheeler” pathway for ascorbic acid synthesis.(38) While plants are largely considered to lack sialic acid(39) and, hence, cytidine 5'-monophosphate (CMP)-sialic acid, they must contain CMP-keto-octulosonic-acid (CMP-KDO), to allow the incorporation of this keto sugar into cell wall polysaccharides.(40) KDO is well-known as a component of the inner core of gram-negative bacterias′ lipopolysaccharides,(41) and hence, e.g., Escherichia coli can be expected to contain CMP-KDO.
For mammalian specimens, the interest in nucleotide sugars arises among other reasons from the influence of protein glycosylation on biological activity, in vivo half-life, and immunogenicity of recombinant protein therapeutics produced in mammalian cell cultures.12,42 Determination of cellular nucleotide levels should not be blemished by coeluting isobaric compounds such as 3′-phosphoadenosine-5′-phosphosulfate (PAPS), which is isobaric with adenosine triphosphate (ATP) to within 0.01 Da. PAPS is the donor substrate for the sulfation of carbohydrates, proteins, and a host of other molecules,(43) but it has received little if any attention in recent papers on nucleotide analysis.
Our recent experience with the analysis of CMP-sialic acid using PGC(32) motivated us to investigate the analysis of other nucleotide sugars and nucleotides by PGC-liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS). The method was applied to nucleotides and activated sugars of animal cells and of green plants, which harbor an especially challenging mixture of nucleotide sugars, e.g., the only recently noticed multitude of UDP-pentoses. To this end, we had to succeed in the “taming of the shrew”, i.e., in achieving proper chromatographic performance of the PGC columns used.
Methods
(See also Supporting Information.)
Metabolite Extraction and Purification
In accordance with recently published experiences,(44) Chinese hamster ovary (CHO) cells (10(6) cells) were quenched by addition of an equal volume of ice-cold phosphate buffered saline to the cell suspension and then centrifuged at 200g for 5 min at 4 °C. No additional washing step was carried out to keep processing time of intact cells to an absolute minimum. Cells were lysed by adding 1 mL of ice-cold 80 mM sodium fluoride. After approximately 10 s of incubation time, 250 μL of this cell lysate was applied to a 25 mg Hypercarb solid-phase extraction (SPE) cartridge (Thermo Scientific) preconditioned by washing with 60% acetonitrile in formate buffer (0.3% formic acid adjusted to pH 9.0 with ammonia) and finally water. After sample application, the cartridges were washed with 1 mL of water followed by 0.3 mL of 60% acetonitrile in water. Phosphorylated analytes were eluted using 60% acetonitrile in said formate buffer. The eluate was dried in a speed-vac concentrator without heating. Samples were taken up in water prior to analysis and 10% aliquots were subjected to LC-MS. The procedure for plant tissues and alternative methods are described in the Supporting Information.
LC-ESI-MS Analysis
Three PGC columns were employed: Column C1 had been in use for about 5 years and subject to numerous acid treatments. Column C3 had experienced two rounds of trifluoroacetic acid (TFA)/HCl treatment, and column C4 had solely been subjected to rinsing with sulfite.
The Hypercarb PGC columns (0.32 × 100 mm, GE Healthcare) were mounted using polyetheretherketone (PEEK) capillaries of 25 μm inner diameter (i.d.) and endowed with grounding clamps. Starting buffer was 0.3% formic acid brought to pH 9.0 with ammonia. The run started with a 20 min gradient from 2 to 15% acetonitrile at a flow rate of 8 μL/min with an Ultimate 3000 (Dionex). Then, following a 1 min ramp, dinucleotides were eluted with 50% acetonitrile for 5 min. Detection was performed by negative mode ESI-MS on a Q-TOF Ultima (Waters-Micromass) with a capillary voltage of −2.7 kV, a cone voltage of 80 V, cone and desolvation gas flows of 75 and 150 L/h, and source and cone temperatures of 100 and 120 °C, respectively. MS/MS experiments were conducted with 20% dissociation energy and argon.
Evaluation of the data sets was performed by means of MassMap, version 2010-06-13 (MassMap GmbH & Co. KG, Wolfratshausen, Germany).
Results
Solid-Phase Extraction of Nucleotides and Nucleotide Sugars Using Carbon Cartridges
While the current work focused on the chromatographic side of the analytical task, it was equally important to optimize the complete sequence of steps from the living cell to the final result. Therefore, we compared two quenching procedures, the methanol/ammonium bicarbonate method(45) and rapid cooling by addition of ice-cold buffer. The latter procedure consistently yielded higher ATP/adenosine diphosphate (ADP) ratios and was, therefore, applied in all further experiments of this study.
