Abstract
microRNAs are ∼22 nucleotide regulatory RNAs that are processed into duplexes from hairpin structures and incorporated into Argonaute proteins. Here, we show that a nick in the middle of the guide strand of an miRNA sequence allows for seed-based targeting characteristic of miRNA activity. Insertion of an inverted abasic, a dye, or a small gap between the two segments still permits target knockdown. While activity from the seed region of the segmented miRNA is apparent, activity from the 3′ half of the guide strand is impaired, suggesting that an intact guide backbone is required for contribution from the 3′ half. miRNA activity was also observed following nicking of a miRNA precursor. These results illustrate a structural flexibility in miRNA duplexes and may have applications in the design of miRNA mimetics.
Keywords: microRNA, miR-124, segmentation, mimetic
INTRODUCTION
microRNAs (miRNAs) are a class of ∼22 nucleotide (nt) noncoding RNAs that play important roles in regulating gene expression in plants and animals. miRNAs are usually produced by a process in which a RNA polymerase II transcript is cut by Drosha to produce a precursor hairpin, which is cut by Dicer in the cytoplasm to produce a double-stranded duplex that is incorporated into Argonaute (Ago) proteins (Kim et al. 2009). After elimination of the passenger strand by cleavage and/or helicase activity, the guide strand can then bind to complementary mRNAs. Studies suggest that the most prevalent form of consequential target recognition occurs through complementary binding to a target 3′ untranslated region (UTR) by the miRNA seed region (positions 2 through 8 at the 5′ end of the guide strand), leading to down-regulation at mRNA and protein levels (Bartel 2009).
Ago2-mediated cleavage of the passenger strand has been found to be important for assembly of small interfering (siRNAs) and some miRNAs (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Leuschner et al. 2006; Diederichs and Haber 2007), and a nicked passenger strand was found to rescue the activity of an siRNA containing chemical modifications that prevented passenger strand cleavage by Ago (Leuschner et al. 2006). This concept has been applied to the design of siRNAs (Bramsen et al. 2007), where passenger segmentation was found to maintain siRNA activity (while eliminating passenger strand activity), while guide segmentation was found to eliminate the desired siRNA activity. However, these experiments did not address whether division of the guide strand of a miRNA would retain the seed-based activity so important for miRNA action. To answer this question, we investigated the effects of strand segmentation on a miR-124 mimic, a miRNA whose targeting activity we have studied previously (Lim et al. 2005).
RESULTS AND DISCUSSION
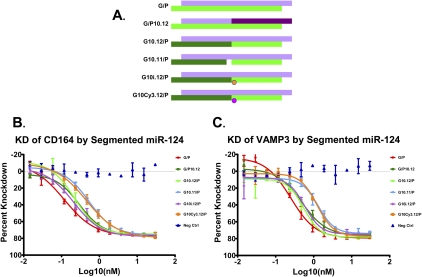
We tested structural variants of a perfectly complementary miR-124 duplex in which a nick was introduced 10 nt from the 5′ end of either the guide (or miRNA) or passenger (or miRNA*) strand (Fig. 1A). Duplexes were transfected into cells and changes in the mRNA levels of miR-124 targets CD164 and VAMP3 were measured. As expected, division of the passenger strand (G/P10.12) had little effect on miR-124 activity (Fig. 1B), leading to a slight increase in EC50 (0.12 nM to 0.29 nM for CD164, 0.25 nM to 0.48 nM for VAMP3). Interestingly, division of the guide strand (G10.12/P) still allowed for miR-124 activity (EC50s of 0.22 nM for CD164 and 0.55 nM for VAMP3), showing that a continuous guide strand is not absolutely required for miRNA RNA-induced silencing complex (RISC) activity. The addition of a one base gap between the guide halves, or capping of the junction with a Cy3 dye or an inverted abasic residue, still gave miR-124 activity, providing evidence that the observed activity was not a result of ligation of the guide halves. Toleration of guide strand segmentation is not a property only of the miR-124 sequence, as division of the guide strand in a miR-34 mimic still allowed for miR-34a activity (Supplemental Fig. 1).
