Abstract
Enzymes generally are thought to derive their functional activity from conformational motions. The limited chemical variation in RNA suggests that such structural dynamics may play a particularly important role in RNA function. Minimal hammerhead ribozymes are known to cleave efficiently only in ∼10-fold higher than physiologic concentrations of Mg2+ ions. Extended versions containing native loop–loop interactions, however, show greatly enhanced catalytic activity at physiologically relevant Mg2+ concentrations, for reasons that are still ill-understood. Here, we use Mg2+ titrations, activity assays, ensemble, and single molecule fluorescence resonance energy transfer (FRET) approaches, combined with molecular dynamics (MD) simulations, to ask what influence the spatially distant tertiary loop–loop interactions of an extended hammerhead ribozyme have on its structural dynamics. By comparing hammerhead variants with wild-type, partially disrupted, and fully disrupted loop–loop interaction sequences we find that the tertiary interactions lead to a dynamic motional sampling that increasingly populates catalytically active conformations. At the global level the wild-type tertiary interactions lead to more frequent, if transient, encounters of the loop-carrying stems, whereas at the local level they lead to an enrichment in favorable in-line attack angles at the cleavage site. These results invoke a linkage between RNA structural dynamics and function and suggest that loop–loop interactions in extended hammerhead ribozymes—and Mg2+ ions that bind to minimal ribozymes—may generally allow more frequent access to a catalytically relevant conformation(s), rather than simply locking the ribozyme into a single active state.
Keywords: conformational change, fluorescence spectroscopy, fluorescence microscopy, molecular dynamics simulation, single molecule FRET
INTRODUCTION
Noncoding RNA plays a central role in a wide variety of cellular processes (Amaral et al. 2008), providing compelling motivation to understand how RNA function arises from structure and dynamics. Modern views of enzyme catalysis encompass notions such as induced fit, conformational capture, active site preorganization, and substrate strain to explain the efficient turnover of a substrate by an enzyme (Fersht 1999; Hammes-Schiffer and Benkovic 2006). These concepts all invoke conformational flexibility (sampling) in both enzyme and substrate, derived from their structural dynamics, or the sum of all thermally activated global and local motions. While quite well established for protein enzymes (Fersht 1999; Hammes-Schiffer and Benkovic 2006), the potentially intricate linkage between function and structural dynamics is still much more tenuous for RNA (Al-Hashimi and Walter 2008).
In recent years, X-ray crystallography has increasingly illuminated the intricate folds associated with the catalytic activity of RNA enzymes, or ribozymes, yet such structural snapshots lack the animation that generally accompanies enzyme action. The hammerhead ribozyme is a self-cleaving small noncoding RNA originally discovered in satellite RNAs associated with plant viruses. More recently, hammerhead motifs have been found in the genomes of the newt, schistosome trematodes, cave crickets, and rodents, suggesting that the motif has arisen multiple times by convergent evolution (Salehi-Ashtiani and Szostak 2001; Martick et al. 2008a). Minimal hammerheads cleave efficiently only in high ionic strength (Murray et al. 1998), while extended versions of the ribozyme containing native, peripheral tertiary interactions show enhanced catalytic activity, particularly at lower, and thus more physiologically relevant, Mg2+ concentrations (De la Peña et al. 2003; Khvorova et al. 2003; Canny et al. 2004). Recent crystal structures of an extended hammerhead ribozyme (Martick and Scott 2006; Chi et al. 2008; Martick et al. 2008b) resolved many long-standing inconsistencies with biochemical data (McKay 1996; Blount and Uhlenbeck 2005; Nelson and Uhlenbeck 2008a), and suggested that the peripheral loop–loop interactions affect cleavage activity at a distance by changing the structure of the catalytic core (Westhof 2007). Since the crystal structures provide only a static picture, however, details of how the loop–loop interactions affect the structural dynamics of the ribozyme and how such effects may relate to catalytic activity have largely remained elusive.
Understanding how dynamics influence function in RNA is crucial; RNA molecules have limited chemical diversity, suggesting their activity in many cases may depend particularly strongly on conformational changes (Al-Hashimi and Walter 2008). Experimental studies on both the minimal and extended forms of the hammerhead, using mutagenesis (Nelson and Uhlenbeck 2008b) and biophysical techniques such as NMR, 2-aminopurine fluorescence, and X-ray crystallography (summarized in Blount and Uhlenbeck 2005) have demonstrated that this ribozyme, and particularly its conserved catalytic core, is dynamic. Whether the dynamic nature of the ribozyme enhances or limits catalysis has been unclear; an attractive model is that the minimal hammerhead is floppy whereas the tertiary contacts in the extended form “lock” it into a catalytically active state. Similarly, while metal ions play key roles in RNA folding and dynamics, there is little information describing how Mg2+ affects the dynamics of the extended hammerhead ribozyme.
To investigate the role of global dynamics in the extended hammerhead ribozyme at varying Mg2+ concentrations, we here have used complementary time-resolved fluorescence resonance energy transfer (tr-FRET) and single-molecule FRET (sm-FRET) techniques to monitor the global structure and dynamics of three hammerheads based on the Avocado Sunblotch Viroid (ASBV) sequence. Their loop sequences represent, respectively, wild-type, partially disrupted, and fully disrupted loop–loop interaction sequences. To monitor structural dynamics at an atomistic level, we furthermore conducted MD simulations on three additional variants, based on the available Schistosoma mansoni crystal structure of the hammerhead ribozyme. These S. mansoni variants are distinct from the ASBV ribozymes, but also represent intact, partially disrupted, and fully disrupted loop–loop interaction sequences. We find that the wild-type loops help populate, at both the global and the local levels, a conformation(s) that harbors catalytic activity. Our results begin to establish a correlation between RNA structural dynamics and function, suggesting that loop–loop interactions in extended hammerhead ribozymes, and Mg2+ ions that bind to minimal ribozymes, allow more frequent access to catalytically relevant conformations, rather than locking the ribozyme into a single catalytically active state.
RESULTS
Design of hammerhead ribozymes for FRET
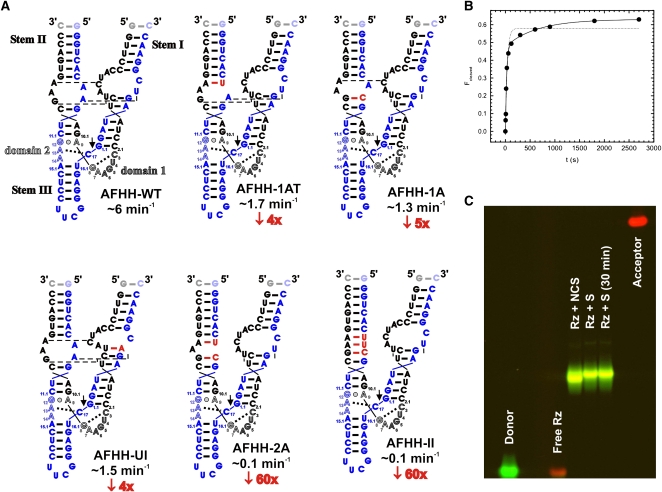
In designing a hammerhead for FRET studies, we tested sequences derived from several species, each containing internal loops in Stems I and II. A sequence with internal loops was found to be ideal, since it allows for labeling the ends of these stems with donor and acceptor fluorophores without interfering with the loop–loop interactions and catalytic activity (notably, internal labeling of the capping Stem II loop of the S. mansoni hammerhead ribozyme [Martick and Scott 2006] led to significant activity loss [data not shown]). Our final “wild-type” sequence was based on the hammerhead ribozyme from the Avocado Sunblotch Viroid (ASBV) (Fig. 1A; Hutchins et al. 1986) (AFHH-WT). Several changes were made to the native ASBV sequence to convert it to a model system suitable for biophysical studies (Figs. 1A, 2A); capping of Stem III with a stable UUCG tetraloop, removal of a 4-nucleotide (nt) bulge in Stem II, and mutation of 4 nt in the ribozyme strand of Stem I increased the cleavage rate; and a base pair flip in Stem I reduced but did not fully eliminate the second phase of otherwise biphasic cleavage kinetics (Fig. 1B), presumably by suppressing misfolding. A nondenaturing, fluorescence detected, electrophoretic mobility shift assay shows a fluorophore labeled version of this hammerhead (Fig. 2A) to only have one major global fold (Fig. 1C). A homology-based atomistic model of this wild-type ribozyme, containing unmodified ASBV loop sequences and termed AFHH-WT, was developed and is shown in Figure 2B. To determine the effect of disrupting the loop–loop interactions on the structure and dynamics of the hammerhead, we generated a series of mutant ribozymes that disrupted cleavage activity to varying extents (Fig. 1A). We chose two mutants to further characterize: The first mutant (AFHH-1A) contains a single A-to-C base mutation in the Stem II loop designed to extend the base pairing in Stem II and so partially disrupt the Stem I–II loop–loop interactions, while the second mutant (AFHH-2A) carries an additional A-to-U mutation in the Stem II loop designed to fully disrupt the Stem I-II interactions (Figs. 1A, 2A). Notably, extending the base pairing from the top of the bulge rather than from the bottom results in nearly the same cleavage activity as for AFHH-1A (Fig. 1A), suggesting that our results do not depend on the specific nucleotide modified.
