Abstract
The plasticity of viral plus strand RNA genomes is fundamental for the multiple functions of these molecules. Local and long-range RNA–RNA interactions provide the scaffold for interacting proteins of the translation, replication, and encapsidation machinery. Using dengue virus as a model, we investigated the relevance of the interplay between two alternative conformations of the viral genome during replication. Flaviviruses require long-range RNA–RNA interactions and genome cyclization for RNA synthesis. Here, we define a sequence present in the viral 3′UTR that overlaps two mutually exclusive structures. This sequence can form an extended duplex by long-range 5′-3′ interactions in the circular conformation of the RNA or fold locally into a small hairpin (sHP) in the linear form of the genome. A mutational analysis of the sHP structure revealed an absolute requirement of this element for viral viability, suggesting the need of a linear conformation of the genome. Viral RNA replication showed high vulnerability to changes that alter the balance between circular and linear forms of the RNA. Mutations that shift the equilibrium toward the circular or the linear conformation of the genome spontaneously revert to sequences with different mutations that tend to restore the relative stability of the two competing structures. We propose a model in which the viral genome exists in at least two alternative conformations and the balance between these two states is critical for infectivity.
Keywords: flavivirus, dengue virus, viral RNA plasticity, long-range RNA–RNA interactions, viral riboswitches
INTRODUCTION
Plus strand RNA viruses are responsible of a plethora of plant, animal, and human illnesses. The plus strand genomes, besides encoding for viral proteins, contain a wide variety of RNA elements that regulate fundamental processes of viral life cycles. These elements function as promoters, enhancers, and repressors of viral translation, transcription, genome replication, and encapsidation (Andino et al. 1990; Song and Simon 1995; McKnight and Lemon 1998; Dreher 1999; Paul et al. 2000; Panavas and Nagy 2003; Pogany et al. 2003; Ranjith-Kumar et al. 2003; Alvarez et al. 2005a; Grdzelishvili et al. 2005; Mir and Panganiban 2005; Lodeiro et al. 2009; Wu et al. 2009). Long-range interactions in viral RNAs explain the remarkable flexibility for the location of these elements throughout the genomes (Kim and Hemenway 1999; Klovins and van Duin 1999; Guo et al. 2001, 2009; Lindenbach et al. 2002; Kim et al. 2003; Fabian and White 2004; Lin and White 2004; Alvarez et al. 2005b; Friebe et al. 2005; Hu et al. 2007; Diviney et al. 2008; You and Rice 2008; Wu et al. 2009). For instance, cis-acting elements required for translation initiation can be found at the 3′ end of the viral RNA, while elements necessary for initiation of RNA synthesis can be located at the 5′ end of the viral genome (Niesters and Strauss 1990; Wang et al. 1997; Gamarnik and Andino 1998; Frolov et al. 2001; Filomatori et al. 2006). In addition, overlapping functions in single RNA structures or overlapping sequences in mutually exclusive RNA elements are strategies to control the utilization of viral genomes (Gamarnik and Andino 2000; Huthoff and Berkhout 2001; D'Souza and Summers 2004; Goebel et al. 2004; Zhang et al. 2006; Zust et al. 2008; Yuan et al. 2009). The molecular bases of how viral RNAs undergo conformational changes during the viral life cycles are still largely unknown. Here, we use dengue virus (DENV) to investigate alternative conformations of the viral genome that are necessary for viral replication in the infected cell.
DENV belongs to the flavivirus genus in the Flaviviridae family and represents the most significant arthropod-borne virus pathogen in humans (WHO 2009). Flavivirus genomes are plus strand RNA molecules of 10–12 kb that share great similarities with respect to the cis-acting signals involved in viral replication (Lindenbach et al. 2007). A conserved feature of these genomes is the presence of inverted complementary sequences at the ends of the RNA that mediate long-range RNA–RNA interactions (Hahn et al. 1987; You and Padmanabhan 1999; Khromykh et al. 2001; Markoff 2003; Thurner et al. 2004; Alvarez et al. 2005b; Gritsun and Gould 2007; Villordo and Gamarnik 2009). Studies from many different laboratories using infectious clones and replicon systems provided compelling evidence for the essential role of genome cyclization during flavivirus replication (Khromykh et al. 2001; Corver et al. 2003; Lo et al. 2003; Alvarez et al. 2005b, 2008; Kofler et al. 2006). Molecular details of viral replication that explain the requirement of circular conformations of flavivirus genomes have been reported (Filomatori et al. 2006). Using DENV, the promoter for RNA replication was identified at the 5′ end of the genome. Genome cyclization was found to be crucial to reposition the promoter–RdRp complex near the 3′ end initiation site for RNA synthesis (Filomatori et al. 2006; Lodeiro et al. 2009). Although circular forms of the flavivirus genomes were proposed to be thermodynamically favored (Hahn et al. 1987; Khromykh et al. 2001; Thurner et al. 2004; Alvarez et al. 2005b; Gritsun and Gould 2007), it is likely that both linear and circular conformations of the RNA exist during viral replication.
To investigate the requirement of the linear conformation of the DENV genome in infected cells, we used a small hairpin (sHP) structure to monitor this RNA conformation. The sequence of the sHP is involved in two mutually exclusive structures. It can form an extended duplex through long-distance base pairing in the circular form of the genome or can fold locally into the sHP in the linear conformation of the RNA. Manipulation of infectious clones indicated that disruption of the stem of the sHP, without altering 5′-3′ complementarity, impairs viral replication. Spontaneous mutations restore both the sHP structure and the viral function. In addition, mutations that stabilize the linear or the circular conformation of the RNA resulted in spontaneous changes that brought the equilibrium near to that observed in the wild-type (WT) sequence. In this regard, second-site mutations were detected in which an increased stability of the sHP was compensated by an increase in the duplex stability, suggesting a genetic link between these two competing structures. Our results indicate that a balance between linear and circular conformations of the viral genome is crucial for DENV replication.
