Abstract
Archaeal and eukaryotic box C/D RNPs catalyze the 2′-O-methylation of ribosomal RNA, a modification that is essential for the correct folding and function of the ribosome. Each archaeal RNP contains three core proteins—L7Ae, Nop5, and fibrillarin (methyltransferase)—and a box C/D sRNA. Base-pairing between the sRNA guide region and the rRNA directs target site selection with the C/D and related C′/D′ motifs functioning as protein binding sites. Recent structural analysis of in vitro assembled archaeal complexes has produced two divergent models of box C/D sRNP structure. In one model, the complex is proposed to be monomeric, while the other suggests a dimeric sRNP. The position of the RNA in the RNP is significantly different in each model. We have used UV-cross-linking to characterize protein–RNA contacts in the in vitro assembled Pyrococcus furiosus box C/D sRNP. The P. furiosus sRNP components assemble into complexes that are the expected size of di-sRNPs. Analysis of UV-induced protein–RNA cross-links revealed a novel interaction between the ALFR motif, in the Nop domain of Nop5, and the guide/spacer regions of the sRNA. We show that the ALFR motif and the spacer sequence adjacent to box C or C′ are important for box C/D sRNP assembly in vitro. These data therefore reveal new RNA–protein contacts in the box C/D sRNP and suggest a role for Nop5 in substrate binding and/or release.
Keywords: box C/D sRNAs, archaea, methylation, RNA–protein interaction, UV-cross-linking, box C/D sRNP
INTRODUCTION
Ribosomal RNA (rRNA) contains many 2′-O-methylated nucleotides that assist in rRNA folding and ribosome function (Decatur and Fournier 2002). The 2′-O-methylation of rRNA is catalyzed by box C/D small nucleolar ribonucleoprotein particles (snoRNPs) and sno-like RNPs (sRNPs) in eukaryotes and archaea, respectively (Dennis and Omer 2005; Reichow et al. 2007). In archaea, the box C/D sRNPs are composed of a box C/D sRNA and the proteins L7Ae, fibrillarin, and Nop5 (Dennis and Omer 2005). Each box C/D sRNA contains two related sequence elements, the box C/D and box C′/D′ motifs (Fig. 1A), which each bind a molecule of L7Ae. The binding of L7Ae to the sRNA is required for the recruitment of Nop5 and fibrillarin (Omer et al. 2002; Tran et al. 2003). The guide region of the sRNA, adjacent to either box D or D′, selects the rRNA modification site by base-pairing with the target sequence (Fig. 1A). Fibrillarin is the methyltransferase subunit (Dennis and Omer 2005) and interacts with the N-terminal domain of Nop5 (Fig. 1B, NTD; Aittaleb et al. 2003; Ye et al. 2009). The coiled-coil domain of Nop5 is required for self-dimerization, effectively linking the RNPs formed at the C/D and C′/D′ boxes and generating a pseudo-symmetrical complex that is essential for methylation activity (Aittaleb et al. 2003; Zhang et al. 2006). The C-terminal region of Nop5 contains the NOP domain, which interacts with both L7Ae and the box C/D sRNA (Ye et al. 2009).
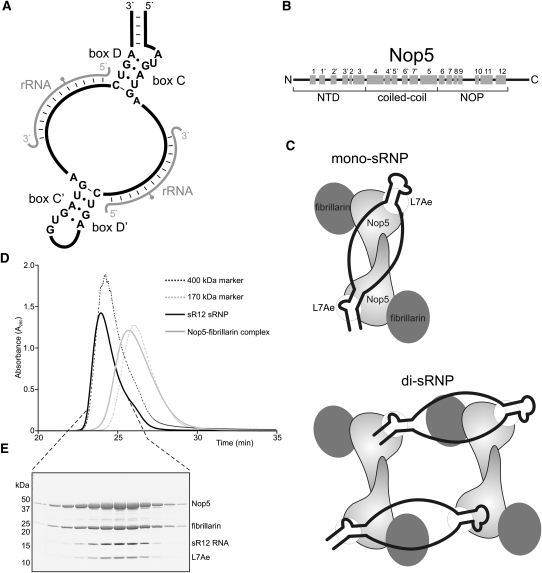
FIGURE 1.