FIGURE 1.
miR-124 activity tolerates segmentation of the guide and the passenger strands. (A) Schematics of the miR-124 duplex designs. The top strand represents the passenger strand (5′ to 3′), while the bottom strand represents the annealed guide strand. Change in color indicates the site of a break in the strand backbone. (Pink circle) A Cy3 label; (orange circle) an inverted abasic modification. Dose-dependent response of CD164 (B) and VAMP3 (C) mRNA in HCT-116 cells, as measured by qRT-PCR. EC50s of the CD164 knockdown curves are: G/P, 0.12 nM; G/P10.12, 0.29 nM; G10.12/P, 0.22 nM; G10.11/P, 0.53 nM; G10i.12/P, 0.21 nM; G10Cy3.12/P, 0.45 nM. EC50s of the VAMP3 knockdown curves are: G/P, 0.25 nM; G/P10.12, 0.48 nM; G10.12/P, 0.55 nM; G10.11/P, 1.32 nM; G10i.12/P, 0.58 nM; G10Cy3.12/P, 1.23 nM.
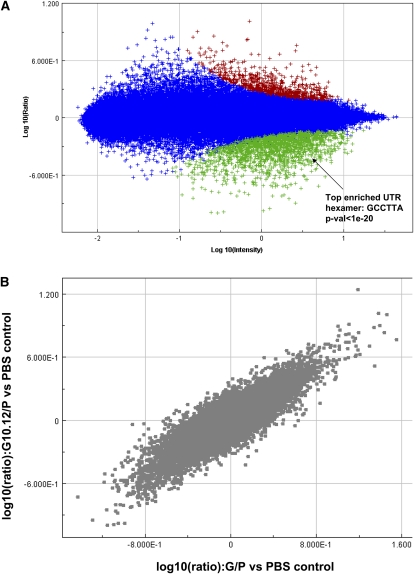
To further confirm the targeting activity of a segmented miRNA duplex, but at a genome-wide level, we used microarrays to profile cells transfected with a miR-124 duplex containing the divided guide strand (Fig. 2A). Analysis of the 3′ UTR sequences of the down-regulated genes shows that the most significantly enriched hexamer is GCCTTA, which corresponds to the seed sequence of miR-124, as would be expected for miR-124 activity. Microarray profiling of the effects of the segmented miR-124 duplex G10.12/P showed striking correlation with the profiling of the effects of the fully intact duplex (G/P), consistent with the preservation of the bulk of miR-124 targeting (Fig. 2B).
FIGURE 2.
Microarray analysis shows seed-based activity from a segmented guide strand. (A) Microarray signature 24 h after transfection of 10 nM G10.12/P into HCT116 dicer hypomorphs, plotted as ratio of probe fluorescence intensity (relative to mock transfection) on the y-axis, and mean fluorescence intensity for the experimental and mock samples on the x-axis. (Green) Significantly down-regulated probes (P < 1 × 10−6); (red) up-regulated probes. Hypergeometric analysis of the hexamer content of the down-regulated UTRs showed that the most significantly enriched hexamer was GCCTTA, corresponding to positions 2–7 of the transfected miR-124. (B) Comparison of gene expression data from G10.12/P transfected cells and G/P transfected cells. Expression ratio of the G/P transfection versus the mock transfection is plotted on the x-axis, and G10.12/P expression ratio (versus mock) is plotted on the y-axis. The correlation coefficient (weighted by microarray error model) was calculated to be 0.9, illustrating the similar effects of both RNA complexes on gene expression.
Although the above analyses highlighted seed-based activity derived from the 5′ half of a divided miRNA, it was unclear whether the 3′ half of the segmented strand was functional. Previous work analyzing microarray profiles has shown that although the preponderance of miRNA targeting is due to seed region activity, a much smaller degree of down-regulation can be attributed to other contributing factors, among them the supplementary binding of positions 13–16 of the miRNA (Grimson et al. 2007). We examined the microarray profiling of miR-124 targets containing supplementary 3′ binding, and did detect a slight shift in down-regulation following guide strand segmentation that was suggestive of a loss of supplementary 3′ binding activity in the divided miRNA (Supplemental Fig. 2).
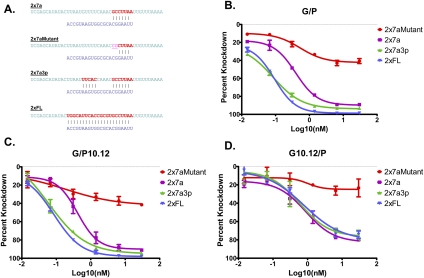
To verify this observation, we measured the activity of segmented miRNAs on luciferase reporter vectors whose 3′ UTRs had been engineered (Fig. 3A) to contain two miRNA-complementary sites that constituted full-length matches (2xFL), seed region matches (2x7a), or matches to the seed region plus supplementary 3′ pairing (2x7a3p). For an intact duplex (G/P), the repressive activity of miR-124 on luciferase activity was highest for 2xFL and followed the order 2xFL∼2x7a3p>2x7a (Fig. 3B). Similar behavior was seen with a segmented passenger strand (G/P10.12, Fig 3C). However, when the guide strand was divided (G10.12/P, Fig 3D), the activities of both the 2xFL and 2x7a3p reporters became equivalent to that of 2x7a, showing that the discontinuity at position 10 of the guide strand prevents productive 3′ binding, while still permitting seed-based activity from the 5′ half.