RESULTS AND DISCUSSION
The sHP structure is essential for DENV replication
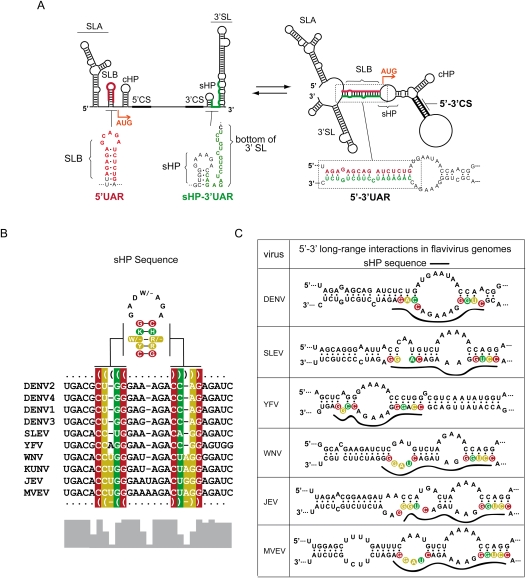
In mosquito-borne flaviviruses, at least two pairs of complementary regions are necessary for genome cyclization (Fig. 1A; for review, see Villordo and Gamarnik 2009). These sequences are known as 5′-3′CS and 5′-3′UAR, of 11 and 16 nucleotides (nt) long, respectively, in DENV. The 5′CS is located inside the ORF, encoding the N terminus of the capsid protein, and the 3′CS is located upstream of the highly conserved 3′SL. The 5′UAR is in the 5′UTR, just upstream of the translation initiator AUG, and the 3′UAR is located within the 3′SL at the 3′ end of the genome. We have recently reported that mutations within UAR that interfere with 5′-3′UAR complementarity impair DENV RNA synthesis, while reconstitution of base pairings with foreign sequences rescue the viral function (Alvarez et al. 2008). In the process of mapping UAR nucleotides involved in the long-range interaction, we found a lethal mutation in 3′UAR that was not rescued by the compensatory mutation at 5′UAR (mutant 7 in Alvarez et al. 2008). Instead, a revertant virus was obtained that restored both 5′-3′UAR complementarity and the structure of a sHP present at the 5′ end of the highly conserved 3′SL. The stem of the sHP overlaps with the 3′UAR sequence. Thus, we hypothesized that at least two alternative conformations of the viral genome (formation of the sHP and the 5′-3′ duplex) are functionally relevant during viral replication (Fig. 1A). In this study, we will refer to the linear form of the RNA as the state in which the ends of the genome are not hybridized, without excluding other possible RNA–RNA interactions along the coding sequence.
FIGURE 1.
Two mutually exclusive structures can be predicted in the DENV genome. (A) Schematic representation of linear and circular conformations of the DENV genome. On the left, the predicted linear conformation of the genome is represented, showing relevant cis-acting elements: the promoter stem–loop A (SLA), stem–loop B (SLB), the capsid region hairpin (cHP), the small hairpin (sHP), the cyclization sequences, the initiator AUG, and the 3′ stem–loop (3′SL). Underneath of this representation, the nucleotide sequences corresponding to the cyclization element 5′UAR and the sHP-3′UAR are shown. On the right is the predicted circular conformation of the genome, showing the hybridized cyclization sequences 5′-3′UAR and the 5′-3′CS. The nucleotide sequence of the 5′-3′UAR is also shown. Note that the predicted SLA and cHP are maintained while the SLB, sHP, and the bottom of the 3′SL change after 5′-3′ hybridization. (B) Multiple alignments showing the sHP sequence of mosquito-borne flaviviruses. Nucleotides in red, yellow, and green indicate the covariation positions with one, two, or three different base pairs, respectively. On the top, the consensus sHP secondary structure is shown. Nucleotides are represented with IUPAC codes: W (A/U), K (G/U), H (not G), D (not C), R (A/G), Y (C/U), or − (deleted). The bottom histogram shows the nucleotide conservation of the indicated region. (C) Schematic representation showing the predicted changes in the sHP structure of different flavivirus genomes upon 5′-3′ end hybridization. The nucleotides corresponding to the stems of the sHP are shown with the same color code indicated in B. In addition, the nucleotides corresponding to the sHP are shown with a solid line on the hybridized conformations.
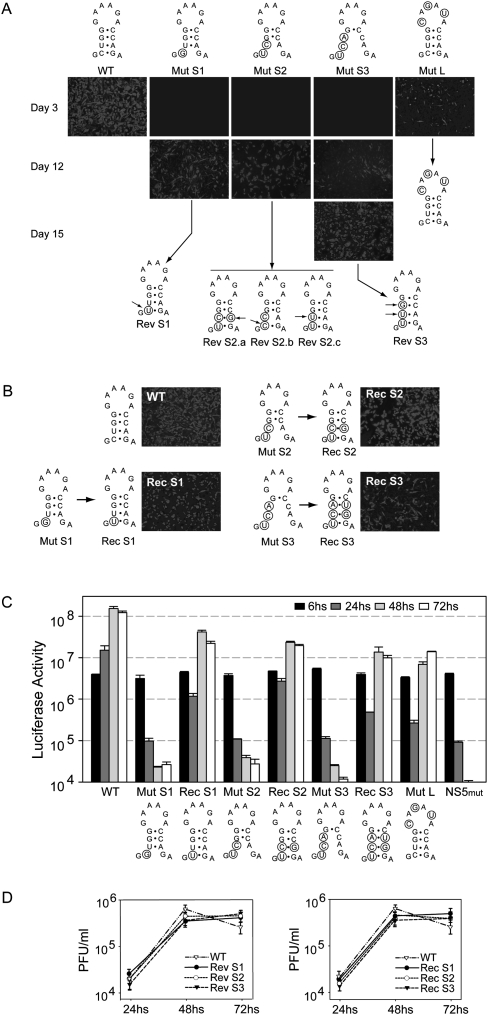
To investigate the relevance of alternative conformations of the viral RNA, we first analyzed the 3′ terminal ∼100 nt of different mosquito-borne flaviviruses using multiple sequence alignments with the RNAz software (Gruber et al. 2007). This study indicated that, in spite of the low nucleotide conservation, the sHP structure was predicted with high probability as a functional RNA element (Fig. 1B). The consensus sHP structure shows covariation in three positions of the stem. Further analysis indicated that, in all the sequences, the sHP region overlapped with nucleotides predicted to be involved in long-range RNA–RNA interactions (Fig. 1C). Previous studies using probing analysis of the 5′ and/or 3′ terminal regions of the flavivirus RNAs support these predictions (Shi et al. 1996; Dong et al. 2008; Polacek et al. 2009). Because the function of the sHP in flavivirus infection has not been previously addressed, the structure was subjected to mutational analysis in the context of DENV infectious RNAs. Substitutions were introduced in the sHP to disrupt gradually the stem and the sequence of the loop, without altering 5′-3′UAR complementarity. Viral RNAs carrying one, two, or three mismatches in the stem of the sHP were transfected into BHK cells (Fig. 2A, Mut S1, Mut S2, and Mut S3, respectively). Viral replication was assessed by indirect immunofluorescence (IF). Transfection of the WT RNA resulted in positive IF in the complete monolayer at day 3. After that time, an extensive cytopathic effect and cell death were observed. For Mut S1 and S2, positive IF in isolated cells was observed after day 6, and viral replication in the