In vitro assembled P. furiosus box C/D sRNPs are dimers. (A) Schematic representation of a box C/D sRNA. The conserved boxes, with consensus sequences, are indicated. (Gray) The rRNA target; (gray circle) the methylation target site. (B) Schematic representation of the structural organization in Nop5. The three domains are indicated below, and α-helices are represented by gray boxes, with the respective numbers above. (C) Schematic representation of the two models of box C/D sRNP architecture. (Black) The sRNA. The positions of the proteins in the two complexes are indicated. (D) Size exclusion analysis of in vitro assembled P. furiosus box C/D sRNP. Box C/D sRNPs were assembled using recombinant, purified P. furiosus proteins and in vitro transcribed sR12 RNA. The assembled complex was then separated on a Superdex 200 column. Protein elution from the column was monitored at 280 nm. The purified Nop5–fibrillarin complex and 400-kDa and 170-kDa markers were run as standards. (E) Fractionated sRNPs (D) were separated by SDS-PAGE and analyzed by staining with Coomassie (proteins) and Toluidine Blue O (RNA). The positions of the proteins and sR12 RNA are indicated on the right. The positions of the protein marker standards are indicated on the left.
The box C/D sRNPs belong to a diverse family of RNPs, found in both archaea and eukaryotes, which contain L7Ae-like proteins. In archaea both the box C/D and H/ACA sRNPs, which direct rRNA pseudouridylation, contain L7Ae (Kuhn et al. 2002; Omer et al. 2002; Rozhdestvensky et al. 2003). In eukaryotes, the family includes box C/D snoRNPs; H/ACA snoRNPs, which are both highly similar to their archaeal counterparts; the spliceosomal U4 small nuclear (sn)RNP; and selenoprotein messenger RNPs (Henras et al. 1998; Watkins et al. 1998, 2000; Nottrott et al. 1999; Allmang et al. 2002). 15.5K (Snu13 in yeast) is the L7Ae-like protein in both the U4 snRNP and the box C/D snoRNPs. The Nop5-like proteins, Nop56 and Nop58 in the snoRNPs and hPrp31 in the U4 snRNP, are specifically recruited to their respective complexes (Nottrott et al. 2002; Watkins et al. 2002; Schultz et al. 2006a,b; Liu et al. 2007).
Two structures of archaeal box C/D sRNPs have recently been published that have given rise to two different models of box C/D sRNP organization (Fig. 1C). One model, based on a structure generated by single-particle electron microscopy, proposes that the sRNP is a dimer, with four copies of each of the proteins and two sRNAs (Bleichert et al. 2009). A second model, based on a crystal structure of a half sRNA, comprised of a C/D motif and a single guide region, bound by L7Ae, Nop5, and fibrillarin, proposes that the sRNP is a monomer containing one sRNA and two copies of each protein (Ye et al. 2009). In this structure, a region of the conserved NOP domain, between α-helices 9 and 10 (Fig. 1B), of Nop5 formed a protuberance that was suggested to be important for the specificity of complex formation and to assist in maintaining the single-stranded guide/spacer regions. The two proposed models disagree in whether the sRNP is a monomer or dimer and, furthermore, in the organization of the sRNA throughout the complex (Fig. 1C). Either of the organization models for the RNA is, however, feasible within the context of a di-sRNP. We therefore set out to define whether sRNPs assembled in vitro using the components derived from other archaeal species form dimers and to use UV cross-linking to characterize RNA–protein contacts in the sRNP.