Several procedures have been developed for cell lysis and extraction of analytes. Many use perchloric acid (PCA) to precipitate proteins and, hence, inactivate phosphatases,12,46 while others use boiling aqueous ethanol for the same purpose.(19) In their study on nucleotide sugars in plant cells, Fry et al. employed potassium fluoride as a phosphatase inhibitor and chloroform/methanol/water two-phase partition to purify the nucleotide sugars.(47) We adopted the addition of hypotonic fluoride solution, which also served to lyse cells, but substituted the other separation steps by solid-phase extraction on carbon. Thus, the total handling time amounted to less than a minute for animal cells. This time increased to 10 min for plant tissues, which require mechanical disruption. An intermediate flush of the cartridge with salt-free water/acetonitrile removed large amounts of phosphate-free contaminants, a step similar to a procedure described earlier, in which, however, triethylamine buffer was used as “ion pair” reagent.13,14
Comparison of this fast and labor saving procedure with PCA or chloroform extraction revealed the NaF-carbon SPE method as superior, as it delivered a higher extraction yield than the chloroform extraction method and less ATP hydrolysis than the PCA method (data not shown). As the carbon SPE step integrates well with the following chromatography on PGC, we refrained from testing any other of the various extraction and purification procedures described.
Chromatography on Porous Graphitic Carbon (PGC) and Column Conditioning
Our recent work on the influence of ionic strength and pH for chromatography on carbon(26) demonstrated that carbon columns could be suitable for nucleotides and related substances. In order to achieve optimal retention and separation of nucleotide sugars and nucleotides, the formate buffer system used recently for oligosaccharides was now adapted for nucleotides. A 0.3% formate solution buffered to pH 9.0 with ammonia promoted elution of the strongly binding compounds ATP, guanosine 5'-triphosphate (GTP), nicotinamide adenine dinucleotide (NAD+), and nicotinamide adenine dinucleotide phosphate (NADP+) as sharp peaks (Figure 1B,E). Lower pH values as well as lower ionic strength led to inferior chromatographic results (data not shown).
Figure 1.
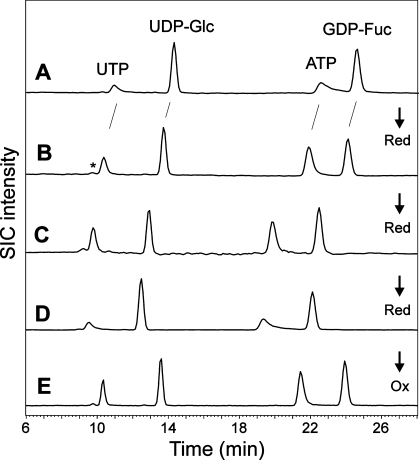
Effect of reduction and oxidation on a new column. (Panel A) A mixture of commercial analytes was subjected to chromatography on a newly purchased PGC column. Note the very broad and tailing peaks for UTP and ATP. (Panels B, C, and D) Chromatography of the same mixture after the column had been rinsed with 100 M sodium thiosulfate for 6 h, an additional 12 h, and an additional 24 h, respectively. (Panel E) Performance of the over-reduced column after 2 h of flushing with hydrogen peroxide. Detection was performed by negative mode ESI-MS. Selected ion chromatograms (SICs) for the annotated compounds are shown. The arrows indicate reductive (Red) or oxidative (Ox) states of the column.
However, regrettably, retention times varied by minutes when the column was used at different times. Even worse, some compounds, especially many triphosphates such as ATP, GTP, deoxythymidin diphosphate (dTTP), deoxyadenosine triphosphate (dATP) and deoxyguanosine triphosphate (dGTP), exhibited unacceptably strong peak tailing, resulting in near to complete loss of detectability. Notably, this occurred especially when new carbon columns had undergone cleaning by boiling in 4 M TFA, a procedure that reduces excessive back pressure. With a very new column, this procedure even led to total disappearance of all nucleotides from a standard mixture (Figure 2A). One essential hint at a solution of this problem came from work on the influence of oxidation and reduction of carbon on its chromatographic properties.27,28 Indeed, treatment of the new carbon column with sodium sulfite resulted in a decrease of retention, improved peak shapes, and stable retention times for nucleotide sugars and nucleotide triphosphates, notably for ATP, GTP, and other purine triphosphates, which are especially prone to peak distortion(24) (Figure 1B,C). On the contrary, hydrogen peroxide led to stronger retention of all analytes and to tailing of, e.g., ATP. Thus, a totally new column must be conditioned by rinsing with 100 mM sodium sulfite for several hours at room temperature in order to obtain an appropriate elution behavior with a tailing factor for ATP of less than 1.3. Noteworthy, extended treatment with reducing agent also resulted in strong peak tailing of purine trinucleotides (Figure 1D). Such reduction beyond the optimum can be reverted by a 30 min flush with hydrogen peroxide (Figure 1E).
Figure 2.