complete monolayer was observed at day 12. Replication of Mut S3 was further delayed, and viral replication in the complete monolayer was observed only at 15 d post-transfection (Fig. 2A). The sequence analysis of the replicating viruses showed spontaneous changes within the sHP structure (Fig. 2A). Revertants of Mut S1 and Mut S2 incorporated nucleotide changes in different positions that partially restored the sHP stem stability (Fig. 2A, Rev S1, Rev S2.a, Rev S2.b, and Rev S2.c). In the case of the revertant for Mut S3, two nucleotide changes were selected in the stem of the sHP (Rev S3). Substitutions in the loop of the sHP yielded viruses that replicated less efficiently than the WT but maintained the input RNA sequence (Fig. 2A, Mut L). These results provide the first evidence for the relevance of the sHP structure in DENV infection.
FIGURE 2.
Relevance of the small hairpin (sHP) in DENV replication. (A) Replication of DENV RNAs carrying substitutions within the sHP. Expression of DENV proteins in BHK cells transfected with full-length WT and mutant (Mut S1, Mut S2, Mut S3, and Mut L) RNAs. Viral replication was monitored by immunofluorescence assay at 3, 6, 9, 12, and 15 d post-transfection using specific anti-DENV antibodies. In each case, the nucleotide sequence of the sHP of replicating viruses is indicated at the bottom. Arrows indicate the spontaneous changes. (B) Expression of DENV proteins in BHK cells transfected with viral full-length RNAs carrying the reconstitutions of the sHP Rec S1, Rec S2, and Rec S3. Viral replication was monitored by immunofluorescence at day 3 post-transfection. (C) The sHP structure is necessary for DENV RNA synthesis. Mutations and reconstitutions within the sHP structure were introduced into a DENV luciferase reported system. Luciferase activity was measured as a function of time after transfection of the RNAs corresponding to the WT, a replication negative control with a mutation in the catalytic site of the NS5 polymerase (NS5 mut), the sHP mutants (Mut S1, Mut S2, Mut S3, and Mut L), and sHP reconstitutions (Rec 1, Rec 2, and Rec 3), as indicated at the bottom. (D) Comparative one-step growth curves of WT, revertants (Rev 1, Rev 2, and Rev 3), and reconstituted (Rec 1, Rec 2, and Rec 3) viruses. Cells were infected at MOI of 0.05 and plaque forming units (PFU) were determined at each time by plaque assay in BHK cells.
In order to confirm the function of the sHP and to eliminate the possibility that other compensatory changes occurred elsewhere in the genome that contributed to the phenotypes observed, new mutants were designed carrying the substitutions observed in the revertant viruses. Mutants Rec S1 and Rec S2 included the substitutions detected in Rev S1 and Rev S2a placed in the WT backbone (Fig. 2B). In addition, a new virus genotype was generated (Rec S3) carrying the same mutations as Mut S3 but including the compensatory changes to reconstitute the stem of the sHP (Fig. 2B). The three Rec mutants replicated slightly less efficiently than did the WT virus. The results indicate that the impaired replication of Mut S1, Mut S2, and Mut S3 can be attributed to the lack of sHP formation and not to the nucleotide changes introduced.
To further investigate the role of the sHP in viral replication, we introduced the mutations and the changes that reconstitute the sHP into a full-length DENV genome carrying a luciferase gene as reporter. We have recently described this reporter system that allows discrimination between DENV translation, RNA amplification, and viral particle formation by measuring luciferase activity post-transfection and post-infection (Samsa et al. 2009). RNAs from WT; a replication negative control with a mutation in the catalytic site of the NS5 polymerase; the sHP mutants Mut S1, Mut S2, Mut S3, and Mut L; and the reconstitutions Rec 1, Rec 2, and Rec 3 were transfected into BHK cells. All the RNAs transfected showed similar luciferase activity at 6 h post-transfection, indicating efficient translation of the input genomes (Fig. 2C). At 24 h, the input RNA of the WT was amplified (Fig. 2C, cf. the WT and NS5mut). The luciferase measured at 24, 48, and 72 h in Mut S1, Mut S2, and Mut S3 was between 1000- and 10,000-fold lower than that for the WT, indicating a profound defect on RNA synthesis. Replication was rescued in Rec 1, Rec 2, and Rec 3 viruses (Fig. 2C). In addition, the mutant in the loop of the sHP (Mut L) showed a delay in RNA synthesis, which explains the delay replication of the virus in the IF assay (Fig. 2A,C, cf. WT and Mut L). The results indicate that the structure of the sHP is dispensable for viral translation but is necessary for DENV RNA synthesis.
To further characterize the phenotype of the sHP revertants and the reconstituted viruses, one-step growth curves were performed. Viral stocks of the replicating Rev S1, Rev S2, Rev S3, Rec S1, Rec S2, and Rec S3 viruses were obtained. Subtle differences in the titers of the WT and the mutants were observed (Fig. 2D). In comparison with the mutants, the WT showed about twofold higher titers at 48 h and a clear cytopathic effect at 72 h, which was not observed with any of the mutants.