RESULTS AND DISCUSSION
In vitro assembled P. furiosus sRNPs are the expected size of a dimer
In order to analyze RNA–protein interactions in the box C/D sRNP, we first needed to establish a system with which to assemble these complexes in vitro. We therefore developed a two-step assembly process to reconstitute box C/D sRNPs using recombinant proteins and the sR12 box C/D RNA from Pyrococcus furiosus (see Materials and Methods). As a first step, L7Ae–sRNA complexes were assembled and purified. The binary complex was then incubated with recombinant Nop5 and fibrillarin, and the assembled complexes were separated on a Superdex 200 gel filtration column (Fig. 1D). The peak fractions were then analyzed by SDS-PAGE, and the RNA and protein components were revealed using Toluidine Blue O and Coomassie, respectively (Fig. 1E). The in vitro assembled P. furiosus sRNP contained all four components and eluted from the size exclusion column in a single peak that eluted slightly before the 400-kDa marker. Similar complexes were also observed with sR2 and sR10 sRNAs from P. furiosus. The in vitro assembled Methanocaldococcus jannaschii box C/D sRNP was shown to be a dimer with two sRNA molecules and four copies of each of the box C/D proteins (Bleichert et al. 2009). The expected size of a dimeric P. furiosus sRNP, containing two molecules of sRNA and four molecules of each protein, is 383 kDa. Our data are therefore consistent with the in vitro assembled P. furiosus sRNP also being a dimer.
The ALFR motif of Nop5 cross-links to the guide/spacer region of the box C/D sRNA
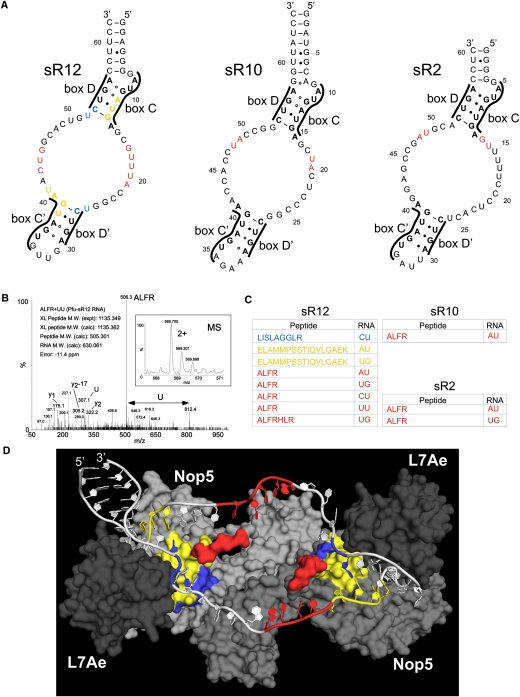
We were next interested in characterizing RNA–protein contacts in the assembled P. furiosus box C/D sRNP complexes. sRNPs, assembled as described above, were UV-irradiated, denatured, and hydrolyzed using ribonucleases and the endoproteinase trypsin, and the cross-linked peptides were enriched using a TiO2 column. Electrospray ionization (ESI)–mass spectrometric analysis of the enriched samples was then used to analyze the RNA–peptide conjugates, as described previously (Luo et al. 2008). For these experiments, three different sRNPs assembled on the sR12, sR10, or sR2 box C/D sRNAs were used (Fig. 2A). RNA–peptide conjugates derived from each assembled sRNP could be detected and their RNA and peptide moieties identified (Fig. 2B). Figure 2B shows an example of the MS-based sequence analysis of a peptide–RNA oligonucleotide derived from in vitro assembled RNP with sR12 RNA. The intact precursor mass (intact mass of the cross-linked conjugate) and the obtained fragment ions (under collision-induced decay [CID]) unambiguously revealed that this particular cross-linked peptide harbors the sequence ALFR, which is cross-linked to a UU dinucleotide. In each case, peptide sequences could be assigned to Nop5, and no peptides were observed from L7Ae or fibrillarin. We believe that UV-light-induced cross-links between L7Ae and the sRNA were most likely not observed because of the double-stranded/highly structured nature of the box C/D motif, to which L7Ae binds. Furthermore, fibrillarin has been proposed to not directly contact the sRNA in the absence of substrate (Bleichert et al. 2009; Ye et al. 2009). Most of the peptides for sR12 sRNP and all of the peptides for the sR10 and sR2 sRNPs contain the ALFR sequence, or a derivative of this region (Fig. 2C). The ALFR peptides, and two further peptides from the sR12 sRNP, were all derived from the protuberance that is comprised of the amino acids found in the loop between α-helices 9 and 10 in the NOP domain (Fig. 1B). This region of the protein was predicted to play a role in keeping the guide/spacer regions of the sRNA single-stranded (Fig. 2D; Ye et al. 2009). Interestingly, a reasonable part of this region was disordered and not visible in the crystal structures published so far.