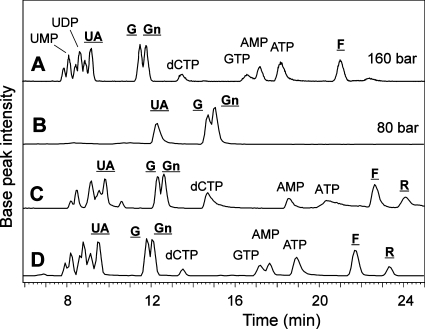
Regeneration of a PGC column by acid treatment. (A) A standard mixture on a rather new column after about 100 injections of diverse extracts. The pressure had risen from 80 bar to about 160 bar. After this rise to about twice the original pressure, columns often were completely clogged within a few injections..(B) The same mixture after boiling the column in 4 M TFA followed by rinsing with sulfite. (C) The same mixture again on the same column after 7 days and several reductive and oxidative treatments. The original separation characteristics had still not be retained. (D) Analysis on the same column after an additional HCl treatment. Underlined labels indicate nucleotide sugars, whereby UA, G, Gn, F, and R stand for UDP-glucuronic acid, UDP-Glc, UDP-GlcNAc, GDP-Fuc, and ADP-Rib.
When clogged carbon columns had undergone cleaning by boiling in 4 M TFA, a procedure that reduces excessive back pressure and re-establishes strong retention of oligosaccharides, it turned out that this had almost the same effect as application of hydrogen peroxide. Brand new columns showed the strongest effect, where almost all nucleotides became invisible (Figure 1A). A comparable increase of retention and improvement of peak shape is, however, also achieved by simply keeping the column in the slightly basic formate buffer for several hours.
For new columns, regeneration by TFA treatment had a drastic effect on column performance that could not be fully reverted even by very long reduction and rinsing periods (Figure 2). Therefore, to regain elution of ATP, etc. after TFA treatment, an additional washing step with HCl was evaluated and was shown to restore proper chromatographic results on the regenerated column (Figure 2).
In summary, our proposed column conditioning scheme for a new column consists in a reduction step only, whereas for used columns it comprises washing with TFA followed by HCl (dispensable for an older column) and finally a 2 h rinse with reducing agent. Variations of this scheme may likewise achieve the same goal. The drastic acid regeneration procedure can be employed when the column exerts excessive back pressure or shows reduced binding affinity after a hundred plus injections of cell extracts. Over time, PGC columns experience a slow loss of affinity. As this decrease of retention times affects all compounds similarly, relative retention times remain essentially constant (Supporting Information, Figure S1).
Analysis of Nucleotide Phosphates with a PGC Column
The next step was to assign the peaks occurring in biological samples (CHO cells, plant tissues, or mouse liver). Peak identity was verified with the help of commercial reference compounds and MS/MS experiments, which were of great value for the discrimination of isobaric constitutional isomers such as ATP, dGTP, PAPS, and 2′-PAPS (iso-PAPS; Table 1, Figure 3). MS/MS, however, cannot define the position of phosphate groups and, thus, the assignment of some metabolites, for which no references were available, remains tentative. This also applies for cyclic adenosine 5'-monophosphate (AMP). In addition to the well-known second messenger 3′,5′-cAMP, which occurs in traces only, a more abundant isomer was found in, e.g., tobacco. It is presumed to represent 2′,3′-cAMP, a product of mRNA degradation (Table 1).(48)
Table 1. List of Identified and Unidentified Analytes in Extracts of Tobacco, A. thaliana, CHO-cells, E. coli, or Commercial Standardsa.
| nucleotide | RRT | nucleotide sugar | RRT |
|---|---|---|---|
| CMP | 0.70 | UDP-GalUA | 0.79 |
| iso-CMP | 1.24 | UDP-GlcUA | 0.81 |
| CDP | 0.76 | UDP-D-Gal | 0.94 |
| CTP | 0.76 | UDP-D-Glc | 1.00 |
| UMP | 0.72 | UDP-Hex-3 | 0.85 |
| iso UMP | 1.24 | UDP-GalNAc | 1.01 |
| iso UMP | 1.40 | UDP-GlcNAc | 1.02 |
| UDP | 0.77 | UDP-HexNAc-3 | 0.86 |
| UTP | 0.80 | UDP-HexNAc-4 | 0.96 |
| dCMP | 0.83 | UDP-Rha | 0.90 |
| dCDP | 0.64 | UDP-S-Quin | 0.98 |
| dCTP | 0.81 | UDP-kd-Glc | 1.10 |
