Abstract
All organisms incorporate post-transcriptional modifications into ribosomal RNA, influencing ribosome assembly and function in ways that are poorly understood. The most highly conserved modification is the dimethylation of two adenosines near the 3′ end of the small subunit rRNA. Lack of these methylations due to deficiency in the KsgA methyltransferase stimulates translational errors during both the initiation and elongation phases of protein synthesis and confers resistance to the antibiotic kasugamycin. Here, we present the X-ray crystal structure of the Thermus thermophilus 30S ribosomal subunit lacking these dimethylations. Our data indicate that the KsgA-directed methylations facilitate structural rearrangements in order to establish a functionally optimum subunit conformation during the final stages of ribosome assembly.
Keywords: ribosome, rRNA modification, RNA structure, Thermus, decoding, antibiotic resistance
INTRODUCTION
All bacteria contain post-transcriptional modifications of their rRNAs that are thought to fine tune ribosome assembly and function. Despite much effort exerted by a number of laboratories over many years, it remains to be determined precisely how rRNA modifications influence ribosome structure, and in turn, how such structural effects manifest themselves biologically. One of the most thoroughly examined rRNA modifications is the N6,N6-dimethylation of A1518 and A1519 (Escherichia coli numbering), in the GGAA tetraloop of 16S rRNA helix 45 (Fig. 1). Both the modification (Van Knippenberg et al. 1984; McCloskey and Rozenski 2005) and the sequence (Woese et al. 1990) of this loop appear to be universally conserved or nearly so, indicative of an important physiological role for this modified structure.
FIGURE 1.
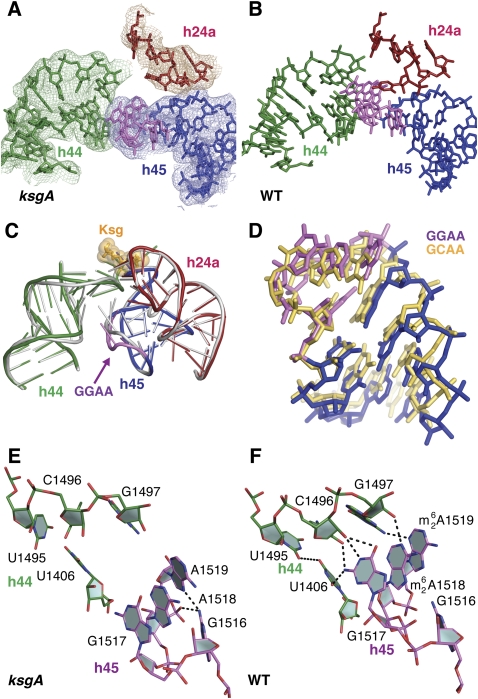
Position of the modified nucleotides A1518 and A1519 in the 30S ribosomal subunit. (A) Secondary structure of T. thermophilus 16S rRNA (Cannone et al. 2002), indicating positions and sequences of helices 6, 24a, 44, and 45 that show structural changes in the unmethylated 30S subunit. (B) Positions of helices 6, 24a, 44, and 45 in the crystal structure of the T. thermophilus 30S subunit (Wimberly et al. 2000). Ribosomal proteins in panel B are omitted for clarity.
Methylation of both bases is catalyzed by a single methyltransferase, KsgA, at a late stage of 30S subunit assembly (Thammana and Held 1974). Lack of methylation due to ksgA mutations confers resistance to the antibiotic kasugamycin (Helser et al. 1971), increases decoding errors during elongation (van Buul et al. 1984), and enhances initiation from non-AUG codons (O'Connor et al. 1997). This pleiotropic phenotype suggests a propagation of structural effects beyond helix 45 and into both the A and P sites of the ribosome.
KsgA and initiation factor 3 (IF3) compete for overlapping binding sites on the 30S subunit, preventing incompletely assembled 30S subunits from engaging in translation initiation prior to methylation by KsgA (Xu et al. 2008). Further, ksgA mutants exhibit cold-sensitive 30S subunit assembly and 16S rRNA processing defects (Connolly et al. 2008). These observations implicate KsgA methylation as a checkpoint for 30S subunit assembly. While a role for KsgA in 30S ribosomal subunit assembly is provided by this model, the structural basis for the effects of methylation deficiency on the subsequent events of protein synthesis have not been determined.