FIGURE 1.
Design and properties of AFHH hammerhead ribozyme and its variants. (A) Cleavage rate constants of AFHH variants with mutations to the loop regions. “Wild-type” AFHH-WT is shown in the top left. For each mutant ribozyme, the nucleotides that have been modified are highlighted in red. Observed cleavage rate constants are shown (blue) along with the relative reduction in rate (red) compared to AFHH-WT. All cleavage assays were conducted under standard conditions: 50 mM Tris-HCl (pH 8.0), 25°C, 1 mM MgCl2. (B) Representative cleavage assay time course of the chemically synthesized AFHH-WT ribozyme under standard conditions: 50 mM Tris-HCl (pH 8.0), 25°C, 1 mM MgCl2. Single (dashed line) and double (solid line) exponential fits are shown, with the double exponential giving a better fit (R2 of 0.99 vs. 0.97). The observed rate constant for the fast phase is 2.4 min−1 (fraction cleaved: 50%), that of the slow phase is 0.09 min−1 (15%). (C) Testing for structural homogeneity of the AFHH-WT ribozyme–substrate complex using a FRET-based electrophoretic mobility shift assay. The ribozyme strand (Rz) and noncleavable (NCS) or cleavable (S) substrate strands were annealed and run on a 10% nondenaturing polyacrylamide gel as described in Materials and Methods. The complex loaded into the “Rz+S (30 min)” lane was incubated for an additional 30 min at 37°C after the standard annealing protocol. Donor emission is shown in green, acceptor emission in red.
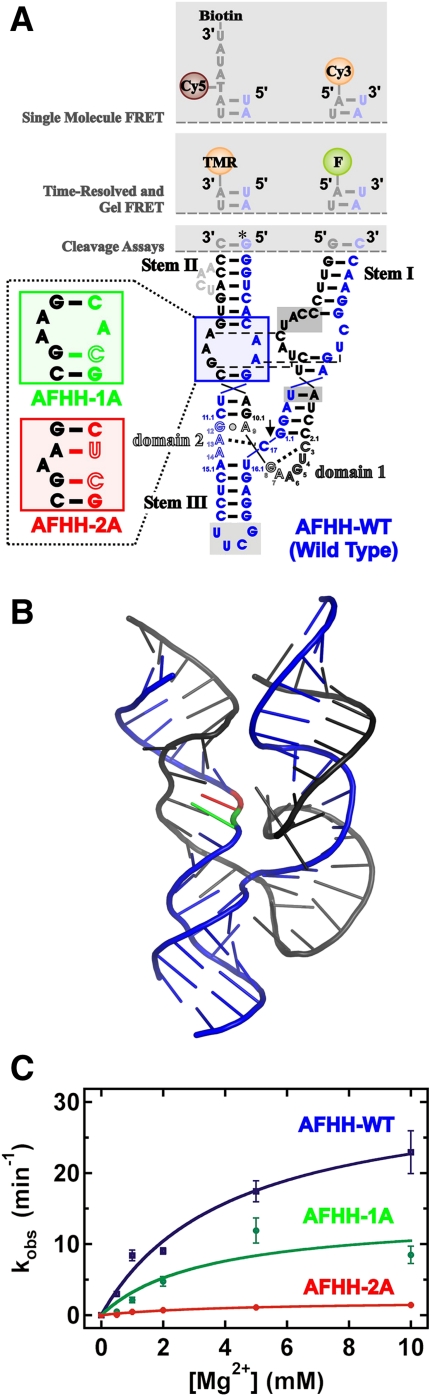
FIGURE 2.
Modifications and cleavage activity of the hammerhead ribozymes used in this study. (A) Secondary structure of the AFHH-WT (AFHH; blue), as well as the partially disrupted (AFHH-1A; green) and fully disrupted (AFHH-2A; red) mutants, based on the sequence from Avocado Sunblotch Viroid (ASBV). Several putative interactions between the internal loops of Stems I and II are indicated as thin dashes. (B) Homology model showing possible orientation of the helices. Colors are the same as in A; the green base is the mutation in AFHH-1A, the red base is the additional mutation in AFHH-2A. (C) Characterization of cleavage rate dependence on Mg2+ concentration under standard conditions (50 mM Tris-HCl [pH 8.0], 25°C).
Wild type yields the highest cleavage activity
To compare the relative activities of our AFHH-WT and mutant ribozymes, we conducted cleavage assays under single-turnover conditions using radiolabeled substrate and obtained the cleavage rate constants of each ribozyme variant as a function of magnesium concentration (Fig. 2C). At near-physiological Mg2+ concentrations (0.5 mM), the AFHH-WT ribozyme cleaves already at 3.0 min−1, faster than the rate constants measured in 10 mM Mg2+ for minimal hammerheads, and comparable to those observed for other extended hammerheads (De la Peña et al. 2003; Khvorova et al. 2003; Canny et al. 2004; Nelson et al. 2005; Weinberg and Rossi 2005; Roychowdhury-Saha and Burke 2006; Boots et al. 2008). In contrast, the fully disrupted AFHH-2A mutant has a rate constant of 0.051 min−1 at 0.5 Mg2+ that saturates at just 1.4 min−1 in 10 mM Mg2+, making it very comparable to the extensively characterized minimal hammerheads (McKay 1996; Stage-Zimmermann and Uhlenbeck 1998). As expected, the observed cleavage rate constants for the partially disrupted AFHH-1A ribozyme fall between those of the AFHH-WT and fully disrupted AFHH-2A mutant at all Mg2+ concentrations (Fig. 2C). Fitting a noncooperative, hyperbolic binding curve to the data yields Mg2+ half-titration points, Mg1/2, of 4.2, 3.3, and 4.3 mM for the AFHH-WT, AFHH-1A, and AFHH-2A mutant ribozymes, respectively, in both the presence and absence of a 100 mM Na+ background (Table 1). The maximum rate constant at saturating Mg2+ concentration of the AFHH-WT ribozyme reaches ∼25 min−1, somewhat faster than that of the extended hammerhead ribozyme from the tobacco ringspot virus satellite (sTRSV) RNA (Kisseleva et al. 2008), but slower than the reported >200 min−1 for the S. mansoni (Mg1/2 > 10 mM Mg2+) (Canny et al. 2004; Kim et al. 2005; Boots et al. 2008) and in vitro selected RzB hammerheads (Mg1/2 ≈ 2 mM Mg2+) (Saksmerprome et al. 2004). Such differences may be due to variations in the protocols used for analyzing cleavage activity (Stage-Zimmermann and Uhlenbeck 1998) and/or be true evidence for the catalytic diversity of extended hammerhead ribozymes (Shepotinovskaya and Uhlenbeck 2008). Regardless, our work now establishes a fourth extended hammerhead particularly suitable for biophysical studies, as demonstrated in the following.