| dTMP | 1.20 | UDP-L-Arap | 0.87 |
| dTDP | 1.18 | UDP-L-Xyl | 1.00 |
| dTTP | 1.16 | UDP-L-Araf | 1.07 |
| GMP | 1.35 | UDP-Pen-4 | 0.93 |
| iso GMP | 1.92 | UDP-Pen-5 | 1.14 |
| iso GMP | 2.19 | UDP-Ara4FN | 0.96 |
| GDP | 1.40 | GDP-D-Man | 1.54 |
| GTP | 1.41 | GDP-L-Gal | 1.64 |
| dGMP | 1.78 | GDP-L-Gul | 1.67 |
| dGDP | 1.81 | GDP-Glc | 1.71 |
| dGTP | 1.78 | GDP-L-Fuc | 1.73 |
| AMP | 1.44 | ADP-D-Rib-P | 1.28 |
| iso AMP | 2.10 | ADP-D-Rib | 1.80 |
| iso AMP | 3.01 | ADP-D-Glc | 1.83 |
| ADP | 1.51 | ADP-Hex-2 | 1.61 |
| 3′,5′-ADP | 0.81 | ADP-Hex-3 | 1.68 |
| 2′,5′-ADP | 1.01 | ADP-gm-Hep | 1.67 |
| ATP | 1.51 | CMP-Neu5Ac | 0.89 |
| dAMP | 1.80 | CMP-Neu5Gc | 0.89 |
| dADP | 1.87 | CMP-KDO | 0.84 |
| dATP | 1.83 | dTDP-L-Rha | 1.42 |
| PAPS | 1.16 | dTDP-dhex-2 | 1.53 |
| 2′-PAPS | 1.16 | dTDP-Fuc4NAc | 1.62 |
| APS | 1.85 | ||
| 2′,3′-cAMP | 2.60 | ||
| 3′,5′-cAMP | 2.65 | ||
| NAD+ | 2.63 | ||
| NADH | 2.35 | ||
| NADP+ | 2.55 |
Clearly assigned compounds are printed in bold. The others were either not or only tentatively assigned. Gul = gulose; S-Quin = sulfoquinovose; kd-Glc = keto-deoxy-glucose; Ara4FN = 4-deoxy-4-formamido-l-arabinose; gm-Hep = glycero-manno-heptose; Fuc4Nac = N-acetyl-fucosamine. Quantitative information is given in the Supporting Table 1, Supporting Information.
Figure 3.
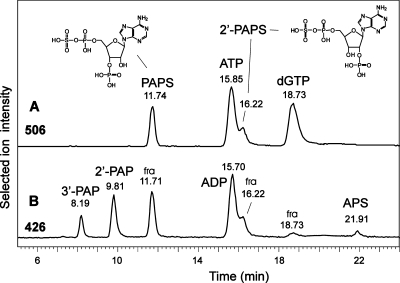
Analytes isobaric to ATP. The traces in panels A and B show the result obtained with a tobacco extract spiked with PAPS and 2′-PAPS. Panel A is the SIC for ATP (505.96−506.02 amu) accompanied by dGTP and the almost isobaric PAPS isomers. Panel B depicts the SIC for ADP (425.98-426.94) with unintended MS fragments (labeled “fra”) of PAPS, 2′-PAPS, and to a lesser degree dGTP. In addition, degradation products present in the PAPS and 2′-PAPS standards tentatively identify the elution position of 3′-phosphoadenosine-5′-phosphate (3′-PAP) and 2′-PAP as well as adenosine-5′-phosphosulfate (APS).
In contrast to ion exchange or ion-pairing separations, the number of phosphate residues on the 5′-position of ribose has little influence on the separation of nucleotides on carbon. Mono-, di-, and triphosphates of a certain nucleoside elute in close proximity. Thus, ADP and ATP usually overlap with the exact sequence of elution depending on the age and pretreatment of the column. ADP elutes in the front of the ATP peak on new columns and in the declining peak on older columns indicating that there is some, albeit small, ion exchange mechanism in effect.
In addition to ATP and dGTP, a small peak for yet another isobaric substance was observed about 3.5 min ahead of ATP in both plant extracts and especially in mouse liver (data not shown). This compound showed a somewhat stronger tendency for fragmentation than ATP and a mass possibly smaller by 0.01 Da, which is at the edge of our instrument’s accuracy. Both features pointed at the presence of sulfate, and hence, we purchased the two available candidate compounds 3′-phosphoadenosine-5′-phosphosulfates (PAPS) and the synthetic analogue 2′-phosphoadenosine-5′-phosphosulfates (2′-PAPS). Remarkably, these compounds eluted far apart from each other, as did the products of partial degradation (Figure 3). The standards also contained degradation products, two unique to either 2′- or 3′-PAPS and one smaller peak occurring in both preparations. This situation promoted the tentative assignment of the elution positions of 2′-PAP, 3′-PAP, and adenosine 5′-phosphosulfate (APS). The cell extracts contained peaks for desulfation products of both PAPS isomers, which raises the question whether 2′-PAPS might be a naturally occurring isomer. APS was not found.
Except for GMP, all nucleotide monophosphates were found as isomer mixtures (Table 1). Adenosine 2′-phosphate (2′-AMP) and adenosine 3′-phosphate (3′-AMP) can result from hydrolysis of adenosine 2′,3′-monophosphate (2′,3′-cAMP) in the course of RNA degradation,48,49 and this may also apply for CMP and UMP isomers.