To define the role of these conserved modifications in ribosome structure and function, we determined the X-ray crystal structure of the 30S subunit from a ksgA deletion mutant of Thermus thermophilus HB8 to 3.7-Å resolution. Our structure indicates that methylation facilitates the formation of a packing interaction between helix 45 and helix 44 in the vicinity of the decoding site. Loss of this packing interaction perturbs surrounding rRNA structure in both the A and P sites of the ribosome, providing a structural basis for the pleiotropic phenotype of ksgA mutants. We conclude that the KsgA methyltransferase acts to establish an active conformation of the 30S ribosomal subunit that is optimized to participate in protein synthesis.
RESULTS AND DISCUSSION
Crystallization of 30S subunits from a ksgA mutant
We previously described a mutant of T. thermophilus HB8 in which the ksgA gene had been deleted and replaced with a kanamycin-resistance gene (Demirci et al. 2009). 30S subunits from this mutant (hereafter referred to as “unmethylated” subunits) were prepared and crystallized as described by others (Wimberly et al. 2000; Clemons et al. 2001). Unmethylated 30S subunits produced tetragonal crystals in the same space group, P41212, that are obtained with fully methylated subunits from wild-type T. thermophilus, but crystallized slowly as small needles over a period of 1 yr, rather than in the 2 mo normally required for 30S subunits. Crystallization of unmethylated subunits was completely inhibited by kasugamycin, suggesting that T. thermophilus ksgA 30S subunits retain the ability to bind kasugamycin, as previously observed with an E. coli ksgA mutant (Schuwirth et al. 2006).
Slow crystallization suggests the existence of structural changes at sites of crystal contacts. In one such contact, 16S rRNA helix 6 (the so-called “spur”) binds to the P site of a symmetry-related subunit in a configuration that closely resembles the binding configuration of peptidyl-tRNA (Wimberly et al. 2000). Kasugamycin destabilizes fMet-tRNAfMet binding to the P site in 30S initiation complexes (Poldermans et al. 1979) and could inhibit crystallization by destabilizing the binding of helix 6 to the P site in an analogous manner. Thus, the crystallization behavior of the ksgA 30S subunit likely reflects physiologically relevant changes occurring in and around the ribosomal P site.
Effect of methylation on helix 45 loop conformation
Electron density in the vicinity of helix 45 was of sufficient quality to allow rRNA backbone phosphates and base positions to be clearly distinguished (Fig. 2A). Significant structural changes resulting from lack of methylation are seen in the loop of helix 45 (Fig. 2B,C, violet). This highly conserved GGAA loop is a member of the GNRA tetraloop family that exhibits a fold characterized by a sheared base pair between the first (G1516) and fourth (A1519) bases, with the second (G1517), third (A1518), and fourth (A1519) bases stacking on the 3′ side of the loop (Heus and Pardi 1991; Jucker et al. 1996). The unmethylated tetraloop in the context of the 30S subunit closely follows the canonical GNRA fold (Fig. 2D). However, in fully methylated (wild type) 30S subunits (Wimberly et al. 2000) the hydrogen bond between the N2 of G1516 and the N7 of A1519, seen in canonical GNRA tetraloops, is absent, and there is a widening of the loop (Fig. 2C, gray).
FIGURE 2.
Structural changes in the unmethylated 30S subunit. (A) Final σA-weighted 2mFo − DFc electron density in the ksgA mutant 30S subunit crystal structure. (Red) 16S rRNA helix 24a (h24a); (green) helix 44 (h44); (blue) helix 45 (h45); (violet) the GGAA tetraloop of helix 45. (B) Contacts between helix 45 and helices 24a and 44. Helices are colored as in A. (C) Bending of the GGAA tetraloop in the unmethylated subunit (violet) is visible. The wild-type 30S subunit structure (PDB entry 1J5E) (Wimberly et al. 2000) is superimposed (gray). Kasugamycin (Ksg, orange spheres) from PDB entry 1VS5 (Schuwirth et al. 2006) is superimposed. (D) Comparison of the helix 45 tetraloop with a GCAA tetraloop as observed in PDB entry 1ZIH (Jucker et al. 1996). (E,F) The ksgA mutation prevents the formation of a hydrogen-bonding network between helices 45 and 44. The helix 45–44 interface in the ksgA mutant (e.g., 30S subunits prior to methylation) (E) and in the wild-type, fully methylated 30S subunit (F). Panels E and F are from identical viewpoints.
These observations are consistent with previous studies demonstrating a thermodynamic destabilization of helix 45 by dimethylation of A1518 and A1519 (Thammana and Cantor 1978; Van Charldorp et al. 1981; Heus et al. 1990; Rife et al. 1998). While an additional methylation at the N2 of G1516 is found in helix 45 of some bacteria but is absent from T. thermophilus (Guymon et al. 2006), it does not alter loop thermal stability or interfere with hydrogen bonding (Rife et al. 1998).