FIGURE 2.
The AFLR motif contacts the guide/spacer regions of the box C/D sRNA. (A) Secondary structures of the P. furiosus sRNAs. The box motifs are indicated. The cross-link sites are colored according to panel C. (B) MS and MSMS spectra of a peptide–RNA cross-link derived from in vitro assembled RNP with sR12 RNA. The figure shows the intact mass and the charge state of the precursor that was selected for MSMS (small box) and the corresponding MSMS spectra that identified the cross-linked peptide and RNA moiety. Y-type fragment ions derived from the cross-linked peptide under CID (collision induced decay) are assigned as well as the m/z values for the nucleotide U in the lower m/z regime. (Arrow) The mass difference that corresponds exactly to a U. The experimentally determined precursor (cross-linked peptide–RNA oligonucleotide), the calculated peptide and RNA mass, and the mass deviation (error) in parts per million (ppm) are listed within the figure. (C) Cross-linked peptides and RNAs. In vitro assembled sRNPs were denatured and digested with trypsin and RNAses. RNA–peptide conjugates were purified and then characterized by ESI-mass spectrometric analysis. The colors correspond to the RNA and protein sites in panels A and D. (D) Structural model of sR12 box C/D sRNP showing RNA and protein contact sites. The structure of the Nop5, L7Ae, sRNA complex published by Ye et al. (2009) was modified to include the sR12 sRNA. Alanines were also used to join the section of the protuberance missing from the crystal structure. A surface view is shown for the proteins with Nop5 (gray and light gray) and L7Ae (dark gray) with the cross-linked peptides colored as in panel C. For clarity, we have not included fibrillarin in the image. The RNA is shown in white, with the cross-linked nucleotides colored as in panels A and C.
Using cross-linking in combination with mass spectrometry, we determined the composition of the nucleotides associated with the peptide (Fig. 2C). It is, however, important to note that we cannot assign a 5′-to-3′ order to them based on the mass spectrometric data. In identifying the position of the nucleotides in each sRNA, we focused on regions that are single-stranded or not involved in Watson-Crick base-pairing. In addition, we also used the published sRNP crystal structure to help localize the cross-linked region of the RNA. The peptides at the base of the protuberance/stalk (Fig. 2C, yellow and blue) cross-linked to sequences in and around stem II of the box C/D or C′/D′ motifs (Fig. 2A,D), supporting the previous observation that stem II is involved in the sequence-specific recruitment of Nop5 (Ye et al. 2009). The RNA coupled to the ALFR and ALFR-containing peptides correlated with the guide/spacer sequences. The variation in cross-link sites on different RNAs may be due to the different guide/spacer sequences in sRNA. This region of the sRNA is, however, single-stranded, with no discernable sequence conservation. It is therefore also possible that Nop5 makes slightly different contacts in each RNP. This may explain why this region of the protein is not resolved in the crystal structure. This is the first demonstration that Nop5 specifically contacts the sRNA outside of the C/D or C′/D′ motifs and supports the previous proposal that this protein plays a role in organizing the structure of the sRNA. This novel RNA–protein interaction may also assist in the assembly and function of the box C/D sRNPs.