FIGURE 3.
Activity of an unsegmented miR-124 guide strand is enhanced by additional complementarity beyond the seed region, but activity of a segmented guide strand is not. (A) Schematic of miR-124 target sites that were duplicated and inserted into a dual luciferase reporter vector. (B) G/P exhibits suppression of activity from a reporter with a seed-region match, which is enhanced by reporters containing additional complementarity to the 3′ end of the guide strand (2x7a3p), or full-length complementarity (2xFL). EC50s for these curves are: 2x7a, 0.39 nM; 2x7a3p, 0.08 nM; 2xFL, 0.09 nM. (C) Segmentation of the passenger strand (G/P10.12) preserves the trends observed in B. EC50s for the curves are: 2x7a, 0.41 nM; 2x7a3p, 0.06 nM; 2xFL, 0.09 nM. (D) When the guide strand is segmented (G10.12/P), activity from reporters with 3′ complementarity is identical to that observed from the seed-only reporter. EC50s for the curves are: 2x7a, 0.89 nM; 2x7a3p, 0.61 nM; 2xFL, 0.80 nM.
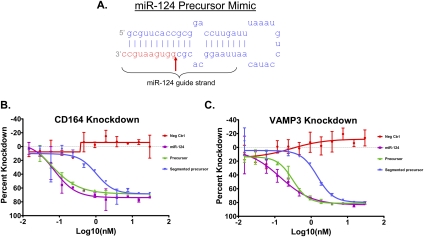
We also evaluated the effect of nicking at various positions in the guide strand besides position 10, and found toleration of 5′ segments of 9 to 12 nucleotides in length (Supplemental Fig. 3), consistent with the notion that backbone discontinuities are tolerated at several positions, as long as they do not disrupt the seed region or the duplex's stability. We also tested activity of a segmented guide strand in the context of a hairpin designed to emulate the natural miR-124 hairpin. Although there was a loss of potency when compared with the intact hairpin, appreciable activity was still observed following guide strand division (Fig. 4), suggesting that processing of a hairpin into an Ago-recognizable duplex can occur in spite of a break in the guide strand.
FIGURE 4.
Activity from a segmented miR-124 hairpin precursor. (A) miR-124 hairpin precursor schematic. This precursor was transfected either fully intact or with a nick at the position indicated with a red arrow. The hairpin precursors, a miR-124 duplex, or a negative control siRNA (guide sequence UACGCGUAGUCGGUCAUACUU, passenger sequence GUAUGACCGACUACGCGUAUU, both 5′ to 3′) were transfected into HCT-116 cells and target knockdown assayed by qRT-PCR. (B) CD164 mRNA dose response. EC50s of the curves shown for CD164 knockdown are: miR-124, 0.07 nM; Precursor, 0.05 nM; Segmented Precursor, 1.02 nM. (C) VAMP3 mRNA dose response. EC50s of the curves shown for VAMP3 knockdown are: miR-124, 0.14 nM; Precursor, 0.31 nM; Segmented Precursor, 1.48 nM.
RNA structures analogous to those described in this paper might occur in vivo, for instance, if endogenous nucleases nicked the miRNA guide strand before or after Ago incorporation. Thus, under certain circumstances, it is conceivable that RNA fragments of only ∼10 nt in length are active components of RISC. It will be interesting to see if actual examples of this can be found in vivo.
Further experiments will also be required to understand why 3′ supplementary binding activity is lost by these structures—it is possible that the break in the guide backbone may lead to improper positioning or even loss of the 3′ half, or perhaps to an inability to carry out conformational changes required for full-length activity (Wang et al. 2009). Finally, these results may be pertinent to development of seed-based miRNA therapeutics, since the toleration of a guide strand break with chemical adducts at this break may allow the creation of miRNA mimics with novel therapeutic properties.