Loss of helix packing interactions
In ribosomes from wild-type cells, there is extensive contact between the helix 45 tetraloop and helix 44, near the decoding site (Wimberly et al. 2000). This interaction is distinct from the GNRA tetraloop–receptor interaction observed in other RNA molecules (Cate et al. 1996). Lack of helix 45 methylation in ribosomes from the ksgA mutant eliminates all of these hydrogen-bonding interactions (Fig. 2E,F).
In the unmethylated 30S subunit, the absence of N6,N6-dimethylation generates a tighter loop structure, with A1519 positioned closer to G1516, establishing the N7–N2 hydrogen bond found in canonical GNRA tetraloops (Fig. 2C). The loop bends significantly, precluding the hydrogen-bonding interactions with helix 44 (Fig. 2C). The N6 atoms of A1518 and A1519 are moved by about 5 Å and 4 Å, respectively, and two hydrogen bonds, between the N1 of G1517 and the 2′-OH of C1496 and between the N2 of G1517 and the O2 of U1406 are absent as these atoms are 8 Å and 7 Å apart, respectively. Also, hydrogen bonds between the N1 of A1518 and the 2′-OH of G1497, and between N3 of A1519 and N2 of G1497, are absent (Fig. 2E,F). Thus, these data indicate that the initial structural role of N6,N6-dimethylation of A1518 and A1519 is to open up the G1516–A1519 base pair and widen the loop, and in so doing, facilitate the subsequent formation of a hydrogen-bonding network that stabilizes the packing interactions between helices 44 and 45.
Structural basis for the ksgA mutant phenotype
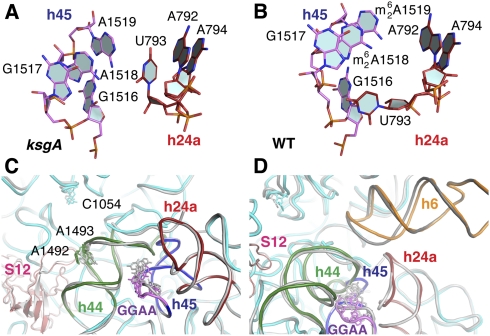
In the absence of this hydrogen-bonding network, small conformational differences are noted in the surrounding structure that may account for resistance to kasugamycin (Fig. 3A,B). In helix 24a, U793 is in the space normally occupied by the methylated adenosines, and the two adjacent bases, A792 and A794, show small shifts. These bases comprise part of the kasugamycin binding site (Schluenzen et al. 2006; Schuwirth et al. 2006), and their movement is consistent with the finding that a base substitution, A794G, confers kasugamycin resistance (Vila-Sanjurjo et al. 1999). Thus, as previously predicted (Schluenzen et al. 2006; Schuwirth et al. 2006), kasugamycin resistance due to lack of methylation most likely results from indirect effects on the conformation of helix 24a in the vicinity of bases directly involved in kasugamycin binding.
FIGURE 3.
Structural readjustments of helices adjacent to the unmethylated GGAA tetraloop. Movement of A793 in the kasugamycin-binding site in unmethylated (A) and fully methylated (Schuwirth et al. 2006) (B) 30S subunits. (C) Close-up view illustrating shifts of A1492, A1493, and C1054 in the decoding site of the unmethylated 30S (cyan) in comparison with the wild-type (gray) subunit (PDB 1J5E) (Wimberly et al. 2000), colored as in Figure 2B. (D) Close-up view illustrating the shift in position of 16S rRNA helix 6 of the unmethylated 30S subunit (orange) in the P site of an adjacent 30S subunit.
The absence of the helix 44–45 packing interaction also influences the position of A1492 and A1493, while a shift of up to 6 Å of the 30S subunit head causes a repositioning of C1054 (Fig. 3C). These three bases participate directly in codon recognition (Ogle et al. 2001), suggesting a basis for the error-prone phenotype of ksgA mutants. Lack of methylation might also influence decoding by perturbing the equilibrium of the global conformational change that occurs upon cognate codon recognition (Ogle et al. 2002). This involves the formation of contacts between ribosomal protein S12 and the side of helix 44 opposite the helix 45 packing site (Fig. 3C). If the loss of the helix 44–45 packing influences the dynamics of this contact, it could reduce the thermodynamic barrier to domain closure and increase incorporation of near-cognate aminoacyl-tRNAs in the A site. We also observed a positional shift of the helix 6 spur of an adjacent 30S subunit in the P site (Fig. 3D). To the extent that this crystal contact mimics P-site tRNA binding (Wimberly et al. 2000), this positional shift suggests that distortions in the P site may be the basis for the observed increase in initiation from non-AUG codons (O'Connor et al. 1997). This most likely is the result of changes in helix 24a.