We define the relevance of a new cis-acting element for DENV RNA replication. In addition, because the sHP is only predicted in the linear form of the RNA, the results support the idea that this conformation of the genome plays a role in viral replication.
The balance between two conformations of the viral genome is crucial for DENV infection
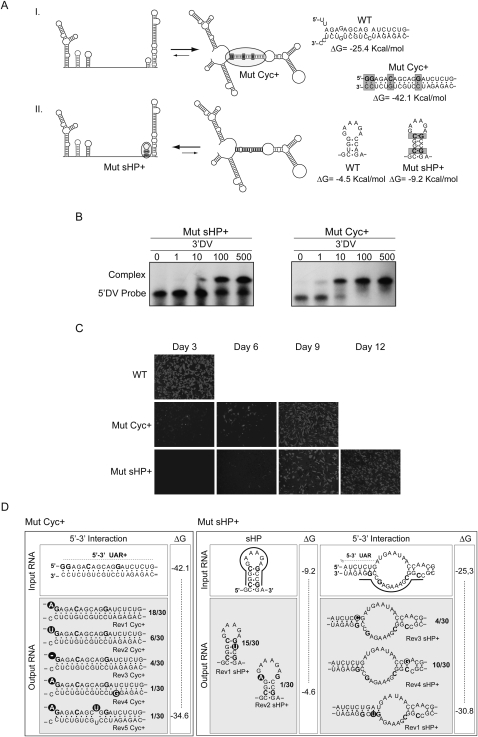
We then sought to investigate the relevance of overlapping sequences in the viral genome involved in alternative structures. To this end, we uncoupled the 3′UAR from the sHP by a 4-nt insertion in the viral 3′UTR (Mut sHP-Dx). A sequence ACAG was inserted at the 3′ end of the sHP structure, which could participate in the 5′-3′UAR interaction without altering the sHP structure (see Materials and Methods). In this mutant, the sHP was predicted in both the linear and circular conformation of the RNA. After RNA transfection, viruses were not recovered (data not shown). One possible explanation for this result is that the nucleotide insertion interferes with the sHP function or the long-range RNA–RNA interaction. In addition, it is possible that the overlapping sequences within the sHP are important to regulate the equilibrium between the two alternative conformations of the genome. To investigate this possibility, we modified the viral RNA in such a way as to favor the circular conformation by increasing the 5′-3′UAR stability (Fig. 3A, Mut Cyc+) or to shift the equilibrium toward the linear form of the RNA by increasing the stability of the sHP (Fig. 3A, Mut sHP+). These mutations were predicted to increase or decrease the stability of the long-range 5′-3′ RNA–RNA interaction. To confirm this prediction, we analyzed the effect of the specific mutations on the RNA–RNA complex formation using EMSA assays. We have previously reported that hybridization of RNA molecules corresponding to the ends of the WT DENV genome was sufficient to form a stable RNA–RNA complex (including 5′-3′UAR, 5′-3′CS, and adjacent complementary nucleotides) with a dissociation constant (Kd) of 8 nM (Alvarez et al. 2005b). Thus, a radiolabeled RNA corresponding to the first 160 nt of the Mut Cyc+ or Mut sHP+ was incubated with a molecule carrying the last 106 nt of each genome. The estimated Kds were 2 and 28 nM for the Mut Cyc+ and Mut sHP+, respectively (Fig. 3B). These results confirmed that the mutations in the Cyc or the sHP regions modified the RNA–RNA complex affinity.
FIGURE 3.
Circular and linear conformations of the DENV genome are crucial for viral infectivity. (A) Schematic representation of mutations that change the thermal stability of the hybridized 5′-3′UAR element (Mut Cyc+) or the sHP structure (Mut sHP+). Nucleotide sequence of the WT and mutants are also shown. (B) Mobility shift assays showing RNA–RNA complex formation. Uniformly labeled 5′DV RNA, corresponding to the first 160 nucleotides (nt) of the Mut Cyc+ or Mut sHP+, was incubated with increasing concentrations of the 3′DV RNA corresponding to the last 106 nt of the respective mutant. The 3′DV was used from 0–500 nM as indicated on the top of the gel. The location of the 5′DV probe and the RNA–RNA complex is also shown. (C) Immunofluorescence of BHK cells transfected with WT, Mut Cyc+, or Mut sHP+ DENV RNAs. Viral replication was monitored using specific anti-DENV antibodies at 3, 6, 9, and 12 d post-transfection. (D) Spontaneous mutations restore the balance between the circular and linear conformations of the RNA. Input RNA sequences Mut Cyc+ or Mut sHP+ are shown together with the nucleotide sequences of the recovered viruses (output RNA). The enthalpy changes (ΔG in Kcal/mol) illustrate the change in stability of the input and output RNAs. In addition, the frequency of each revertion is indicated for each case.
Next, we constructed full-length RNA genomes corresponding to the Mut Cyc+ and Mut sHP+. Transfection of these RNAs into BHK cells took 9–12 d to produce viruses, suggesting that the mutations impair viral replication (Fig. 3C). To determine the sequence of the replicating viruses, the RNAs were purified; the ends of the molecules were ligated after decapping and used to sequence both ends of the genome. Thirty individual clones were sequenced for each mutant. In the case of Mut Cyc+, a variety of spontaneous mutations were selected. Deletions and nucleotide changes were observed at the 5′ and/or 3′UAR sequences (Fig. 3D, left panel). All the selected nucleotide changes decreased the predicted stability of the hybridized 5′-3′UAR sequence. When the input was the Mut sHP+ RNA, two kinds of spontaneous mutations were observed. In the first case, the stability of the sHP was reduced by selection of viruses carrying changes in the stem of the sHP (Fig. 3D, Rev1 sHP+ and Rev2 sHP+). In the second kind, instead of decreasing the stability of the sHP, the spontaneous mutations were obtained in the 5′ end of the RNA, which included new GC base pairs that increase the potential for 5′-3′ interaction (Fig. 3D, Rev3 sHP+ and Rev 4 sHP+). These second-site mutations revealed a genetic link between the two competing structures. In addition, this observation shows the importance of the relative stability between the sHP and the duplex structures, rather than the absolute stability of each structure.