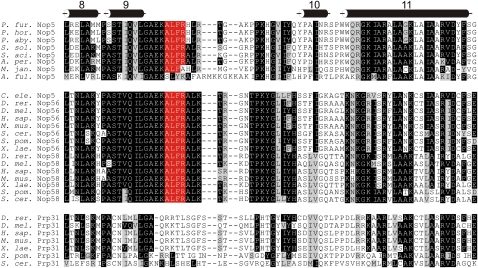
The ALFR motif is conserved in archaeal Nop5 and eukaryotic Nop56 and Nop58 but not in hPrp31
If the ALFR motif is important for box C/D sRNP assembly, we would expect this motif to be highly conserved in both the archaeal and eukaryotic box C/D NOP-domain proteins. We therefore examined the evolutionary conservation of the ALFR motif. The NOP domain is the most conserved region of this family of proteins and is predicted to make sequence and/or structure-specific contacts in each complex (Liu et al. 2007; Ye et al. 2009). Sequences of the NOP-domain of archaeal Nop5 and eukaryotic Nop56, Nop58, and Prp31 were aligned based on the available crystal structures of Nop5 and hPrp31 (Fig. 3; Aittaleb et al. 2003; Liu et al. 2007; Oruganti et al. 2007; Ye et al. 2009). This revealed that the ALFR region is highly conserved in the archaeal (Nop5) and eukaryotic (Nop56 and Nop58) box C/D proteins. The sequence of this region of the eukaryotic U4 snRNP protein Prp31 is also conserved, but the sequence is different from that seen in the sRNP/snoRNP proteins (Fig. 3). This is consistent with the idea that the ALFR motif plays an important role in box C/D sRNP formation and/or structure. Furthermore, this also fits with the proposal that the respective regions play an important role in differentiating between binding of hPrp31 and Nop56/58 proteins to the U4 snRNA and to the box C/D snoRNAs in eukaryotes, respectively (Liu et al. 2007; Ye et al. 2009).
FIGURE 3.
The ALFR motif is highly conserved in archaeal Nop5 and eukaryotic Nop56 and Nop58. Sequences of archaeal Nop5 and eukaryotic Nop56, Nop58, and Prp31 were aligned. The region encompassing α-helices 8 though 11 of the NOP domain is shown. The positions of the α-helices are indicated at the top. Highly conserved amino acids are shown in white with a black background. Conserved amino acids are shown in black with a gray background. The ALFR motif is shown in white with a red background.
The ALFR-motif–sRNA interaction is essential for efficient box C/D sRNP formation
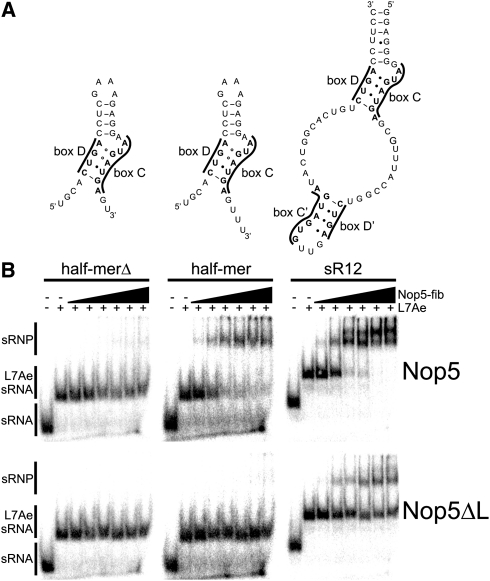
Having revealed that the Nop5 ALFR motif binds the box C/D sRNA and that this region of the protein is highly conserved, we were next interested in determining whether this interaction was important for sRNP formation in vitro. To test this possibility, a mutant Nop5 was generated in which the ALFR loop was deleted (Nop5ΔL). In addition, we wanted to test whether the single-stranded RNA region contacted by the ALFR sequence was also important for RNP formation. We therefore examined protein-binding to a full-length sR12, an sR2 half-mer sRNA, and an sR2 half-mer sRNA in which most of the sequence downstream from box C had been deleted (half-merΔ) (Fig. 4A). The half-mer sRNAs were comprised of a box C and D sequence joined by a tetraloop. Radiolabeled RNAs were generated corresponding to the wild-type and mutant sRNAs and incubated with the recombinant proteins. The complexes formed were then analyzed by electrophoretic gel mobility shift assay.