NAD+ and NADP+ were found in all biological samples studied. The reduced forms NADH and NADPH eluted somewhat earlier. Both NADH and NADPH were found in some extracts. To our surprise, both the commercial standards as well as the natural samples exhibited several peaks for these substances. Given this and their redox sensitivity, their quantitative analysis requires a closer examination, which goes beyond the scope of this manuscript. A large peak for a compound with all characteristics of nicotinic acid adenine dinucleotide (NAAD), the precursor of NAD+ biosynthesis, was observed in all samples (Table 1). Taken together, some 30 different nucleotides, many of them isomers, could be separately detected by PGC-LC-MS in one or more of the four samples considered.
Analysis of Nucleotide Sugars with PGC-LC
The PGC-LC system exhibited excellent selectivity for nucleotide sugars, as can be seen from the analysis of wild-type tobacco leaves and selected standards (Supporting Information, Figure S1). The assignment of the peaks for UDP-Glc, UDP-Gal, UDP-GlcUA, UDP-Xylp, UDP-Arap, GDP-Man, and GDP-Fuc was based on commercial reference compounds as well as MS/MS. The identification of yet other compounds is described below. UDP-GalNAc and UDP-GlcNAc coelute on PGC, even though double peaks were obtained on the new column. These UDP-hexosamines are also difficult to separate by ion exchange HPLC,4,6 and they appear to coelute on a HILIC column.(11) They could, however, be separated by IP-RP with UV-detection.12,13
In extracts of mammalian cells, CMP-activated sialic acid was found (Supporting Information, Figure S2). Mouse liver contained a compound eluting at almost the same time as CMP-Neu5Ac but was larger by 16 Da. This additional mass resided in the sugar moiety, and thus, the compound could be assigned as CMP-Neu5Gc. The same elution time was found for CMP-Neu5Gc generated with the help of recombinant human CMP-sialic acid synthase (C. Jin, unpublished results). Some CMP-Neu5Gc was also seen in CHO extracts (Supporting Information, Figure S2).
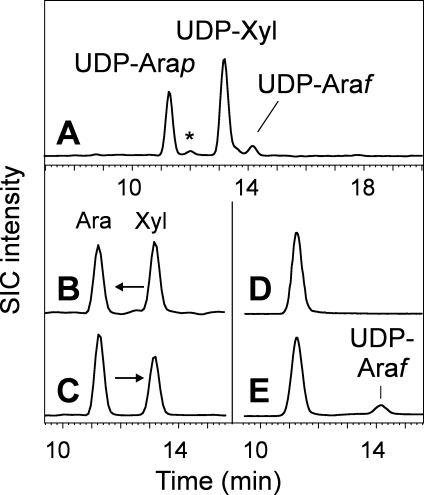
Plant tissues, as expected, contained UDP-xylose and UDP-arabinose (mainly UDP-Arap) (Figure 4). CHO cells likewise contained some UDP-xylose and, surprisingly, also UDP-Arap or at least a substance indistinguishable by chromatography and MS/MS (data not shown). GDP-l-galactose and other special cases are considered in the Supporting Information.
Figure 4.
UDP-pentoses in plants. Panel A shows the SIC for UDP-pentoses (536.00-526.08) in tobacco leaves. The peak labeled * is an unknown UDP-pentose. Panels B and C show the results of testing UDP-xylose epimerase activity in microsomes prepared from mung bean sprouts with either UDP-Xyl (UX in panel B) or UDP-Arap (UAp in panel C) as the substrate. Panels D and E exemplify the results obtained when UDP-arabinopyranose was incubated without (panel D) or with (panel E) recombinant UDP-arabinose mutase. Both enzyme reactions were conducted such that they essentially reached equilibrium.
Quantitation and Method Validation
Quantitation of individual compounds is usually performed by MS/MS.3,12,16,17,23 For this explorative work, we wanted to be able to revisit the data on the search for additional metabolites and unexpected isobaric compounds. Therefore, quantitation of peaks was performed on full scan MS data by means of the expert software MassMap. Settings for this program were chosen such that no real peak was missed. False identifications were prevented by the user assessment for each peak or by the retention times and mass errors of the compounds. Comparing selected results with manual integration indicated a correct depiction of the peak intensities by MassMap even for peaks of widely differing intensity. Thus, MassMap was used for the quantitative evaluations (Supporting Information, Tables S1 and S2).
The focus of this work was on the separation of nucleotides and related substances on PGC columns and not its application to a specific biological system. Hence, we limited the method validation to the bare essentials. The relative molar response of selected substances (UDP-GlcNAc, AMP, ADP, ATP, UDP-Glc, GDP-Fuc, and UDP-GlcUA) was found to be similar to within ±20%. Using a capillary format, the lower limit of detection for all four compounds was about 25 fmol (dissolved in 5 μL = 5 nmol/L, as far as this figure makes sense in the context of a solid-phase extraction method). The upper limit of detection is dictated by the Q-TOF′s detector and the deterioration of peak shapes on the carbon column and lies at about 25 pmol on column. In the medium zone of this range, the relative standard deviation (RSD%) of peak volumes in replicate analyses (n = 3) ranged from 1 to 5% for the test compounds, which compares well with IP-RP or HILIC methods.11−13,16
The reproducibility of retention times was checked with three different columns and three injections each of a tobacco leaf extract (Supporting Information, Figure S1). Despite the differences in absolute retention times arising from column history, the relative retention times varied with a mean standard deviation of less than 1%.