Recognition of 30S subunits by the KsgA methyltransferase
Although the in vivo substrate for KsgA is not known, methylation occurs at a late stage of 30S subunit assembly (Thammana and Held 1974), subsequent to incorporation of almost the full complement of ribosomal proteins, but prior to the establishment of a final active 30S subunit conformation (Desai and Rife 2006). Ribosomal protein S21 is inhibitory to methylation by KsgA, although T. thermophilus lacks this protein (Henne et al. 2004). The conformation suitable for methylation can be mimicked by reducing the concentration of either magnesium (Demirci et al. 2009) or monovalent cation (Zamir et al. 1969; Desai and Rife 2006). The latter condition induces altered reactivity of 16S rRNA to base-specific chemical probes (Moazed et al. 1986) and also causes the dimethylated adenosines A1518 and A1519 to become accessible to antibodies (Thammana and Cantor 1978). Thus, KsgA would appear to recognize a nonnative conformation of the 30S subunit that can in some way be mimicked by conditions that perturb or destabilize ribosome structure. While the checkpoint model proposes that KsgA prevents 30S subunits from entering the translating pool prior to final activation (Xu et al. 2008), the structure described here suggests that methylation by KsgA plays an additional direct role in establishing a fully active 30S subunit conformation.
The importance of packing interactions between helices 44 and 45 has been suggested previously by mutagenesis experiments (Vila-Sanjurjo and Dahlberg 2001), but it is the structural data presented here that resolve the long-standing paradox as to why a GGAA tetraloop sequence is so highly conserved in helix 45 while its fold is incompatible with optimum ribosome function. Studies using oligoribonucleotide analogs of helix 45 indicate that methylation influences loop thermal stability (Van Charldorp et al. 1981; Heus et al. 1990; Rife et al. 1998) and fold (Rife and Moore 1998). Thermodynamically stable GNRA tetraloops serve as nucleating sites for RNA folding (Heus and Pardi 1991; Jucker et al. 1996), and this stabilization may be utilized to prevent misfolding of the terminal helix of 16S rRNA during transcription. Subsequent destabilization and distortion of the tetraloop by methylation would then facilitate the formation of a more stable and functional tertiary structure in the final stages of 30S subunit assembly.
MATERIALS AND METHODS
30S ribosomal subunits from the Thermus thermophilus HB8 ksgA deletion mutant (Demirci et al. 2009) were purified, crystallized, and cryoprotected essentially as described (Clemons et al. 2001). Diffraction data were collected from a single crystal at beamline ID-24-C at the Advanced Photon Source in Argonne, Illinois. Diffraction data were processed with the HKL2000 package (Table 1; Otwinowski and Minor 1997). Coordinates from the wild-type 30S subunit structure (PDB entry 1J5E) (Wimberly et al. 2000) were used for initial rigid-body refinement with PHENIX (Adams et al. 2002). After simulated-annealing refinement, individual coordinates, two group B factors per residue, and TLS parameters were refined against the 3.7-Å data set. Potential positions of magnesium or potassium ions were compared with those in a high-resolution 2.5-Å 30S subunit structure (PDB entry 2VQE) (Kurata et al. 2008) in Coot (Emsley and Cowtan 2004), and positions with strong difference density were retained. All figures were produced with PyMOL (DeLano 2002). Coordinates and structure factors have been deposited in the Protein Data Bank under accession number 3OTO.
TABLE 1.