FIGURE 4.
The ALFR motif and the guide/spacer region are important for efficient box C/D sRNP formation. (A) Schematic representation of sR12 and the half-mer sRNAs. The conserved boxes are indicated. (B) Electrophoretic gel mobility shift analysis. sRNAs were assembled with L7Ae and then incubated with increasing amounts of Nop5–fibrillarin complex (25 nM, 50 nM, 100 nM, 250 nM, 500 nM, and 1 μM). The Nop5–fibrillarin complex contained either wild-type or mutant Nop5, in which the ALFR sequence had been deleted (Nop5ΔL), as indicated on the right. The assembled complexes were then separated on a native polyacrylamide gel and visualized using a PhosphorImager. The RNA used and the proteins added are indicated at the top. The positions of the free sRNA, the L7Ae-sRNA complex, and assembled sRNP are indicated on the right.
The three RNAs efficiently formed a stable complex with L7Ae. When Nop5 and fibrillarin were added to this binary complex, the full-length and half-mer RNAs both efficiently formed sRNP complexes. In contrast, the shorter half-mer RNA (half-merΔ), lacking the sequence downstream from box C, did not bind Nop5 and fibrillarin. This region of the sRNA therefore plays an important role in RNP formation. It is important to note that while this RNA contains both sR2 cross-link sites, we believe that the contact site extends beyond this and that more single-stranded sequence is important for the Nop5 interaction with this RNA. The mutant Nop5, lacking the ALFR motif, was unable to support sRNP formation with either of the half-mer RNAs. Furthermore, the mutant Nop5 showed, relative to the wild-type protein, reduced binding to the full-length sRNA-L7Ae complex and never achieved >50% complex formation. A double band was seen for the assembled RNP with the wild-type Nop5. In contrast, only a single RNP band was seen for the mutant Nop5, suggesting that the complexes formed with the mutant protein differ from the wild-type complex. This therefore implies that the interaction between the ALFR motif and the region of the sRNA downstream from box C/C′ is required for efficient box C/D sRNP formation. This region is not conserved between the different sRNAs, and it is likely that it is recognized as a single-stranded region adjacent to the C and C′ boxes.
CONCLUSIONS
We have shown a novel interaction between Nop5 and the guide/spacer region of the box C/D sRNA in Archaea that is important for sRNP formation. The region of Nop5 involved in this interaction is part of a protrusion found between helices 9 and 10 of the NOP domain (Fig. 1B) that was previously predicted to be important for the organization of the guide regions (Ye et al. 2009). Our data support a role for Nop5 in presenting the guide region for substrate binding and possibly directly in sRNA–rRNA base-pairing. Nop5 binding involves specific contacts to both stem II in the box C/D and box C′/D′ motifs and the single-stranded guide/spacer regions. This indicates that Nop5 plays an important role in sRNP formation and function. We have used the mono-sRNP model (Ye et al. 2009) to interpret the cross-link data, and it is very likely that the box C/D and C′/D′ motifs would be bound in the same way in a di-sRNP (Bleichert et al. 2009). The key difference would be in the positioning of the guide/spacer regions. The in vitro assembled P. furiosus sRNPs appear to be di-sRNPs, and in this complex both RNA organization models are feasible. Unfortunately, based on our cross-link data, we are unable to determine whether the mono-sRNP RNA organization model is correct, and we cannot comment on how this fits to the di-sRNP model until a higher-resolution model is available.