A major aim in devising sample workup steps is to prevent degradation of labile sugar−phosphates and triphosphates. We verified the extent of degradation by addition of 13C-ATP to the cell suspensions prior to extraction. Two percent of the 13C-nucleotide appeared as ADP. This is hardly more than the original content of 13C-ADP in the 13C-ATP employed (1.8%). The possible interference by “in-source” fragments is an important point, as the peaks of AMP, ADP, and ATP may overlap. Using freshly HPLC-purified ATP, the ESI-induced decay of ATP to ADP amounted to about 1.3%. For precise determinations of ADP levels, this unintended fragmentation of ATP may have to be considered.
Discussion
About 70 different nucleotides or nucleotide sugars were detected by the LC-ESI-MS method present herein. The principle aptitude of PGC-ESI-MS for nucleotide analysis has been demonstrated earlier.22,24,26 However, these publications did not put emphasis on how control over the delicate character of PGC columns can be gained. The starting point of this work was the frustrating fact that despite measures such as column grounding and high ionic strength of the eluent, the use of columns at different stages of their working life resulted in a highly inconsistent performance regarding retention times and peak shapes up to the point where nucleotides were not eluted at all. An invaluable clue toward answering this problem came from a paper of the Törnkvist-Nyholm group, who studied the influence of reducing and oxidizing agents on the carbon surface.(28) This influence also applies to oligosaccharides.(29)
The increase of retention strength following acidic or oxidative treatment may be explained by the exposure or generation of charged groups.(27) The strong peak tailing obtained with new, with acid washed, or with oxidized columns and especially the fact that a similarly strong tailing occurred upon excessive reduction, is, however, not easily reconciled with a simple ion exchange mechanism, especially, as the eluent presents considerable ionic strength. At least under this condition, ion exchange appears not to be the dominant mechanism as indicated by the similar retention of mono-, di-, and triphosphates. Triphosphates were, however, more susceptible to peak tailing and even total peak vanishing as it was observed after TFA treatment especially of newer columns (Figure 2). This problem could be overcome only by repeated reduction and “soothing” sequences over many days or by a brief HCl treatment. We can only speculate whether these phenomena root in chemical modifications of the carbon phase itself, in modifications of noncarbon constituents, which can be slowly washed out of the column (which would explain the gradual decrease of retention), in the nature of the counterion (formate, chloride, TFA or peptides) neutralizing the few possibly present ionic groups, or in compounds otherwise adsorbed to the PGC surface. However, the column treatment steps proposed in this study permitted one to use brand new and moderately used as well as rather old columns for this especially demanding application. Unlike with other types of stationary phases, a very harsh and effective column cleaning procedure can be applied. While this will, over time, lead to some loss of binding affinity resulting in an essentially parallel forward shift of all peaks, the column will in return loose much of its adverse peculiarities and, thus, PGC columns, like red wine, become even better with age, which translates into many years of productive column life.
PGC separates nucleotides in a manner virtually orthogonal to IP-RP HPLC, as the nature of the base as well as the attached sugar influence retention much more than the number of phosphates. The linkage of phosphates, however, appears to matter for PGC as is exemplified by the occurrence of two distinct cAMP peaks representing the well-known messenger 3′,5′-cAMP and, most probably, the mRNA degradation product 2′,3′-cAMP.(48)
A number of nucleotide sugars could be found in A. thaliana or tobacco leaves that, to the best of the authors' knowledge, have not yet been directly determined. Similarly, CMP-Neu5Ac was hardly (and CMP-Neu5Gc not at all) considered in metabolomic analysis conducted with MS detection, whereas a few papers report on UV detection after anion exchange(6) or IP-RP chromatography.13,14
The specificity of MS detection obviates lengthy sample preparation procedures. This helps to preserve the levels of the very sensitive CMP nucleotides. Despite the extremely short half-life of CMP-KDO of about half an hour at neutral pH and room temperature, where CMP-KDO is 265 times less stable than CMP-Neu5Ac,(50) it could be found in an E. coli extract (Supporting Information, Figure S2). The levels of CMP-KDO in plants may be considerably lower, as we could never find it there. CMP-KDO has recently been determined by IP-RP-HPLC after enzymatic in vitro synthesis, where large amounts of a pure analyte accrue.(51)
In contrast to ion pairing reversed-phase HPLC, PGC does not require buffer components that are difficult to remove from the LC-MS system and tend to form unwanted adducts with other ionic analytes. This advantage counts most on an instrument used for varying purposes. With an electrically grounded column, with peek-tubing, and with proper conditioning, PGC columns constitute a highly useful and reproducible separation device.