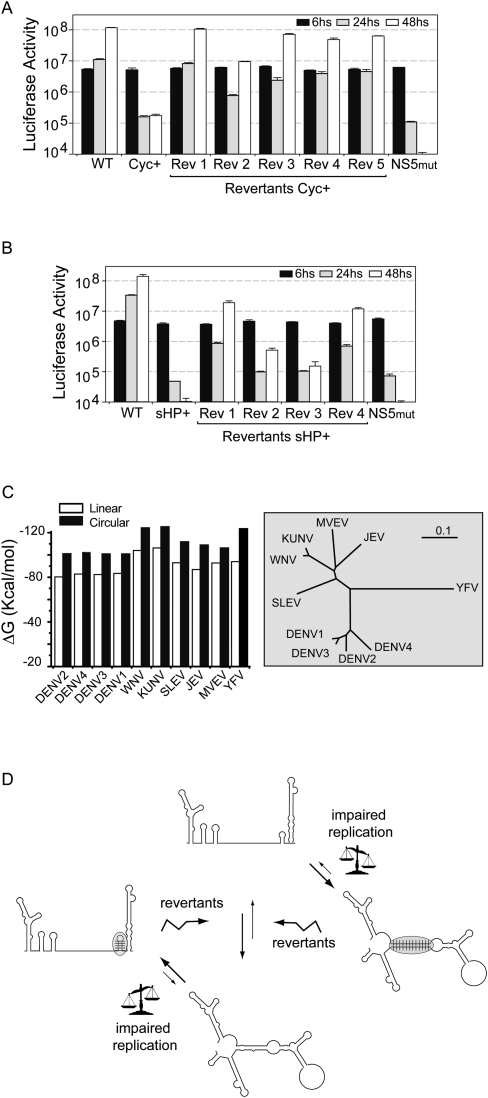
To confirm that the specific nucleotide changes detected in the viruses recovered in cell culture were responsible for the phenotypes observed, we engineered each of the spontaneous mutations of Figure 3D into the DENV reporter system. Genome-length viral RNAs carrying the Cyc+ mutation, each of the Mut Cyc+ revertions (Rev 1, Rev 2, Rev 3, Rev 4, and Rev 5), the sHP+ mutation, or each of the Mut sHP+ revertions (Rev 1, Rev 2, Rev 3, and Rev 4) were designed and transfected into BHK cells. All the cells transfected with the viral RNAs displayed high levels of luciferase activity at the early time points, indicating efficient translation (Fig. 4A,B). However, the Mut Cyc+ and the Mut sHP+ showed profound defects on viral RNA synthesis, in agreement with the IF data (Fig. 3C). All the viruses carrying the nucleotide changes found in the revertants rescued the viral function to different degrees (Fig. 4A,B). In the case of the revertant viruses from the Mut Cyc+, Rev 1 was the most frequently recovered sequence (61%) (Fig. 3D), and the virus replicated similarly to the WT (Fig. 4A). For the Mut sHP+ revertants, the spontaneous changes found in Rev 1 and Rev 4 were the most frequent ones (50% and 35%) (Fig. 3D), and this correlated with higher levels of RNA amplification compared with the other sHP+ revertant viruses (Fig. 4B). In summary, the spontaneous nucleotide changes identified were responsible for increasing between 100- and 1000-fold viral RNA amplification.
FIGURE 4.
Spontaneous mutations restored both the balance between the duplex and the sHP structures and viral RNA replication. (A,B) Luciferase activity measured as a function of time after transfection of full-length DENV luciferase reporter system of Mut Cyc+, revertants Cyc+ (Rev 1, Rev 2, Rev 3, Rev 4, and Rev 5), Mut sHP+, revertants sHP+ (Rev 1, Rev 2, Rev 3, and Rev 4), WT, and a replication negative control with a mutation in the catalytic site of the NS5 polymerase (NS5 mut), as indicated at the bottom. In addition, the frequency of each revertion is indicated at the bottom of each plot. (C) The stability difference of the ends of the genome folded in the lineal or hybridized conformation is highly conserved among mosquito-borne flaviviruses. On the left, a histogram illustrates the thermal stabilities calculated for the linear and circular forms of viral RNAs. On the right, an un-rooted neighbor-joining tree shows the relationship between different flavivirus sequences using the 5′ and 3′ terminal nucleotides of the genomes. The tree was constructed using the program ClustalX2 (Larkin et al. 2007) and drawn using TreeView (Page 1996). The scale indicates the number of nucleotide substitutions per site. (D) Representation of a model showing the requirement of a balance between circular and linear conformations of DENV genome. Mutations that increase the stability of the circular or the linear form spontaneously revert to sequences that resemble the parental stability.
In most of the revertant viruses, small changes in the thermodynamic stability of the sHP or the 5′-3′ double helix region were sufficient to restore viral replication, suggesting a great vulnerability in the balance between the two structures. It is worth noting that the predicted stability for the hybridized ends of the genome exceeds that predicted for the terminal 5′ and 3′ end structures. A difference in stability of the two alternative conformations was found to be conserved among mosquito-borne flavivirus genomes (Fig. 4C). Previous studies using different flaviviruses provide evidence for the relevance of the bottom half of the stem of the 3′SL, which also overlaps with 3′UAR (Fig. 1A; Tilgner and Shi 2004; Song et al. 2008; Teramoto et al. 2008). These observations obtained studying a structure that is downstream of the sHP are in agreement with our results. In the Japanese encephalitis virus, three discontinuous complementary RNA sequences were reported (Song et al. 2008). These sequences correspond to the 5′-3′UAR and the bottom stem of the 3′SL reported for dengue and West Nile virus genomes (Khromykh et al. 2001; Corver et al. 2003; Lo et al. 2003; Alvarez et al. 2005b, 2008; Kofler et al. 2006; Zhang et al. 2008). Furthermore, a recent report suggested a role of an additional complementary region adjacent to 5′-3′UAR in the DENV genome, named DAR (Friebe and Harris 2010). This element also overlaps with the sHP structure. In this regard, the reconstituted and revertant viruses Rec 3 and Rev 1 sHP+ include spontaneous changes that restored the sHP stability and the viral function, while tolerating mismatches in the DAR region. These observations are in agreement with the flexibility to nucleotide changes in the DAR element previously reported (Friebe and Harris 2010). In addition, mutations disrupting the stem of the sHP that did not alter the 5′-3′ DAR stability (Mut S1 and Mut S2) (Fig. 2) impaired viral replication, confirming the essential role of the sHP structure.