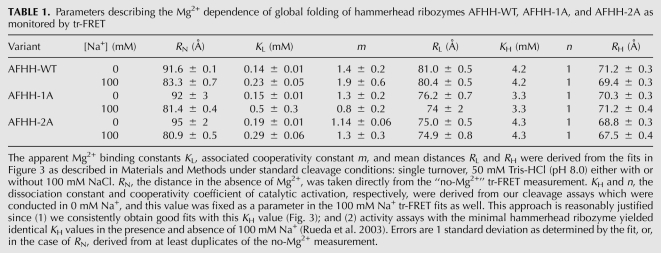
TABLE 1.
Parameters describing the Mg2+ dependence of global folding of hammerhead ribozymes AFHH-WT, AFHH-1A, and AFHH-2A as monitored by tr-FRET
AFHH-WT shows the broadest Stem I–II distance distribution
We used tr-FRET to monitor the global structure distribution of our three hammerhead ribozymes. This approach allows us to derive equilibrium distance distributions between the donor and acceptor fluorophores at the ends of Stems I and II as a function of Mg2+ concentration (Rueda et al. 2003). Figure 3 shows the measured mean distance and full width at half-maximum (FWHM) of each distance distribution plotted against the Mg2+ concentration in either the absence or presence of a 100 mM Na+ background (which ensures proper secondary structure formation even at low ionic strength). The primary effect of the sodium background is a slightly smaller initial mean distance, indicating that some tertiary folding of the ribozymes occurs in the presence of monovalents alone, similar to our previous observations on minimal hammerheads (Rueda et al. 2003).
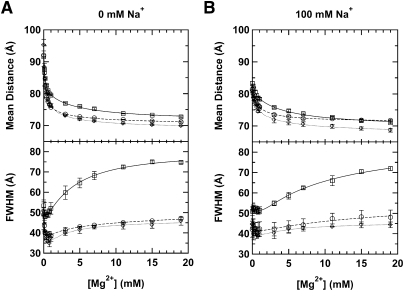
FIGURE 3.
Distance distributions between stems I and II of the AFHH-WT (solid line), as well as partially disrupted (AFHH-1A; dashed line) and fully disrupted (AFHH-2A; dotted line) hammerhead mutants monitored by tr-FRET. (A) Mean distance and full width at half-maximum (FWHM) of the distance distributions (modeled as a single Gaussian) as a function of Mg2+ concentration under standard conditions (50 mM Tris-HCl [pH 8.0], 25°C). (B) tr-FRET measurements as in A, but the buffer included 100 mM Na+.
Under all conditions, we observed a two-phase decrease in the mean distance accompanied by first a decrease then an increase in FWHM with increasing Mg2+ concentration, which we interpret as a (at least) two-step folding process, similar to our previous results on minimal hammerheads (Rueda et al. 2003). We fit the Mg2+ dependences of distance and FWHM in the absence of Na+ to a two-step Mg2+ binding model, and obtained Mg1/2 values of 0.14, 0.15, and 0.19 mM for the first step of folding for the AFHH-WT, AFHH-1A, and AFHH-2A mutant ribozymes, respectively (with cooperativity coefficients of 1.1–1.4, close to unity) (Table 1). These apparent Mg2+ affinities are approximately five- to 10-fold tighter (lower Mg1/2) than those previously observed for the catalytically slower minimal ribozymes. (As expected, a background of 100 mM competing Na+ reduces the Mg2+ affinity, by approximately two- to threefold) (Table 1). These observations are generally consistent with previous steady-state FRET measurements on extended hammerheads (Penedo et al. 2004; Boots et al. 2008) and suggest that all three of our ribozymes fold at lower ionic strength than their minimal counterparts (Rueda et al. 2003). Just as with the minimal hammerhead ribozymes (Rueda et al. 2003), however, our combined cleavage and tr-FRET assays reveal a lower-affinity, noncooperative folding step of the extended ribozymes that coincides with their catalytic activation in the low-millimolar Mg2+ range (3–4 mM) (Table 1). This observation may be consistent with observations of the Mg2+-induced catalytic activation (Canny et al. 2004) and folding of the Schistosoma hammerhead as monitored by steady-state FRET (Boots et al. 2008). Boots et al. report a single phase of folding with half-titration points of ∼1 mM Mg2+ (in 100 mM NaCl) as monitored by steady-state FRET. Their data also show a rise in FRET beyond this initial transition at tens of millimolar Mg2+, which could represent the second folding step that is observed here by tr-FRET and is related to catalytic activity. Our more detailed tr-FRET experiments show that during this second folding step the Stem I–II distance slightly decreases further, whereas the FWHM increases dramatically for AFHH-WT. We note that despite this qualitative agreement, our wild-type hammerhead ribozyme exhibits Mg2+ half-titration points for the first and second steps of folding of 0.14–0.19 mM (depending on monovalent background) and 4.2 mM, respectively (Table 1), that are approximately five- to 20-fold tighter than those reported for the previously characterized extended hammerhead ribozymes from sTRSV and S. mansoni (Canny et al. 2004; Kim et al. 2005; Boots et al. 2008) and appear more similar to those of the selected RzB hammerhead (Saksmerprome et al. 2004). We conclude that our wild type is a hammerhead with comparably high Mg2+ affinity, and therefore well suited for biophysical studies. Importantly, we find our observations for the wild type to be less pronounced for the AFHH-1A and AFHH-2A mutants with partially and fully disrupted loop–loop interaction sequences, respectively (Fig. 3). The relative increase of the FWHM in AFHH-WT over that of the mutants suggests that in AFHH-WT the Stem I–II distance distribution is the broadest.
AFHH-WT exhibits the most sm-FRET states and the greatest interstem dynamics
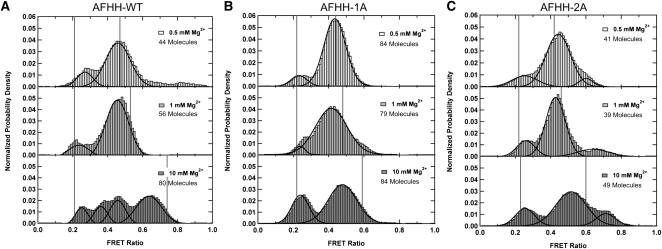
To determine the source of the broad FWHMs measured by tr-FRET at low millimolar Mg2+ concentrations and probe dynamics of individual molecules over time, we used total internal reflection fluorescence (TIRF)-based sm-FRET. In contrast to the similarly small hairpin ribozyme and an aptazyme based on the minimal hammerhead, which both show dynamics between only two sm-FRET states (Zhuang et al. 2002; Ditzler et al. 2008; de Silva and Walter 2009), we observed at least four distinguishable states in our extended hammerhead ribozymes (Fig. 4A). The individual FRET time traces within a set of conditions showed considerable heterogeneity with multiple types of dynamics between subsets of states; examples for the AFHH-WT as well as AFHH-1A and AFHH-2A mutants are found in Figure 5A–C. “Heterogeneity” refers here to a lack of interconversion, within the accessible observation window, between kinetically distinct molecular species, also observed for other RNAs such as the hairpin (Zhuang et al. 2002; Bokinsky et al. 2003; Rueda et al. 2004; Ditzler et al. 2008) and group I intron ribozymes (Solomatin et al. 2010), as well as the tetraloop–receptor tertiary interaction (Fiore et al. 2009). To analyze our data, we first plotted aggregate FRET histograms for each set of conditions and fit them with Gaussians to identify FRET states (Fig. 4A–C). These histograms define multiple FRET states under all conditions, with more FRET states appearing and the distribution among states becoming more even as the Mg2+ concentration increases. Most significantly, we observed a high FRET state of ∼0.7 that becomes increasingly populated at 10 mM Mg2+, particularly in the catalytically most active AFHH-WT, suggesting that enhanced activity correlates with more frequent, yet transient visits to this state.