Data collection and refinement statistics
ACKNOWLEDGMENTS
This work was supported by grants GM19756 and GM19756-37S1 from the National Institutes of Health to A.E.D. We thank Aiko and Kenji for numerous discussions; Jason Rife, Gloria Culver, Stephen Douthwaite, Jennifer Carr, Anton Vila-Sanjurjo, and James Dahlberg for critical reading of the manuscript; Michael Bender for helpful discussions; and Bruce Fitzgerald and Rick Cole for excellent technical assistance. The authors dedicate this paper to the memory of Warren DeLano and Peter Van Knippenberg.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2357210.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA 1996. RNA tertiary structure mediation by adenosine platforms. Science 273: 1696–1699 [DOI] [PubMed] [Google Scholar]
- Clemons WM Jr, Brodersen DE, McCutcheon JP, May JL, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V 2001. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Purification, crystallization and structure determination. J Mol Biol 310: 827–843 [DOI] [PubMed] [Google Scholar]
- Connolly K, Rife JP, Culver G 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- Demirci H, Belardinelli R, Seri E, Gregory ST, Gualerzi C, Dahlberg AE, Jogl G 2009. Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5′-methylthioadenosine. J Mol Biol 388: 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PM, Rife JP 2006. The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit. Arch Biochem Biophys 449: 57–63 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Guymon R, Pomerantz SC, Crain PF, McCloskey JA 2006. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: The complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry 45: 4888–4899 [DOI] [PubMed] [Google Scholar]
- Helser TL, Davies JE, Dahlberg JE 1971. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol 233: 12–14 [DOI] [PubMed] [Google Scholar]
- Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, et al. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22: 547–553 [DOI] [PubMed] [Google Scholar]
- Heus HA, Pardi A 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194 [DOI] [PubMed] [Google Scholar]
- Heus HA, Formenoy LJ, Van Knippenberg PH 1990. Conformational and thermodynamic effects of naturally occurring base methylations in a ribosomal RNA hairpin of Bacillus stearothermophilus. Eur J Biochem 188: 275–281 [DOI] [PubMed] [Google Scholar]
- Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J Mol Biol 264: 968–980 [DOI] [PubMed] [Google Scholar]
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T 2008. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U·G wobble pairing during decoding. J Biol Chem 283: 18801–18811 [DOI] [PubMed] [Google Scholar]
- McCloskey JA, Rozenski J 2005. The Small Subunit rRNA Modification Database. Nucleic Acids Res 33: D135–D138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Van Stolk BJ, Douthwaite S, Noller HF 1986. Interconversion of active and inactive 30S ribosomal subunits is accompanied by a conformational change in the decoding region of 16S rRNA. J Mol Biol 191: 483–493 [DOI] [PubMed] [Google Scholar]
- O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE 1997. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res 25: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Poldermans B, Goosen N, Van Knippenberg PH 1979. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16S ribosomal RNA of Escherichia coli. I. The effect of kasugamycin on initiation of protein synthesis. J Biol Chem 254: 9085–9089 [PubMed] [Google Scholar]
- Rife JP, Moore PB 1998. The structure of a methylated tetraloop in 16S ribosomal RNA. Structure 6: 747–756 [DOI] [PubMed] [Google Scholar]
- Rife JP, Cheng CS, Moore PB, Strobel SA 1998. N2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res 26: 3640–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, Nierhaus KH, et al. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13: 871–878 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JH, Vila-Sanjurjo A 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 13: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammana P, Cantor CR 1978. Studies on ribosome structure and interactions near the m62Am62A sequence. Nucleic Acids Res 5: 805–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammana P, Held WA 1974. Methylation of 16S rRNA during ribosome assembly in vitro. Nature 251: 682–686 [DOI] [PubMed] [Google Scholar]
- van Buul CP, Visser W, van Knippenberg PH 1984. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett 177: 119–124 [DOI] [PubMed] [Google Scholar]
- Van Charldorp R, Heus HA, Van Knippenberg PH, Joordens J, De Bruin SH, Hilbers CW 1981. Destabilization of secondary structure in 16S ribosomal RNA by dimethylation of two adjacent adenosines. Nucleic Acids Res 9: 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Knippenberg PH, Van Kimmenade JM, Heus HA 1984. Phylogeny of the conserved 3′ terminal stucture of the RNA of small ribosomal subunits. Nucleic Acids Res 12: 2595–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Sanjurjo A, Dahlberg AE 2001. Mutational analysis of the conserved bases C1402 and A1500 in the center of the decoding domain of Escherichia coli 16S rRNA reveals an important tertiary interaction. J Mol Biol 308: 457–463 [DOI] [PubMed] [Google Scholar]
- Vila-Sanjurjo A, Squires CL, Dahlberg AE 1999. Isolation of kasugamycin resistant mutants in the 16S ribosomal RNA of Escherichia coli. J Mol Biol 293: 1–8 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR 1990. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops.” Proc Natl Acad Sci 87: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, O'Farrell HC, Rife JP, Culver GM 2008. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol 15: 534–536 [DOI] [PubMed] [Google Scholar]
- Zamir A, Miskin R, Elson D 1969. Interconversion between inactive and active forms of ribosomal subunits. FEBS Lett 3: 85–88 [DOI] [PubMed] [Google Scholar]