In summary, our results support a model in which the DENV genome adopts two alternative conformations, and highlight the relevance of a balance between two competing structures (Fig. 4D). In this regard, we speculate that the stabilities of RNA structures would function as regulators of the circular/linear transition.
Unlike well-studied riboswitches in cellular RNAs, the importance of conformational changes in viral RNAs during infection is a topic that has been recently recognized (Simon and Gehrke 2009). Here, we report that the DENV genome acquires different conformations during infection. How and why the viral RNA switches from one to an other structure is still unclear. It is possible that evolving overlapping functions in the viral RNA provides a mechanism for organizing the multiple functions of the viral genome. It is likely that the interaction of viral and cellular proteins with the viral genome stabilize certain conformation of the RNA to promote or repress a viral function. Several cellular proteins have been reported to interact with flavivirus genomes (Blackwell and Brinton 1997; Ta and Vrati 2000; De Nova-Ocampo et al. 2002; Li et al. 2002; Yocupicio-Monroy et al. 2003, 2007; Garcia-Montalvo et al. 2004; Paranjape and Harris 2007; Vashist et al. 2009; Agis-Juarez et al. 2009). In addition, viral proteins with RNA helicase and RNA chaperone activities could be responsible for modulating viral RNA plasticity. For DENV, we have previously demonstrated that only circular genomes are competent for viral RNA replication in infected cells (Filomatori et al. 2006). The model for minus strand RNA synthesis involves cyclization of the genome to locate the SLA–NS5 complex near the 3′ end of the RNA. However, it is still unknown if this conformation of the RNA is also required for plus strand RNA synthesis. It is possible that switching from circular to linear conformations inhibit minus strand RNA synthesis, providing a mechanism to control the 100:1 ratio of plus versus minus strand RNA found in infected cells. The same genomic RNA must also function as template for translation and encapsidation. The interaction of the 5′UTR with translation initiation factors could destabilize the hybridized 5′-3′ ends of the genome, favoring a linear structure of the RNA; however, this possibility has not been experimentally tested. Encapsidation is perhaps one of the most obscure steps of the flavivirus replication cycle (Samsa et al. 2009). It is still unclear how and where the capsid protein recruits the viral genome for assembly. However, it is possible that a certain conformation of the RNA is necessary for viral capsid recognition; as it was previously demonstrated for retroviruses (Huthoff and Berkhout 2001; D'Souza and Summers 2004). Future studies should investigate the role of alternative conformations of the RNA in each stage of flavivirus replication. Defining the molecular mechanisms that control viral riboswitches will raise our understanding of flavivirus life cycles.
MATERIALS AND METHODS
Construction of recombinant DENVs
Mutations were introduced in the full-length cDNA of dengue type 2 pD2/IC AflII/NotI previously described (Samsa et al. 2009) replacing the SacI-NotI (5′ mutations) or AflII-XbaI (3′ mutations) fragments of the WT plasmid with the respective fragment derived from overlapping PCRs. The overlapping PCRs were performed with the common outside oligonucleotides AVG-194 (GGAATTCGAGCTCCGCGGACGCGTAAATTTAATACGAC) and AVG-7 (GTGGGTTCGAAAGTGAGAATCTCTTTGTCAGCT) for the 5′ mutants. In the case of 3′ mutants, the outside oligonucleotides were AVG-90 (TAGAAAGCAAAACTTAAGATGAAAC) and AVG-263 (GCTTATCATCGATAAGCTTG). In the mutant sHP-Dx, the sequence ACAG was inserted in the position 10645, and the changes C10631G and G10644C were made to generate the following nucleotide sequence 10630-GTGGGAAAGACCACACAGAGA-10648.
The full-length DENV luciferase reporter system mDV-R recently described (Samsa et al. 2009) was used to analyze the replication of viruses carrying mutations, revertions, and reconstitutions. The mutations in the mDV-R were introduced using a similar approach as the one described above. The overlapping PCRs for the 5′ mutants were performed with the common outside oligonucleotides AVG-273 (GAATTCGAGCTCACGCGTAAATTTAATACGACTCACTAT AAGTTGTTAGTCTACGTGG) and AVG-7 (GTGGGTTCGAAAGTGAGAATCTCTTT GTCAGCT). In the case of the mutants at the 3′ end of the genome, the outside oligonucleotides AVG-90 (TAGAAAGCAAAACTTAAGATGAAAC) and AVG-1034 (CAGCTTATCATCGATAAGCTTGCGCGATGTCGACTCTAGAGAACCTGTTGATTC) were used.
RNA transcription and transfection
WT or recombinant plasmid DNAs were linearized by digestion with XbaI restriction enzyme and used as templates for transcription by T7 polymerase Plus (Ambion) in the presence of m7GpppA cap structure analog (New England Biolabs). After a 90-min incubation at 37°C, the transcripts were treated with a DNase I RNAase-free kit (Ambion Inc). The RNA transcripts were transfected with Lipofectamine 2000 (Invitrogen) into BHK-21 cells grown in 60-mm-diameter tissue culture dishes. The transfected cells were trypsinized on day 3 post-transfection, and two-thirds of total cells were reseeded. This procedure was repeated every 3 d for 3 wk. Supernatants derived from transfected BHK-21 cells were harvested at 3, 6, 9, 12, and 15 d post-transfection and used to search for infectious DENV.