FIGURE 4.
Single molecule FRET histograms, each constructed from 50–100 single molecule time traces prior to photobleaching of the acceptor under standard conditions (50 mM Tris-HCl [pH 8.0], 25°C, with varying Mg2+ concentration). Vertical solid gray lines are FRET ratios corresponding to the tr-FRET derived FWHMs. (A) AFHH-WT ribozyme; (top) 0.5 mM Mg2+, (middle) 1 mM Mg2+, (bottom) 10 mM Mg2+. (B) Partially disrupted mutant AFHH-1A; Mg2+ concentrations as in A. (C) Fully disrupted mutant AFHH-2A; Mg2+ concentrations as in A.
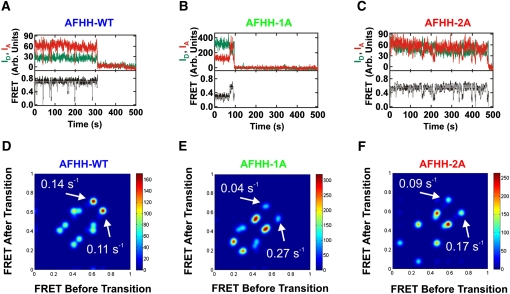
FIGURE 5.
Single molecule FRET analysis. (A–C) Exemplary single molecule time traces showing donor (green) and acceptor (red) intensities, and the corresponding FRET ratio (black) with HaMMy fit (gray). Conditions are, 50 mM Tris-HCl (pH 8.0), 10 mM Mg2+, at 25°C. (A) AFHH-WT ribozyme. (B) Partially disrupted AFHH-1A ribozyme. (C) Fully disrupted AFHH-2A ribozyme. (D–F) Corresponding transition density plots for the same conditions as in A–C, showing the FRET states, transitions between those states, and specific transition rate constants determined by fitting time traces with the software HaMMy (McKinney et al. 2006) for AFHH-WT AFHH (80 molecules; D), partially disrupted mutant AFHH-1A (84 molecules; E) and fully disrupted mutant AFHH-2A (49 molecules; F).
For a relative comparison, we mapped the distances derived from the tr-FRET analysis onto the FRET ratios of our sm-FRET derived histograms, using similar strategies and assumptions as described previously (Pereira et al. 2008). This comparison shows the general consistency between the two types of data. At increasing Mg2+ concentrations, we observed broader tr-FRET distance distributions with smaller mean Stem I–II distances, which were correlated with broader sm-FRET histograms and enhanced occurrence of high FRET states (Fig. 4A–C). This comparison also illustrates the difficulty of ensemble approaches to discern multiple overlapping distance distributions, which appear only as a broad range of tr-FRET derived distances but are readily dissected by complementary sm-FRET analyses.
To more quantitatively characterize the dynamics of single hammerhead ribozymes, we used hidden Markov modeling (HMM, as implemented in the software HaMMY [McKinney et al. 2006]) to determine the most probable path traversed in each sm-FRET trajectory among up to four FRET states (Fig. 5A–C). HMM also yields transition density contour maps that we find to be highly symmetric with respect to the main diagonal for all ribozymes and Mg2+ concentrations, indicative of the fact that most transitions occur reversibly between pairs of FRET states (Fig. 5D–F) as expected for an RNA at thermal equilibrium (Abelson et al. 2010). In addition, the maps reveal more frequent (color scale in Fig. 5D–F) and faster (AFHH-WT, 0.14 sec−1; AFHH-1A, 0.04 sec−1; AFHH-2A, 0.09 sec−1) transitions of the AFHH-WT ribozyme to the 0.7 FRET state compared to its two mutant counterparts, while the mutants show more frequent and faster (AFHH-WT, 0.11 sec−1; AFHH-1A, 0.27 sec−1; AFHH-2A, 0.17 sec−1) transitions away from the 0.7 FRET state, further supporting the notion that the most active ribozyme transiently accesses a state(s) with diminished Stem I–II distance(s) most frequently and for the longest time period.
Taken together, our experimental data consistently suggest that the wild-type loop sequences specifically lead to frequent, transient encounters of Stems I and II, as well as enhanced catalytic activity of the AFHH-WT ribozyme. Both interstem encounters and catalytic activity are significantly reduced by even a single nucleotide mutation designed to disrupt the loop–loop interactions. Increasing Mg2+ concentrations also enhance both the frequency of Stem I–II encounters and catalytic activity, but cannot fully compensate for the loss of specific interstem interactions. These global dynamics, however, cannot explain how loop–loop interactions between Stems I and II impact the cleavage site over a distance of >10 Å (Fig. 2B).
MD simulations show more active site dynamics in the wild type
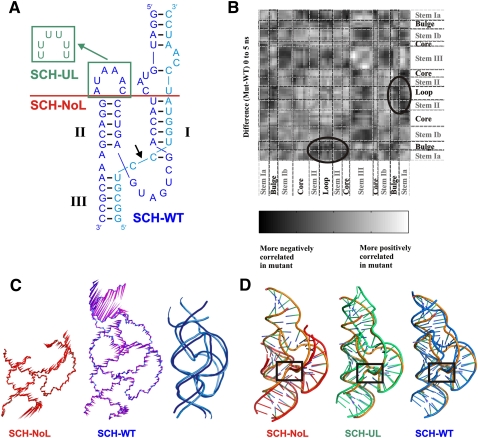
To examine whether the patterns of hammerhead global dynamics observed by FRET are reflected at the active site, we conducted MD simulations on three variants also representing intact, partially disrupted, and fully disrupted loop–loop interaction sequences (Fig. 6A). Since MD simulations require an accurate starting geometry as provided by a high-resolution crystal structure (McDowell et al. 2007), we chose the S. mansoni hammerhead ribozyme structure (PDB ID 2GOZ) as our wild type (SCH-WT) (Martick and Scott 2006). Our partially disrupted loop mutant is based on this SCH-WT, but we replaced each nucleotide in the Stem II loop with uracil (Fig. 6A), creating a mutant (SCH-UL) shown experimentally to have significantly reduced self-cleavage activity (Osborne et al. 2005). Finally, our fully disrupted loop mutant (SCH-NoL) was created by deleting the loop–loop regions (all nucleotides of the Stem II loop as well as all bases of Stem I above the A1.6–U2.6 base pair) (Fig. 6A), leaving essentially a minimal hammerhead.
FIGURE 6.
Molecular dynamics simulations of three S. mansoni hammerheads reveal differences in global structural dynamics. (A) Secondary structure of the three variants representing SCH-WT (crystal structure sequence [Martick and Scott 2006]), partially disrupted (SCH-UL; green), and fully disrupted mutants (SCH-NoL; red). (B) Global residue correlation difference map (SCH-UL minus SCH-WT) for the first 5 nsec of simulation time. Circled areas show where correlation between the loop and the bulge is lost for the U-loop mutant, SCH-UL. (C) Normal mode analysis. Porcupine plots (visualized using VMD) depict the motion of each backbone atom for the first normal mode of SCH-NoL (left) and SCH-WT (right) over the first 10 nsec of MD simulation. Arrows vectorially represent the magnitude and direction of movement. The right-most panel shows snapshots of the Pymol movie visualizing the first normal mode of SCH-WT; the two shades of blue represent the two extremes of the motion. (D) Alignment of simulated variants to the crystal structure (orange). The simulated structures are averages over the time period of 10.5–10.6 nsec. The core residues are aligned to the crystal structure core using Pymol. SCH-WT, blue; SCH-UL, green; SCH-NoL, red.