Acknowledgments
This work was in part funded by the Austrian Science Fund (Grant Nos. P19172 and P20132). We are grateful to Pia Gattinger and Richard Strasser for allowing us to snip small pieces from their tobacco plants and to Thomas Dalik for various technical help.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Reiter W. D. Curr. Opin. Plant Biol. 2008, 11, 236–243. [DOI] [PubMed] [Google Scholar]
- Lehmann R.; Huber M.; Beck A.; Schindera T.; Rinkler T.; Houdali B.; Weigert C.; Haring H. U.; Voelter W.; Schleicher E. D. Electrophoresis 2000, 21, 3010–3015. [DOI] [PubMed] [Google Scholar]
- Feng H. T.; Wong N.; Wee S.; Lee M. M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2008, 870, 131–134. [DOI] [PubMed] [Google Scholar]
- Ritter J. B.; Genzel Y.; Reichl U. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2006, 843, 216–226. [DOI] [PubMed] [Google Scholar]
- Sweeney C.; Mackintosh D.; Mason R. M. Biochem. J. 1993, 290 (Pt 2), 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiya N.; Ailor E.; Lawrence S. M.; Betenbaugh M. J.; Lee Y. C. Anal. Biochem. 2001, 293, 129–137. [DOI] [PubMed] [Google Scholar]
- Jansen R. S.; Rosing H.; Schellens J. H.; Beijnen J. H. J. Chromatogr., A 2009, 1216, 3168–3174. [DOI] [PubMed] [Google Scholar]
- Veltkamp S. A.; Hillebrand M. J.; Rosing H.; Jansen R. S.; Wickremsinhe E. R.; Perkins E. J.; Schellens J. H.; Beijnen J. H. J. Mass Spectrom. 2006, 41, 1633–1642. [DOI] [PubMed] [Google Scholar]
- Antonio C.; Larson T.; Gilday A.; Graham I.; Bergstrom E.; Thomas-Oates J. Rapid Commun. Mass Spectrom. 2008, 22, 1399–1407. [DOI] [PubMed] [Google Scholar]
- Bajad S. U.; Lu W.; Kimball E. H.; Yuan J.; Peterson C.; Rabinowitz J. D. J. Chromatogr., A 2006, 1125, 76–88. [DOI] [PubMed] [Google Scholar]
- Preinerstorfer B.; Schiesel S.; Lammerhofer M.; Lindner W. J. Chromatogr., A 2010, 1217, 312–328. [DOI] [PubMed] [Google Scholar]
- Kochanowski N.; Blanchard F.; Cacan R.; Chirat F.; Guedon E.; Marc A.; Goergen J. L. Anal. Biochem. 2006, 348, 243–251. [DOI] [PubMed] [Google Scholar]
- Nakajima K.; Kitazume S.; Angata T.; Fujinawa R.; Ohtsubo K.; Miyoshi E.; Taniguchi N. Glycobiology 2010, 20, 865–871. [DOI] [PubMed] [Google Scholar]
- Rabina J.; Maki M.; Savilahti E. M.; Jarvinen N.; Penttila L.; Renkonen R. Glycoconjugate J. 2001, 18, 799–805. [DOI] [PubMed] [Google Scholar]
- Cai Z.; Qian T.; Yang M. S. Se Pu 2004, 22, 358–360. [PubMed] [Google Scholar]
- Cordell R. L.; Hill S. J.; Ortori C. A.; Barrett D. A. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2008, 871, 115–124. [DOI] [PubMed] [Google Scholar]
- Klawitter J.; Schmitz V.; Klawitter J.; Leibfritz D.; Christians U. Anal. Biochem. 2007, 365, 230–239. [DOI] [PubMed] [Google Scholar]
- Ramm M.; Wolfender J. L.; Queiroz E. F.; Hostettmann K.; Hamburger M. J. Chromatogr., A 2004, 1034, 139–148. [DOI] [PubMed] [Google Scholar]
- Seifar R. M.; Ras C.; van Dam J. C.; van Gulik W. M.; Heijnen J. J.; van Winden W. A. Anal. Biochem. 2009, 388, 213–219. [DOI] [PubMed] [Google Scholar]
- Turnock D. C.; Ferguson M. A. Eukaryotic Cell 2007, 6, 1450–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuytten R.; Lemiere F.; Dongen W. V.; Esmans E. L.; Slegers H. Rapid Commun. Mass Spectrom. 2002, 16, 1205–1215. [DOI] [PubMed] [Google Scholar]
- Agrofoglio L. A.; Bezy V.; Chaimbault P.; Delepee R.; Rhourri B.; Morin P. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 1523–1527. [DOI] [PubMed] [Google Scholar]
- Wang J.; Lin T.; Lai J.; Cai Z.; Yang M. S. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2009, 877, 2019–2024. [DOI] [PubMed] [Google Scholar]