The ALFR motif is also conserved in the eukaryotic box C/D snoRNP proteins Nop56 and Nop58. Earlier work showed that a minimal single-stranded sequence was needed between boxes C and D for snoRNP formation/snoRNA accumulation in Xenopus (Watkins et al. 1996). This implies that, like the archaeal complexes, a single-stranded region of the snoRNA is required for RNP formation and suggests that the ALFR–RNA interaction, between Nop56/Nop58 and the snoRNA, occurs in the eukaryotic complexes. The equivalent region of hPrp31 was shown to assist RNA-binding specificity and to discriminate between different 15.5K-containing complexes (Liu et al. 2007; Ye et al. 2009). It is therefore likely that the ALFR motif of eukaryotic Nop56 and Nop58 helps these proteins specifically bind snoRNP complexes.
MATERIALS AND METHODS
Cloning, mutagenesis, and protein production
Coding sequences of L7Ae, fibrillarin, and Nop5 were PCR-amplified from P. furiosus genomic DNA and cloned into pET-M11 (EMBL), pGEX-6p-1 vector (GE Healthcare), and pET28a (Novagen), respectively. To generate the Nop5 loop mutant, Nop5ΔL, the nucleotides encoding amino acids 293–300 (ALFR) were deleted using the Phusion site-directed mutagenesis system (Finnzymes).
Escherichia coli BL21 (DE3) cells expressing hexahistidine-tagged L7Ae were lysed in 50 mM Tris (pH 7.5), 500 mM NaCl, and 2 mM β-mercaptoethanol using a microfluidizer (Microfluidics), and the protein was purified using Ni2+-NTA beads. To remove the nucleic acid contaminants, the beads were incubated in 50 mM Tris (pH 7.5), 2 M LiCl, and 2 mM β-mercaptoethanol. The protein was eluted using imidazole and dialyzed against 20 mM Tris (pH 7.5), 150 mM NaCl, and 2 mM DTT containing TEV protease to remove the hexahistidine tag. The cleaved tag and protease were subsequently removed using Ni2+-NTA beads. The flow-through was concentrated and then further purified using a Superdex-75 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2 mM DTT.
Nop5/Nop5ΔL and fibrillarin were coexpressed in E. coli Rosetta2 (DE3) cells. The cells were lysed in 50 mM Tris (pH 8.0), 1 M NaCl, 2 mM DTT, 5% glycerol complemented with 10 μg/mL DNaseI, 3 mM MgCl2, and 100 μg/mL lysozyme using a microfluidizer. The Nop5–fibrillarin complex was then purified using gluthathione-Sepharose (GE Healthcare) and eluted using reduced L-gluthathione. The protein was dialyzed against 50 mM Tris (pH 7.5), 1 M NaCl, 5% glycerol, 0.5 mM EDTA, and 2 mM DTT with PreScission protease (GE Healthcare). The cleaved GST moiety and the protease were removed using gluthatione-Sepharose, and the complex was purified using a Superdex-200 column (GE Healthcare).
Electrophoretic gel mobility shift assays
Half-mer sRNAs were synthesized by Dharmacon and labeled using [γ-32P]ATP. The full-length box C/D RNAs were synthesized by in vitro transcription using T7 RNA polymerase and PCR-amplified templates from P. furiosus genomic DNA. Labeled RNAs were mixed with 500 nM L7Ae and varying concentrations of Nop5–fibrillarin complex (25 nM to 1 μM) in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM DTT, and 0.5 mg/mL tRNA. Reactions were incubated for 15 min at 70°C followed by 30 min of incubation at 20°C. Protein–RNA complexes were then resolved on a 6% non-denaturing polyacrylamide gel at 4°C and visualized using a PhosphorImager.