Standard MD simulations were performed on these three ribozyme variants in explicit solvent over 20 nsec each, using AMBER8 and the parm99 Cornell et al. force field as previously described (McDowell et al. 2007; Sefcikova et al. 2007). Cross-correlation matrices were derived from AMBER for every 5-nsec segment of each trajectory, highlighting both correlated and anti-correlated motions among individual nucleotides across the entire RNA, as previously described (Rhodes et al. 2006). A difference map of these global residue cross-correlations of our wild-type (SCH-WT) and U-loop mutant (SCH-UL) simulations suggests that disruption of the loop–loop interaction reduces long-range structural correlations, especially between the residues of the Stem I and Stem II loops, already over the first 5 nsec of simulation time (observed as the circled dark regions in Fig. 6B). Complementary normal mode analysis was performed (calculating the eigenvectors from a covariance matrix) for each simulation to highlight the dominant global motions in each RNA molecule. In this analysis, large-scale (global) motions are represented by the lowest-frequency eigenvectors. For example, inspection of “porcupine” plots (representing the vectorial directions of motion as arrow heads) of the first normal modes over the first 10 nsec of the SCH-WT and SCH-NoL simulations indicates a similar pattern of global motion in these molecules as experimentally observed by sm-FRET in our AFHH sequences. In particular, comparing the first normal modes of these variants shows large-scale motion in Stem I of the SCH-WT, while the motion in Stems I and II of the SCH-NoL is relatively dampened (Fig. 6C). These large-scale motions of Stem I of SCH-WT represent a bending of the whole helix, rather than a fraying of individual base pairs, and are therefore consistent with being on-path to the dissociation of the loop–loop interactions, which must occur given the transient nature of the Stem I–II interactions observed in our experimental sm-FRET studies. Despite this apparent correspondence of the relative global motions in our MD simulations and experimental studies it has to be noted, however, that the timescales at which these motions occur are very different (nanoseconds for the MD simulations, >100 msec for the sm-FRET studies). The fast MD simulation timescale therefore has to be thought of as exploring local fluctuations within a minimum of the folding free energy landscape, whereas the much slower experimental timescale captures much larger segments of the landscape.
Overlays comparing the simulated structures (averaged over the time period of 10.5–10.6 nsec) to the crystal structure show that while the overall global fold of each simulation remains similar to the starting structure, some differences do appear (Fig. 6D). In particular, in the absence of loop–loop interactions, the relative helical twists of the three stems of the SCH-NoL mutant significantly change relative to the starting crystal structure and the simulated SCH-WT structure (Fig. 6D, cf. SCH-NoL and SCH-WT).
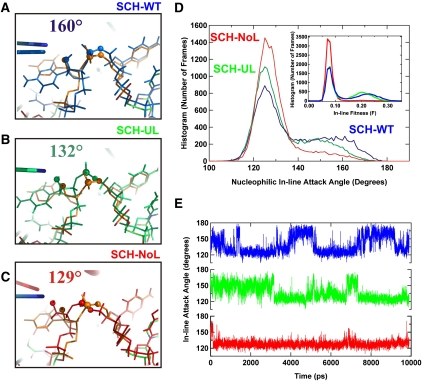
The cleavage reaction of the hammerhead ribozyme proceeds through in-line nucleophilic attack by the 2′-oxygen of C17 on the phosphorous of C1.1 (van Tol et al. 1990). The ideal nucleophilic in-line attack angle (IAA, 2′O-P-5′O) for this reaction is 180° (Soukup and Breaker 1999). An alignment of the active sites of the simulated structures (averaged over the time period of 10.5–10.6 nsec in about the middle of the trajectory) to the crystal structure (Fig. 7A–C) highlights the significant effect of the loop–loop interactions on the IAA at long range; while the SCH-WT cleavage site remains in a geometry similar to that in the crystal structure, with a relatively favorable IAA of 160° (Fig. 7A), modification of the loop region leads to more frequent adoption of less favorable IAAs. In the snapshots shown in Figure 7B,C, the SCH-UL and SCH-NoL mutants adopt IAAs of 132° and 129°, respectively. Figure 7D shows histograms of nucleophilic IAAs for the SCH-WT, SCH-UL, and SCH-NoL variants constructed from the angles measured every 100 psec over 10 nsec of simulation time. Each distribution shows a dominant peak around a low IAA of 125°, but also has a shoulder consisting of IAAs closer to the ideal 180°. Strikingly, access to the higher, catalytically most favorable IAA angles is enhanced in the SCH-WT with wild-type loop sequences, and least prevalent in the SCH-NoL mutant, with the partially disrupted SCH-UL mutant falling in between (Fig. 7D). The same trend is seen in the in-line fitness parameter (Soukup and Breaker 1999; Lee et al. 2008), which includes both the IAA and the O2′-P attack distance (Fig. 7D, inset). Finally, Figure 7E reveals the dynamic nature of the IAA distribution by plotting time courses of nucleophilic IAAs over the first 10 nsec. Taken together, our observations support the notion that wild-type Stem I–II loop–loop interactions lead to both global and local structural dynamics that are correlated with increased catalytic activity.
FIGURE 7.
MD simulations also reveal differences in local structural dynamics. (A–C) Close-up view of the alignment of simulated variants to the crystal structure. Boxed regions from Figure 6D are shown in more detail to highlight the cleavage site; (A) SCH-WT, blue; (B) SCH-UL, green; (C) SCH-NoL, red. (D) Nucleophilic in-line attack angle (IAA) for SCH-WT (blue), SCH-UL (green) and SCH-NoL (red) mutants. Histograms were constructed from angles measured every 100 psec over 10 nsec of simulation time. (Inset) The in-line fitness parameter was calculated as described (Soukup and Breaker 1999) and is plotted as a histogram. (E) IAAs for SCH-WT (blue), SCH-UL (green), and SCH-NoL (red) over 10 nsec of simulation time.
DISCUSSION
Here we have combined cleavage assays, Mg2+ titrations monitored by tr- and sm-FRET, and MD simulations to probe the global and local structural dynamics of extended hammerhead ribozymes with wild-type, partially, or fully disrupted Stem I–II loop–loop interaction sequences. We find that wild-type loop sequences and increased Mg2+ concentrations act to enhance both cleavage activity and structural dynamics and heterogeneity of the ribozyme. It has been appreciated since 2003 that the peripheral loop–loop interactions increase ribozyme activity (De la Peña et al. 2003; Khvorova et al. 2003). Our results are consistent with the notion that the Stem I–II distance in our wild-type hammerhead ribozyme is surprisingly dynamic. These motions help populate catalytically relevant in-line attack angles at the distal cleavage site. Our observations therefore indicate that the extended hammerhead ribozyme is not restricted rigidly to a single (or few) catalytically relevant conformation(s).
Our tr-FRET monitored Mg2+ titrations show a biphasic decrease in mean distances (Fig. 3), consistent with our previous results on minimal hammerheads (Rueda et al. 2003), revealing significant similarities between both forms of the ribozyme. Similarly, recent mechanistic studies comparing minimal and extended hammerheads have suggested fundamental similarities between the two ribozyme forms as their catalytic cores are in a dynamic equilibrium between similar inactive and active conformations, albeit with different bias (Scott 2007; Nelson and Uhlenbeck 2008a,b). Furthermore, our tr-FRET results reveal that the mean Stem I–II distances of the AFHH-WT ribozyme are slightly larger than those of the mutants, inconsistent with the wild-type loops simply holding Stems I and II closer together (Fig. 3). We generally observed broad tr-FRET derived distribution widths (FWHM values) that may reflect either global conformational dynamics (rapid conformational isomerization) or heterogeneity (distinct, slowly interconverting conformers), or both. All our measured widths increase at high Mg2+ concentrations (Fig. 3), consistent with both our previous work on minimal hammerheads (Rueda et al. 2003) and with data on a Diels-Alderase ribozyme that likewise showed increased widths of intramolecular distance distributions with increasing Mg2+ concentration (Kobitski et al. 2007). Notably, the distribution of distances measured for the AFHH-WT hammerhead is significantly wider than those of the mutants (Fig. 3). The fact that the fluorophore attachment sites are identical for all three variants and distal from the mutations suggests that these differences are significant. Our tr-FRET data are contrary to the hypothesis that the hammerhead loop–loop interactions lock the ribozyme in a single (or few) active structure(s) (Martick and Scott 2006).