Inmunofluorescence
Transfected cells with WT and mutated full-length DENV RNA were used for IF assay. BHK-21 cells were grown in 60-mm-diameter tissue culture dishes containing a 1-cm2 coverslip inside. The coverslips were removed and directly used for IF analysis. The transfected cells were trypsinized at day 3, and two-thirds of total cells were reseeded to a 60-mm-diameter tissue culture dish containing a new coverslip inside. This procedure was repeated every 3 d for 15 d. At each time point, a 1:200 dilution of murine hyperimmune ascitic fluid against the DENV type 2 in phosphate buffered saline (PBS) 0.2% gelatin was used to detect viral antigens. Cells were fixed for 10 min in cold methanol. Alexa fluor 488 rabbit anti-mouse immunoglobulin G conjugate (Molecular Probes) was used as detector antibody at 1:500 dilution. Photomicrographs (200× magnification) were acquired with an Olympus BX60 microscope coupled to a CoolSnap-Pro digital camera (Media cybernetics), and analyzed with Image-PRO PLUS software.
One-step growth curves
The supernatants of WT and mutant viruses were used to obtain viral stocks. The stocks were quantified by plaque assays in BHK cells as previously described (Alvarez et al. 2005a). For one-step growth curves, subconfluent BHK-21 cells in a six-well plate were infected with equal amounts of WT and mutant viruses. A multiplicity of infection (MOI) of 0.05 in 500 μL of PBS was used. After 1 h of adsorption, the cells were washed three times with PBS, and 2 mL of growth media was added. At each time point after infection, cell supernatants were collected. For virus quantification, supernatants were serially diluted and plaque assays performed in BHK-21 cells.
Extraction of viral RNA and sequencing
After clarification of supernatants at 2500g for 5 min at 4°C, viral RNA was TRIZOL extracted from 250 μL aliquots. RNA was treated with Tobacco acid pyrophosphatase (Epicenter) to remove the cap structure, and then 3′ and 5′ ends of the RNA were ligated with T4 RNA ligase (Epicenter) as described previously (Mandl et al. 1991). After phenol-chloroform extraction and ethanol precipitation, the pellet was resuspended in 20 μL of RNAse free water. Five microliters of this solution was used for RT-PCR using random primers (Invitrogen) and primers designed to amplify the 5′-3′ joined ligation including both cyclization regions. The RT-PCR products were directly sequenced to determine the consensus sequence and were subcloned into pGEM-T Easy plasmid (Promega). At least 20 independent clones were sequenced for each PCR product.
RNA binding assays
RNA–RNA interactions were analyzed by electrophoretic mobility shift assays. Uniformly 32P-labeled RNA probes corresponding to the first 160 nt of the viral genomes were obtained by in vitro transcription using T7 RNA polymerase and were purified on 5% polyacrylamide and 6 M urea gels. The binding reaction mixtures contained 5 mM HEPES (pH 7.9), 100 mM KCl, 5 mM MgCl2, 3.8% glycerol, 2.5 μg tRNA, 5′DV probe (0.1 nM, 30,000 cpm), and 0, 1, 10, 100 or 500 nM of 3′DV RNA (corresponding to the last 106 nt of the genome) in a final volume of 30 μL. RNA samples were heat denatured for 5 min at 85°C and slow cooled to room temperature. RNA–RNA complexes were analyzed by electrophoresis through native 5% polyacrylamide gels supplemented with 5% glycerol. Gels were prerun for 30 min at 4°C at 150V, and then 25 μL of sample was loaded and electrophoresis was allowed to proceed for 4 h at constant voltage. Gels were dried and visualized by autoradiography or exposed on a PhosphorImager plate.
Sequence analysis and RNA structure prediction
For the analysis of sequence and structure conservation of the sHP, we used the 3′ end nucleotides of DENV1 (GenBank accession no. NC_001477), DENV2 (NC_001474), DENV3 (NC_001475), DENV4 (NC_002640), SLEV (NC_007580), YFV (NC_002031), KUNV (L24512), WNV (NC_001563), JEV (NC_001437), and MVEV (NC_000943). The sequences were grouped according to their serological classification and then aligned using ClustalX2 software (Larkin et al. 2007) and subsequently corrected manually. The RNAz web server was used to detect thermodynamically stable and evolutionary conserved RNA secondary structures from multiple sequences alignments (Gruber et al. 2007), and a consensus sequence was elaborated using the RNA alifold server (Hofacker 2007). The RNA secondary structure modeling and stability calculus of WT, mutants, and revertants viruses were made with the Zuker and Turner Mfold software, version 3.2 (Zuker 2003). The RNA stabilities presented in Figure 4 were calculated using the mosquito-borne flavivirus sequences listed above. For the linear conformation, the predictions were calculated for the 5′ and the 3′ ends of the respective genome, and the stabilities were added. The 5′ end used included from the first nucleotide of the genome to the end of the predicted 5′CS complementary sequence for each flavivirus; the 3′ end, included from the 3′CS sequence to the last nucleotide of each genome. To predict the stability of the circular conformation of each RNA, the hybridized ends were predicted by Mfold linking the 5′ and 3′ regions with a poli(A) spacer as previously described (Khromykh et al. 2001).
ACKNOWLEDGMENTS
We thank Richard Kinney for DENV cDNA infectious clone. We also thank Dr. Andrew White and members of Gamarnik's laboratory for helpful discussions concerning the manuscript. This work was supported by grants from HHMI, DENFRAME, and PICT-2003. A.V.G. is a member of the Argentinean Council of Investigation (CONICET). D.E.A. and S.M.V. were granted CONICET fellowships.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2120410.