MATERIALS AND METHODS
Segmented miRNA synthesis and sequence
Oligonucleotides were synthesized at Sigma-Aldrich or Merck & Co. All oligonucleotides were unphosphorylated. Annealing was accomplished by mixing single strands at 10 μM in 10 mM pH7.5 Tris-HCl/50 mM NaCl and heating at 95°C for 2 min before slowly cooling to 37°C over the course of 1 h.
The RNA oligonucleotides used were as follows (5′ to 3′ orientation, “iB” denotes an inverted abasic, while “Cy3” denotes a Cy3 fluorescent dye molecule): G, UUAAGGCACGCGGUGAAUGCCA; P, GCAUUCACCGCGUGCCUUAAAU; G10, UUAAGGCACG; G.10, CGGUGAAUGCCA; G.9, GGUGAAUGCCA; P10, GCAUUCACCG; P.10, CGUGCCUUAAAU; G10i, UUAAGGCACG-iB; G10Cy3, UUAAGGCACG-Cy3; 58-mer, GCGUUCACCGCGGACCUUGAUUUAAAUGUCCAUACAAUUAAGGCACGCGGUGAAUGCC; 48-mer, GCGUUCACCGCGGACCUUGAUUUAAAUGUCCAUACAAUUAAGGC ACGC; 10-mer, GGUGAAUGCC. A nontargeting RNA duplex with guide sequence UACGCGUAGUCGGUCAUACUU (5′ to 3′) and passenger sequence GUAUGACCGACUACGCGUAUU (5′ to 3′) was used as a negative control.
qPCR measurements
Transfection and qPCR was carried out as described (Kenski et al. 2010). Briefly, HCT-116 cells were cultured in McCoy's 5A Medium (Mediatech Inc.) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin, plated in 96-well culture plates at a density of 6000 cells/well 24 h prior to transfection, and then transfected using Opti-MEM I Reduced Serum Media (GIBCO) and Lipofectamine RNAiMax (Invitrogen), with a final concentration ranging from 30 nM down to 0.01 nM along a 12-point titration curve. Twenty-four hours after transfection, cells were washed with phosphate-buffered saline and processed using the Cells-to-CT Kit (Applied Biosystems). qRT-PCR was carried out using TaqMan gene-specific probes (Applied Biosystems) on an ABI Prism 7900HT Sequence Detector. Reverse transcription conditions were as follows: 60 min at 37°C followed by 5 min at 95°C. qRT-PCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, and 1 min at 60°C. GUSB mRNA levels were used for data normalization.
Luciferase reporter measurements
Transfection was carried out as described above, except cells were plated at a density of 25,000 cells/well, and Lipofectamine 2000 was used instead of RNAiMAX. miRNAs were cotransfected with siCHECK2 vectors (Genscript) containing cloned target inserts (see Fig. 3) consisting of a tandem repeat of a seed match to miR-124 (2x7a), a seed match containing additional 3′ complementarity to positions 13–17 of miR-124 (2x7a3p), or a full-length match to miR-124 (2xFL). A seed match with a two-base mutation (2x7aMutant) was used as a control. Twenty-four hours after transfection, transfection medium was replaced with fresh growth medium. Forty-eight hours after transfection, cells were lysed and both firefly and Renilla luciferase activity were measured using the Dual-Glo Luciferase Assay System (Promega) on a Wallac EnVision 2103 Multilabel Reader (Perkin-Elmer). Transfections using an siRNA to GAPDH (guide sequence AAGUUGUCAUGGAUGACCUUU, passenger sequence AGGUCAUCCAUGACAACUUTT, both 5′ to 3′) were used to normalize luciferase data.
Microarray analysis
HCT-116 Dicer hypomorph cells were transfected with 10 nM miRNA duplex as described previously (Jackson et al. 2006). RNA was extracted using RNeasy (Qiagen), amplified using the Ovation protocol (Nugen), and profiled on custom Affymetrix arrays (Rosetta Custom Human 2.0, Affymetrix). Array signals were analyzed with Affymetrix GeneChip Operating Software and Affymetrix Power Tools. UTR hexamer enrichment was analyzed as described previously (Jackson et al. 2006).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Francis Gosselin and Mark Levorse for oligonucleotide synthesis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2296210.
REFERENCES
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J 2007. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res 35: 5886–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenski DM, Cooper AJ, Li JJ, Willingham AT, Haringsma HJ, Young TA, Kuklin NA, Jones JJ, Cancilla MT, McMasters DR, et al. 2010. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5′-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res 38: 660–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]