Our sm-FRET approach reports on the detailed structural dynamics underlying the broad FWHM values observed in tr-FRET, indicating that both Mg2+ and wild-type loop–loop interaction sequences bias the folding free energy landscape toward more disparate stem–stem distances by making new conformations accessible and/or by populating previously rare conformations (Fig. 4). Because the ribozyme more frequently accesses and spends more time in the ∼0.7 FRET state under conditions and in variants that show higher catalytic activity (Figs. 4, 5), this state exhibits properties expected for an active conformation. Conversion of the distance distributions from tr-FRET to apparent sm-FRET ratios finds good agreement in the general trends (Fig. 4, solid vertical lines), but suggests that high FRET states remain relatively underestimated by the tr-FRET approach. Recent work has suggested that relating sm-FRET values to absolute distances (as derived by tr-FRET) can be complicated by the potential of Cy3 and Cy5 to stack on opposite ends of a single nucleic acid helix (Iqbal et al. 2008). In our labeling scheme, Cy5 is internally attached, the two fluorophores are coupled to different helices (Stems I and II) (Fig. 2A), and long linkers are used on both fluorophores, suggesting that helix-end stacking should be less dominant. In addition, such complications should not affect relative comparisons of our three variants given that they are all labeled in the same manner. Clearly, tr-FRET and sm-FRET complement and cross-validate each other, further supporting the notion that our wild-type loops enhance, rather than quench, the conformational sampling of the helical stems that bracket the catalytic core.
While our FRET techniques allow us to experimentally measure global dynamics between the interacting Stems I and II, they can report only on specific fluorophore distances. MD simulations allow us to calculate complementary, more detailed dynamic information, albeit at a much shorter timescale. Normal mode analysis of the SCH-WT and SCH-NoL simulations shows increased motion between Stems I and II of the wild-type hammerhead relative to a loop-disrupted mutant (Fig. 6). Thus our FRET-based observations are echoed in these simulations, using distinct hammerhead sequences and a very different methodology. MD simulations allow us to focus on local motions, and suggest that the loop–loop interactions have a significant long-range effect on the cleavage site. More specifically, we observe that the presence of wild-type loop sequences biases the nucleophilic IAA toward more frequent and extended visits to catalytically favorable values near 180°, while mutations that disrupt the Stem I–II interactions disfavor catalytically relevant IAAs proportionally to their experimentally observed adverse impact on catalysis (Fig. 7). Again, the loop–loop interactions do not lock the hammerhead active site into a single conformation but instead lead to dynamics that sample favorable IAAs. These observations are consistent with the notion that distal tertiary interactions and global dynamics couple with local dynamics and catalytic function of the hammerhead ribozyme. Given that the timescales of our MD and FRET approaches are very different (10 nsec and 100 msec, respectively), exploring different fractions of the folding free energy landscapes (Rhodes et al. 2006; McDowell et al. 2007; Ditzler et al. 2010), the consistency of observing specific differences between wild-type and mutant ribozymes is remarkable.
Thus, with our specific wild-type loop–loop interactions we observe extensive global and local dynamics in the hammerhead, with different patterns of dynamics for each ribozyme variant and at each magnesium concentration. While it is not likely that all dynamically visited states are “on-pathway” for catalysis, at both the global and local scales we find structural dynamics enhanced in parallel with ribozyme activity. A favorable IAA has been predicted to enhance the rate of ribozyme catalysis between 10- and 100-fold (Emilsson et al. 2003; Min et al. 2007), consistent with our observations. More specifically, we measure a 16-fold enhancement of AFHH-WT cleavage rate constants over our fully disrupted mutant in 10 mM Mg2+ (Fig. 2C), and our computational results suggest that the SCH-WT simulation samples IAAs with values >170° ∼12-times more frequently than does the minimal hammerhead simulation (Fig. 7D). Even keeping in mind the limitations posed by their disparate timescales, our FRET and MD data thus jointly support the notion that the peripheral loop–loop interactions exert a long-range effect on active site dynamics of the hammerhead ribozyme.
In summary, our results provide evidence for the notion that the dynamic nature of at least the hammerhead ribozymes studied here is conducive, rather than disruptive, to catalysis. Conformational flexibility and sampling of the folding free-energy landscape is altered in the presence of peripheral loop–loop interactions and/or increasing Mg2+ concentrations, in a way that populates catalytically relevant in-line attack angles in the distal active site. Our findings are consistent across the two hammerhead species studied here, and are consistent with observations from other RNAs, providing a potentially generalizable model for how peripheral structures influence RNA dynamics and function. NMR studies reporting on both global and local dynamics of HIV TAR (Al-Hashimi et al. 2003) show that increasing Mg2+ concentrations lead to a stabilization of global helix stacking, but in parallel increase local motions. We observe a similar phenomenon here, where an increase in Mg2+ concentration favors an overall global structure while promoting local structural “relaxation,” with conformational states sampled more broadly, populating states favorable to in-line nucleophilic attack of a scissile phosphate. Similarly, the hairpin ribozyme has been observed to populate more catalytically relevant “docked” states in the presence of increasing Mg2+ concentrations and stabilizing architectural elements (Zhuang et al. 2002; Bokinsky et al. 2003; Tan et al. 2003). A picture emerges wherein the concepts of induced fit, conformational capture, active site preorganization, and substrate strain (Fersht 1999) begin to gain a dynamic dimension for an RNA enzyme, suggesting that specific ribozyme variants can, to varying degrees, bias molecular motions toward catalytically active conformations, analogous to current notions in protein enzymology (Hammes-Schiffer and Benkovic 2006). Future work needs to examine the mechanism of the observed linkage of global and local dynamics, as well as ask whether global collective motions are directly coupled to the enzymatic function of RNA, as is thought to be the case in protein enzymes (Hammes-Schiffer and Benkovic 2006; Henzler-Wildman et al. 2007) and as has been proposed for the hairpin ribozyme (Rhodes et al. 2006). To this end, a true integration of experimental and computational single molecule studies into a single conceptual framework, as foreshadowed here, will likely be necessary.
MATERIALS AND METHODS
Preparation of RNA oligonucleotides
For cleavage assays, RNA oligonucleotides were transcribed in vitro using T7 RNA polymerase (Milligan and Uhlenbeck 1989). Deprotected RNA was purified as previously described (Walter 2002). 5′-Fluorescein, Cy3, and biotin were attached to the RNA by the manufacturer, while 3′-tetramethylrhodamine and Cy5 were incorporated post-synthetically as previously described (Walter 2002). RNA concentrations were calculated from absorption at 260 nm and, in the case of the tr-FRET HHRz strand, corrected for the additional absorption of fluorescein and tetramethylrhodamine by using the relations A260/A492 = 0.3 and A260/A554 = 0.49, respectively. The following sequences were used: (1) HHRz strand: 5′-GGUUCCCAUCAUCUAUCCCUGAAGAGACGAAGUGACCC-3′; (2) HHS strand: GGGUCACAAGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACC; (3) HH3′A strand: GGGUCACACGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACC; and (4) HH2A strand: GGGUCACUCGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACC. For tr-FRET and sm-FRET, RNA oligonucleotides were purchased with 2′-protection groups from the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource RNA Laboratory at the Yale University School of Medicine and were deprotected as suggested by the manufacturer (http://keck.med.yale.edu/oligos/). The following sequences were used: (1-a) tr-FRET HHRz strand: 5′-(F-AU)GUUCCCAUCAUCUAUCCCUGAAGAGACGAAGUGACC(UA–TMR)-3′; (1-b) sm-FRET HHRz strand: 5′-(Cy3-AU)GUUCCCAUCAUCUAUCCCUGAAGAGACGAAGUGACC(UAT(Cy5)AUAU-biotin)-3′; (2) HHS strand: UAGGUCACAAGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACAU; (3) HH3′A strand: UAGGUCACACGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACAU; and (4) HH2A strand: UAGGUCACUCGUCGAAACUCCUUCGGGAGUCGGAUAGUCGGAACAU. F represents 5′-fluorescein and TMR 3′-tetramethylrhodamine; to obtain noncleavable substrate analogs the underlined C nucleotide was modified with a 2′-methoxy group.