Analysis of RNA–protein cross-links
Assembly of box C/D sRNPs was done in a stepwise manner. Equimolar amounts of sRNA and L7Ae were incubated in 10 mM HEPES (pH 7.3), 100 mM NaCl, and 2 mM DTT for 15 min at 70°C, and the complex was purified using a Superdex-75 column (GE Healthcare) and concentrated. Purified Nop5–fibrillarin complex was slowly added to the concentrated binary complex, and the assembled complex was purified using a Superdex-200 gel filtration column (GE Healthcare). Fractions containing the sRNP were pooled and concentrated.
Reconstituted sRNP complexes (1 mg in 100 μL) were UV-irradiated at 254 nm, dried, and dissolved in 4 M urea, 50 mM Tris-HCl (pH 7.5). After dilution to 1 M urea using 50 mM Tris-HCl (pH 7.5), the RNA was digested using RNase A and RNase T1, followed by digestion of the protein moiety using trypsin (Urlaub et al. 2000, 2002; Kuhn-Holsken et al. 2005). RNA-cross-linked peptides were enriched using a titanium dioxide (TiO2) column, as described (Larsen et al. 2005). Enriched tryptic peptides were purified using a C18 trap column and then separated on an analytical C18 capillary column coupled with an API-Q-TOF mass spectrometer, as described previously.
ACKNOWLEDGMENTS
We thank Reinhard Lührmann for generous support, Elke Penka for skilled technical assistance, and other members of the groups for helpful discussions. Thanks also to Kenneth McKeegan for critically reading the manuscript and to Claudia Schneider for help with the figures. H.G. was supported by a PhD scholarship from the International Max Planck Research School (IMPRS) for molecular biology. H.U. was supported by a EURASNET YIP grant within the 6th EU framework. N.J.W was supported by a grant from the BBSRC.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2380410.
REFERENCES
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol 10: 256–263 [DOI] [PubMed] [Google Scholar]
- Allmang C, Carbon P, Krol A 2002. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Gagnon KT, Brown BA II, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ 2009. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science 325: 1384–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Dennis PP, Omer A 2005. Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694 [DOI] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JF, Tran EJ, Maxwell ES 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res 30: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Holsken E, Lenz C, Sander B, Lührmann R, Urlaub H 2005. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA 11: 1915–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ 2005. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 4: 873–886 [DOI] [PubMed] [Google Scholar]
- Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, Carlomagno T, Wahl MC 2007. Binding of the human Prp31 Nop domain to a composite RNA–protein platform in U4 snRNP. Science 316: 115–120 [DOI] [PubMed] [Google Scholar]
- Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC 2008. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol Cell 32: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R 1999. Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J 18: 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Urlaub H, Lührmann R 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J 21: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H, Robinson H, Terns MP, Terns RM, Yang W 2007. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol 371: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G 2007. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A, Nottrott S, Hartmuth K, Lührmann R 2006a. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem 281: 28278–28286 [DOI] [PubMed] [Google Scholar]
- Schultz A, Nottrott S, Watkins NJ, Lührmann R 2006b. Protein–protein and protein–RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol Cell Biol 26: 5146–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Maxwell ES 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J 22: 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Kostka S, Grelle G, Lührmann R 2000. A general approach for identification of RNA–protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA–protein contacts in native U1 and U4/U6·U5 snRNPs. J Biol Chem 275: 41458–41468 [DOI] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Lührmann R 2002. A two-tracked approach to analyze RNA–protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26: 170–181 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Leverette RD, Xia L, Andrews MT, Maxwell ES 1996. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2: 118–133 [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Lührmann R 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol 22: 8342–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L 2009. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci 106: 13808–13813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Champion EA, Tran EJ, Brown BA II, Baserga SJ, Maxwell ES 2006. The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA 12: 1092–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]