- Xing J.; Apedo A.; Tymiak A.; Zhao N. Rapid Commun. Mass Spectrom. 2004, 18, 1599–1606. [DOI] [PubMed] [Google Scholar]
- Cohen S.; Megherbi M.; Jordheim L. P.; Lefebvre I.; Perigaud C.; Dumontet C.; Guitton J. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2009, 877, 3831–3840. [DOI] [PubMed] [Google Scholar]
- Pabst M.; Altmann F. Anal. Chem. 2008, 80, 7534–7542. [DOI] [PubMed] [Google Scholar]
- Shibukawa M.; Terashima H.; Nakajima H.; Saitoh K. Analyst 2004, 129, 623–628. [DOI] [PubMed] [Google Scholar]
- Törnkvist A.; Markides K. E.; Nyholm L. Analyst 2003, 128, 844–848. [Google Scholar]
- Melmer M.; Stangler T.; Premstaller A.; Lindner W. J. Chromatogr., A 2010, 1217, 6092–6096. [DOI] [PubMed] [Google Scholar]
- Pauly M.; Keegstra K. Plant J. 2008, 54, 559–568. [DOI] [PubMed] [Google Scholar]
- Rosti J.; Barton C. J.; Albrecht S.; Dupree P.; Pauly M.; Findlay K.; Roberts K.; Seifert G. J. Plant Cell 2007, 19, 1565–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A.; Pabst M.; Leonard R.; Veit C.; Altmann F.; Mach L.; Glossl J.; Strasser R.; Steinkellner H. Plant Physiol. 2008, 147, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G. J.; Roberts K. Annu. Rev. Plant Biol. 2007, 58, 137–161. [DOI] [PubMed] [Google Scholar]
- Konishi T.; Ono H.; Ohnishi-Kameyama M.; Kaneko S.; Ishii T. Plant Physiol. 2006, 141, 1098–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T.; Takeda T.; Miyazaki Y.; Ohnishi-Kameyama M.; Hayashi T.; O’Neill M. A.; Ishii T. Glycobiology 2007, 17, 345–354. [DOI] [PubMed] [Google Scholar]
- Hantus S.; Pauly M.; Darvill A. G.; Albersheim P.; York W. S. Carbohydr. Res. 1997, 304, 11–20. [DOI] [PubMed] [Google Scholar]
- Reuhs B. L.; Glenn J.; Stephens S. B.; Kim J. S.; Christie D. B.; Glushka J. G.; Zablackis E.; Albersheim P.; Darvill A. G.; O’Neill M. A. Planta 2004, 219, 147–157. [DOI] [PubMed] [Google Scholar]
- Wolucka B. A.; Van Montagu M. J. Biol. Chem. 2003, 278, 47483–47490. [DOI] [PubMed] [Google Scholar]
- Zeleny R.; Kolarich D.; Strasser R.; Altmann F. Planta 2006, 224, 222–227. [DOI] [PubMed] [Google Scholar]
- Vidal S.; Doco T.; Williams P.; Pellerin P.; York W. S.; O’Neill M. A.; Glushka J.; Darvill A. G.; Albersheim P. Carbohydr. Res. 2000, 326, 277–294. [DOI] [PubMed] [Google Scholar]
- Gronow S.; Xia G.; Brade H. Eur. J. Cell Biol. 2010, 89, 3–10. [DOI] [PubMed] [Google Scholar]
- Jefferis R. Trends Pharmacol. Sci. 2009, 30, 356–362. [DOI] [PubMed] [Google Scholar]
- Klaassen C. D.; Boles J. W. FASEB J. 1997, 11, 404–418. [DOI] [PubMed] [Google Scholar]
- Oldiges M.; Lutz S.; Pflug S.; Schroer K.; Stein N.; Wiendahl C. Appl. Microbiol. Biotechnol. 2007, 76, 495–511. [DOI] [PubMed] [Google Scholar]
- Sellick C. A.; Hansen R.; Maqsood A. R.; Dunn W. B.; Stephens G. M.; Goodacre R.; Dickson A. J. Anal. Chem. 2009, 81, 174–183. [DOI] [PubMed] [Google Scholar]
- Zur Nedden S.; Eason R.; Doney A. S.; Frenguelli B. G. Anal. Biochem. 2009, 388, 108–114. [DOI] [PubMed] [Google Scholar]
- Fry S. C.; Northcote D. H. Plant Physiol. 1983, 73, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.; Mi Z.; Stewart N. A.; Jackson E. K. J. Pharmacol. Exp. Ther. 2009, 328, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. E.; Venegas F. D.; Raines R. T. Biochemistry 1994, 33, 7408–7414. [DOI] [PubMed] [Google Scholar]
- Lin C. H.; Murray B. W.; Ollmann I. R.; Wong C. H. Biochemistry 1997, 36, 780–785. [DOI] [PubMed] [Google Scholar]
- Misaki R.; Kajiura H.; Fujii K.; Fujiyama K.; Seki T. J. Biosci. Bioeng. 2009, 108, 527–529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.