Cleavage reactions
All cleavage reactions were conducted under single-turnover (pre-steady-state) conditions in standard buffer (50 mM Tris-HCl, pH 8.0) and analyzed as previously described (Rueda et al. 2003; Sefcikova et al. 2007). 5′-32P-labeled substrates were prepared by phosphorylation with T4 polynucleotide kinase and [γ-32P]ATP. Standard buffer was 50 mM Tris-HCl (pH 8.0). Mg2+ concentrations were varied as shown in Figure 2. Final RNA concentrations were 1 μM for the Rz strand, and trace amounts (<1 nM) of the radiolabeled substrates. Error bars stem from at least two independent cleavage assays. Time traces of product formation were fit with a double-exponential equation y(t) = y0 + A1[1 – e^(−kobs1t)] + A2[1 − e^(−kobs2t)], employing Marquardt-Levenberg nonlinear least-squares regression (Igor Pro), where A1 and A2 are the extents of cleavage and kobs1 and kobs2 are observed rate constants. The first phases of the time traces were dependent on Mg2+ concentration, and a set of cleavage assay time courses (typically four) under the same conditions were globally fit to obtain a single fast-phase cleavage rate constant (first-phase amplitudes and second-phase rates and amplitudes were free parameters for each time course). The second phases were not dependent on Mg2+ concentration, were generally at least an order of magnitude slower, contributed typically less than one-fourth of the total substrate cleaved, and were not further analyzed. Mg2+ dependencies of the kobs1 rate constant were fit with the binding equation kobs = ksat(Mgn)/[(Mgn) + (kHn)] to yield the cleavage rate constant ksat under saturating Mg2+ conditions, the apparent metal ion dissociation constant KH, and the cooperativity coefficient n.
FRET electrophoretic mobility shift assays
FRET electrophoretic mobility shift (gel FRET) assays were conducted as previously described (Pereira et al. 2002), to test the homogeneity of ribozyme–substrate complexes. Nondenaturing gel electrophoresis was performed in 10% (w/v) polyacrylamide (acrylamide:bisacrylamide 19:1), 50 mM Tris-HOAc (pH 8.0) and either 1 or 10 mM Mg(OAc)2. Ten pmol doubly fluorophore labeled ribozyme strand and 30 pmol unlabeled substrate strand were annealed in 50 mM Tris-HOAc (pH 8.0) and either 1 mM or 10 mM Mg(OAc)2, and 10% (v/v) glycerol, by heating for 2 min to 90°C and cooling to room temperature. The electrophoresis unit was assembled and pre-equilibrated at 4°C, the samples were loaded onto the gel, and an electric field of 6 V/cm was immediately applied. After electrophoresis overnight, the gel was scanned between its low-fluorescence glass plates in a FluorImager SI fluorescence scanner with ImageQuant software (Molecular Dynamics). RNAs labeled with only fluorescein and only tetramethylrhodamine were included as color calibration standards.
Time-resolved FRET
The global structure of the hammerhead ribozyme was studied as a function of Mg2+ concentration by tr-FRET analysis of ribozyme complexes doubly labeled with fluorescein and tetramethylrhodamine as previously described (Rueda et al. 2003). The preannealed ribozyme-noncleavable substrate complex (80 μL; 1 μM Rz and 3 μM S) was incubated at 25°C for at least 15 min in standard buffer either with or without 100 mM NaCl prior to collection of time-resolved emission profiles of the donor fluorescein using time-correlated single-photon counting similar to previously described procedures (Rueda et al. 2003). Mg2+ was titrated by incremental additions of 0.5 μL aliquots of appropriate MgCl2 stock solutions in standard buffer, taking into account the volume change. Fluorescein emission decays were analyzed as previously described, with distance distributions modeled as single Gaussians (Rueda et al. 2003).
Single-molecule FRET
Single-molecule experiments were performed as previously described (Rueda et al. 2004; Pereira et al. 2008; de Silva and Walter 2009). The biotinylated and Cy3-Cy5-labeled ribozyme and noncleavable substrate strands (Fig. 2A) were annealed at a concentration of 1 μM each in a buffer containing 50 mM Tris-HCl (pH 8.0) and 1% (v/v) 2-mercaptoethanol. The annealing solution was heated to 90°C for 2 min before cooling to room temperature over >5 min. The annealed complex was diluted to <20 pM and bound to a streptavidin-coated quartz slide surface via the biotin–streptavidin interaction. The donor (ID) and acceptor (IA) fluorescence signals of optically resolved single molecules (characterized by single-step photobleaching) were detected on a total internal reflection fluorescence microscope as described (Rueda et al. 2004; Pereira et al. 2008). The FRET ratio [defined as IA/(IA + ID)] was followed in real time for each individual molecule. Measurements were performed under standard buffer conditions (50 mM Tris-HCl [pH 8.0] and either 0.5, 1, or 10 mM MgCl2) at 25°C, with an oxygen scavenging system consisting of 10% (w/v) glucose, 2% (v/v) 2-mercaptoethanol, 750 μg/mL glucose oxidase, and 90 μg/mL catalase to reduce photobleaching. Traces that showed very high FRET (∼0.9) were not analyzed, since this FRET level corresponded to an enzyme strand that did not form a complex with a substrate (data not shown). sm-FRET intensity traces were analyzed using a nonlinear filter (Haran 2004), and fit with HaMMy software (McKinney et al. 2006), which uses hidden Markov modeling to determine FRET states and transitions between states. HaMMy fits were also used to generate transition density plots (McKinney et al. 2006).
Comparison of tr-FRET distances to sm-FRET ratios
The apparent FRET efficiency was calculated using a scaling factor c = 0.68 as in Sabanayagam et al. (2005), then the FRET ratio was calculated from this apparent FRET efficiency (Lakowicz 2006), in an analogous manner to previously described calculations (Pereira et al. 2008). Since our observed sm-FRET ratio is ∼0.2 in the absence of acceptor fluorophore, this is the lower limit of our calculations.
Homology modeling
The homology model in Figure 2B was constructed based on models of Stems I and II kindly provided by Dr. John SantaLucia (Wayne State University) using his structural bioinformatics approaches. These stems were appended to the core of the extended hammerhead crystal structure (Martick and Scott 2006) using ERNA-3D software (Pentafolium-Soft), after which the model was minimized using AMBER 8 (Case et al. 2004).
MD simulations
Explicit solvent MD simulations using AMBER8 with the parm99 Cornell et al. force field were conducted as previously described (Sefcikova et al. 2007). Initial structures were based on the full-length crystal structure of the hammerhead ribozyme, PDB ID 2GOZ (Martick and Scott 2006). The SCH-WT structure was changed only by removing the bromine atoms and replacing the 2′-O-methyl at the cleavage site with a 2′-OH. The partially disrupted U-Loop mutant, SCH-UL, was constructed from the SCH-WT structure by using InsightII to replace each of the six bases in the Stem II loop with a U. The fully disrupted SCH-NoL mutant was constructed by simply deleting the six nucleotides of the Stem II loop and all nucleotides of Stem I above the A1.6–U2.6 base pair. Explicit solvent MD simulations, using particle mesh Ewald electrostatics with a 9 Å cutoff, were conducted using AMBER8 (Case et al. 2004) with the parm99 Cornell et al. force field as previously described (Sefcikova et al. 2007). Each of the three variants (SCH-WT, SCH-UL, and SCH-NoL) was simulated for 20 nsec. MD trajectories were analyzed using the ptraj module of AMBER8, cross-correlation matrices (Rhodes et al. 2006) were plotted using MATLAB7.5, structures were visualized using PyMOL (DeLano Scientific), and histograms were created in IGOR Pro (WaveMetrics). Normal mode analysis was conducted using software written and developed by Meyer et al. (2006) and Haider et al. (2008). Normal modes were calculated for the first 10 nsec of simulations using the backbone atoms P, O5′, C5′, C4′, C3′, and O3′. Movies were visualized using PyMOL (DeLano Scientific), and porcupine plots representing these global motions were created using VMD (University of Illinois).