REFERENCES
- Agis-Juarez RA, Galvan I, Medina F, Daikoku T, Padmanabhan R, Ludert JE, del Angel RM 2009. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J Gen Virol 90: 2893–2901 [DOI] [PubMed] [Google Scholar]
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV 2005a. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339: 200–212 [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV 2005b. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79: 6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, Filomatori CV, Gamarnik AV 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375: 223–235 [DOI] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Baltimore D 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63: 369–380 [DOI] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA 1997. Translation elongation factor-1α interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol 71: 6433–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 77: 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM 2002. Translation elongation factor-1α, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295: 337–347 [DOI] [PubMed] [Google Scholar]
- Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol 82: 9008–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381: 123–135 [DOI] [PubMed] [Google Scholar]
- Dreher TW 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol 37: 151–174 [DOI] [PubMed] [Google Scholar]
- D'Souza V, Summers MF 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431: 586–590 [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA 2004. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mrna: A potential common mechanism for tombusviridae. J Biol Chem 279: 28862–28872 [DOI] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20: 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Harris E 2010. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J Virol 84: 6103–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol 79: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Hardy R, Rice CM 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7: 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12: 2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol 74: 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montalvo BM, Medina F, del Angel RM 2004. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of dengue 4 virus RNA. Virus Res 102: 141–150 [DOI] [PubMed] [Google Scholar]
- Goebel SJ, Hsue B, Dombrowski TF, Masters PS 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J Virol 78: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grdzelishvili VZ, Garcia-Ruiz H, Watanabe T, Ahlquist P 2005. Mutual interference between genomic RNA replication and subgenomic mRNA transcription in brome mosaic virus. J Virol 79: 1438–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun TS, Gould EA 2007. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology 366: 8–15 [DOI] [PubMed] [Google Scholar]
- Gruber AR, Neubock R, Hofacker IL, Washietl S 2007. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res 35: W335–W338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell 7: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Guo R, Lin W, Zhang J, Simon AE, Kushner DB 2009. Structural plasticity and rapid evolution in a viral RNA revealed by in vivo genetic selection. J Virol 83: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 198: 33–41 [DOI] [PubMed] [Google Scholar]
- Hofacker IL 2007. RNA consensus structure prediction with RNAalifold. Methods Mol Biol 395: 527–544 [DOI] [PubMed] [Google Scholar]
- Hu B, Pillai-Nair N, Hemenway C 2007. Long-distance RNA-RNA interactions between terminal elements and the same subset of internal elements on the potato virus X genome mediate minus- and plus-strand RNA synthesis. RNA 13: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7: 143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75: 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Hemenway CL 1999. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA 5: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Lee SH, Kim CS, Seol SK, Jang SK 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9: 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J, van Duin J 1999. A long-range pseudoknot in Qbeta RNA is essential for replication. J Mol Biol 294: 875–884 [DOI] [PubMed] [Google Scholar]
- Kofler RM, Hoenninger VM, Thurner C, Mandl CW 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J Virol 80: 4099–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol 76: 11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, White KA 2004. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J 23: 3365–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Sgro JY, Ahlquist P 2002. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J Virol 76: 3905–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM 2007. Flaviviridae: The viruses and their replication. In Fields virology, pp. 1101–1152 Lippincott-Raven, Philadelphia [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol 77: 10004–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodeiro MF, Filomatori CV, Gamarnik AV 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Heinz FX, Puchhammer-Stockl E, Kunz C 1991. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. Biotechniques 10:484, 486 [PubMed] [Google Scholar]
- Markoff L 2003. 5′ and 3′ NCRs in flavivirus RNA. In The flaviviruses: Advances in virus research, pp. 177–228 Elsevier Academic Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KL, Lemon SM 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J Virol 79: 1824–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters HG, Strauss JH 1990. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J Virol 64: 4162–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD 1996. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Panavas T, Nagy PD 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J Virol 77: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape SM, Harris E 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem 282: 30497–30508 [DOI] [PubMed] [Google Scholar]
- Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74: 10359–10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Fabian MR, White KA, Nagy PD 2003. A replication silencer element in a plus-strand RNA virus. EMBO J 22: 5602–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek C, Foley JE, Harris E 2009. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J Virol 83: 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Zhang X, Kao CC 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J Virol 77: 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5: e1000632 doi: 10.1371/journal.ppat.1000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Brinton MA, Veal JM, Zhong YY, Wilson WD 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35: 4222–4230 [DOI] [PubMed] [Google Scholar]
- Simon AE, Gehrke L 2009. RNA conformational changes in the life cycles of RNA viruses, viroids, and virus-associated RNAs. Biochim Biophys Acta 1789: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Simon AE 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol 254: 6–14 [DOI] [PubMed] [Google Scholar]
- Song BH, Yun SI, Choi YJ, Kim JM, Lee CH, Lee YM 2008. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for Flavivirus RNA replication. RNA 14: 1791–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta M, Vrati S 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J Virol 74: 5108–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto T, Kohno Y, Mattoo P, Markoff L, Falgout B, Padmanabhan R 2008. Genome 3′-end repair in dengue virus type 2. RNA 14: 2645–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurner C, Witwer C, Hofacker IL, Stadler PF 2004. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol 85: 1113–1124 [DOI] [PubMed] [Google Scholar]
- Tilgner M, Shi PY 2004. Structure and function of the 3′ terminal six nucleotides of the west nile virus genome in viral replication. J Virol 78: 8159–8171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Anantpadma M, Sharma H, Vrati S 2009. La protein binds the predicted loop structures in the 3′ non-coding region of Japanese encephalitis virus genome: role in virus replication. J Gen Virol 90: 1343–1352 [DOI] [PubMed] [Google Scholar]
- Villordo SM, Gamarnik AV 2009. Genome cyclization as strategy for flavivirus RNA replication. Virus Res 139: 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J 16: 4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2009. Disease outbreak news. http://www.who.int/csr/don/archive/disease/dengue_fever/en/
- Wu B, Pogany J, Na H, Nicholson BL, Nagy PD, White KA 2009. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog 5: e1000323 doi: 10.1371/journal.ppat.1000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocupicio-Monroy RM, Medina F, Reyes-del Valle J, del Angel RM 2003. Cellular proteins from human monocytes bind to dengue 4 virus minus-strand 3′ untranslated region RNA. J Virol 77: 3067–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocupicio-Monroy M, Padmanabhan R, Medina F, del Angel RM 2007. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 357: 29–40 [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem 274: 33714–33722 [DOI] [PubMed] [Google Scholar]
- You S, Rice CM 2008. 3′ RNA elements in hepatitis C virus replication: Kissing partners and long poly(U). J Virol 82: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Shi K, Meskauskas A, Simon AE 2009. The 3′ end of turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA 15: 1849–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 373: 1–13 [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhang J, George AT, Baumstark T, Simon AE 2006. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA 12: 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Miller TB, Goebel SJ, Thiel V, Masters PS 2008. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J Virol 82: 1214–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]