ACKNOWLEDGMENTS
We thank Dr. John SantaLucia, Wayne State University, for modeling Stems I and II of our extended hammerhead ribozyme and Dr. Anastasia Khvorova (Dharmacon Inc. and Thermo Fisher Scientific) for the original sequence of the ASBV hammerhead.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1829110.
REFERENCES
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, et al. 2010. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol 17: 504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Walter NG 2008. RNA dynamics: It is about time. Curr Opin Struct Biol 18: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi H, Pitt S, Majumdar A, Xu W, Patel D 2003. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J Mol Biol 329: 867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS 2008. The eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 415–440 [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X 2003. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci 100: 9302–9307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots JL, Canny MD, Azimi E, Pardi A 2008. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA 14: 2212–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A 2004. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc 126: 10848–10849 [DOI] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. 2004. AMBER 8. University of California, San Francisco [Google Scholar]
- Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH 2008. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol 6: e234 doi: 10.1371/journal.pbio.0060234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva C, Walter NG 2009. Leakage and slow allostery limit performance of single drug-sensing aptazyme molecules based on the hammerhead ribozyme. RNA 15: 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG 2008. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res 36: 7088–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Otyepka M, Sponer J, Walter NG 2010. Molecular dynamics and quantum mechanics of RNA: Conformational and chemical change we can believe in. Acc Chem Res 43: 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson GM, Nakamura S, Roth A, Breaker RR 2003. Ribozyme speed limits. RNA 9: 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A 1999. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding. Freeman, New York [Google Scholar]
- Fiore JL, Kraemer B, Koberling F, Edmann R, Nesbitt DJ 2009. Enthalpy-driven RNA folding: Single-molecule thermodynamics of tetraloop–receptor tertiary interaction. Biochemistry 48: 2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Parkinson GN, Neidle S 2008. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys J 95: 296–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Benkovic SJ 2006. Relating protein motion to catalysis. Annu Rev Biochem 75: 519–541 [DOI] [PubMed] [Google Scholar]
- Haran G 2004. Noise reduction in single-molecule fluorescence trajectories of folding proteins. Chem Phys 307: 137–145 [Google Scholar]
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D 2007. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 450: 913–916 [DOI] [PubMed] [Google Scholar]
- Hutchins CJ, Rathjen PD, Forster AC, Symons RH 1986. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res 14: 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM 2008. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci 105: 11176–11181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Kim NK, Murali A, DeRose VJ 2005. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J Am Chem Soc 127: 14134–14135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva N, Khvorova A, Westhof E, Schiemann O, Wolfson AD 2008. The different role of high-affinity and low-affinity metal ions in cleavage by a tertiary stabilized cis hammerhead ribozyme from tobacco ringspot virus. Oligonucleotides 18: 101–110 [Google Scholar]
- Kobitski A, Nierth A, Helm M, Jaschke A, Nienhaus G 2007. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res 35: 2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR 2006. Principles of fluorescence spectroscopy. Springer, New York [Google Scholar]
- Lee TS, Silva Lopez C, Giambasu GM, Martick M, Scott WG, York DM 2008. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Am Chem Soc 130: 3053–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Scott WG 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG 2008a. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Lee TS, York DM, Scott WG 2008b. Solvent structure and hammerhead ribozyme catalysis. Chem Biol 15: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SE, Spackova N, Sponer J, Walter NG 2007. Molecular dynamics simulations of RNA: An in silico single molecule approach. Biopolymers 85: 169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DB 1996. Structure and function of the hammerhead ribozyme: An unfinished story. RNA 2: 395–403 [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T 2006. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91: 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Ferrer-Costa C, Prez A, Rueda M, Bidon-Chanal A, Luque FJ, Laughton CA, Orozco M 2006. Essential dynamics: A tool for efficient trajectory compression and management. J Chem Theory Comput 2: 251–258 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Min D, Xue S, Li H, Yang W 2007. ‘In-line attack’ conformational effect plays a modest role in an enzyme-catalyzed RNA cleavage: A free energy simulation study. Nucleic Acids Res 35: 4001–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol 5: 587–595 [DOI] [PubMed] [Google Scholar]
- Nelson JA, Uhlenbeck OC 2008a. Hammerhead redux: Does the new structure fit the old biochemical data? RNA 14: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JA, Uhlenbeck OC 2008b. Minimal and extended hammerheads utilize a similar dynamic reaction mechanism for catalysis. RNA 14: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JA, Shepotinovskaya I, Uhlenbeck OC 2005. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry 44: 14577–14585 [DOI] [PubMed] [Google Scholar]
- Osborne EM, Schaak JE, Derose VJ 2005. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA 11: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MJ, Harris DA, Rueda D, Walter NG 2002. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: A conformational change accompanies catalysis. Biochemistry 41: 730–740 [DOI] [PubMed] [Google Scholar]
- Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG 2008. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol 382: 496–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes MM, Reblova K, Sponer J, Walter NG 2006. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc Natl Acad Sci 103: 13381–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury-Saha M, Burke DH 2006. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA 12: 1846–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D, Wick K, McDowell SE, Walter NG 2003. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry 42: 9924–9936 [DOI] [PubMed] [Google Scholar]
- Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG 2004. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc Natl Acad Sci 101: 10066–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam C, Eid J, Meller A 2005. Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: Corrections due to nonideal transfer. J Chem Phys 122: 061103 doi: 10.1063/1.1854120 [DOI] [PubMed] [Google Scholar]
- Saksmerprome V, Roychowdhury-Saha M, Jayasena S, Khvorova A, Burke DH 2004. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA 10: 1916–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84 [DOI] [PubMed] [Google Scholar]
- Scott WG 2007. Morphing the minimal and full-length hammerhead ribozymes: Implications for the cleavage mechanism. Biol Chem 388: 727–735 [DOI] [PubMed] [Google Scholar]
- Sefcikova J, Krasovska M, Sponer J, Walter N 2007. The genomic HDV ribozyme utilizes a previously unnoticed U-turn motif to accomplish fast site-specific catalysis. Nucleic Acids Res 35: 1933–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepotinovskaya IV, Uhlenbeck OC 2008. Catalytic diversity of extended hammerhead ribozymes. Biochemistry 47: 7034–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomatin SV, Greenfeld M, Chu S, Herschlag D 2010. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature 463: 681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann TK, Uhlenbeck OC 1998. Hammerhead ribozyme kinetics. RNA 4: 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci 100: 9308–9313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol H, Buzayan JM, Feldstein PA, Eckstein F, Bruening G 1990. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res 18: 1971–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NG 2002. Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET). Curr Protoc Nucleic Acid Chem 11.10: 11.10.11–11.10.23 [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Rossi JJ 2005. Comparative single-turnover kinetic analyses of trans-cleaving hammerhead ribozymes with naturally derived non-conserved sequence motifs. FEBS Lett 579: 1619–1624 [DOI] [PubMed] [Google Scholar]
- Westhof E 2007. A tale in molecular recognition: The hammerhead ribozyme. J Mol Recognit 20: 1–3 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S 2002. Correlating structural dynamics and function in single ribozyme molecules. Science 296: 1473–1476 [DOI] [PubMed] [Google Scholar]