Abstract
Several mitochondrial mRNAs of the trypanosomatid protozoa are edited through the post-transcriptional insertion and deletion of uridylates. The reaction has provided insights into basic cellular biology and is also important as a potential therapeutic target for the diseases caused by trypanosomatid pathogens. Despite this importance, the field has been hindered by the lack of specific inhibitors that could be used as probes of the reaction mechanism or developed into novel therapeutics. In this study, an electrochemiluminescent aptamer-switch was utilized in a high-throughput screen for inhibitors of a trypanosomatid RNA editing reaction. The screen identified GW5074, mitoxantrone, NF 023, protoporphyrin IX, and D-sphingosine as inhibitors of insertion editing, with IC50 values ranging from 1 to 3 μM. GW5074 and protoporphyrin IX are demonstrated to inhibit at or before the endonuclease cleavage that initiates editing and will be valuable biochemical probes for the early events of the in vitro reaction. Since protoporphyrin IX and sphingosine are both naturally present within the trypanosomatids, their effectiveness as in vitro inhibitors is also suggestive of the potential for in vivo modulatory roles.
Keywords: trypanosomatid, RNA editing, inhibitors, protoporphyrin IX, D-sphingosine, suramin
INTRODUCTION
The Trypanosoma and Leishmania genera of trypanosomatid protozoa include pathogens that are causative agents of several distinct diseases including Chagas disease, human African trypanosomiasis, and leishmaniasis. Collectively, the diseases cause about 100,000 annual deaths with more than 20 million people currently infected and another 500 million at risk for infection. There is a major need for new therapeutics as resistance has developed to several of the current first-line drugs, and many of the available alternatives have severe side effects and are not very effective (for review, see Stuart et al. 2008).
A mitochondrial RNA editing reaction that is essential and unique to the trypanosomatid parasites is an attractive target for new drug development (Schnaufer et al. 2001). The proteins that interact with the RNA and catalyze the editing reaction are part of a core complex of ∼1600 kDa that should present a large drug-binding landscape for targeting (Rusche et al. 1997; Panigrahi et al. 2001, 2003; Aphasizhev et al. 2003). The editing proceeds through reactions that are initiated at an editing site by an endonucleolytic cleavage producing a 5′-fragment with a 3′-OH (Blum et al. 1990; Kable et al. 1996; Seiwert et al. 1996). U-addition by a terminal uridylyltransferase activity (TUTase) or U-deletion by an exonuclease activity at the 3′-OH followed by ligation of the 5′- and 3′-fragments completes one editing cycle. Guide RNAs (gRNAs) contain sequence complementary to specific blocks of edited mRNA and direct the location of the U-insertions and deletions (Blum et al. 1990). Although at least 20 different proteins have been identified that are involved with the core editing activities, the mechanism through which these proteins initially recognize and interact with an editing site is still largely unknown.
There are currently no drugs available that specifically inhibit the editing reaction. In silico screening is a highly promising strategy previously used to identify novel drug-like compounds that can be docked to the crystal structure of an RNA ligase from the editing complex (Amaro et al. 2008). However, it is limited by the availability of crystal structures and by differences in structure or drug accessibility that can result from the study of individual proteins outside the context of the intact multi-protein complex. High-throughput screening (HTS) of large chemical libraries is an alternative strategy to identify inhibitors, but it only recently became feasible with the development of an assay with the required fidelity, sensitivity, and format (Liang and Connell 2009). The novel assay exploits an electrochemiluminescent (ECL) aptamer switch, and we describe here its application in an HTS of a chemical library. Five inhibitors were identified from the screen, and two are demonstrated to act predominantly at or prior to the endonuclease cleavage that initiates editing. The study provides valuable biochemical probes for early events of the in vitro editing reaction and is suggestive of potential modulatory mechanisms for in vivo editing.
RESULTS AND DISCUSSION
A high-throughput screen for inhibitors of editing
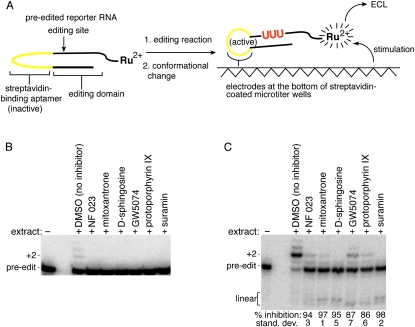
A novel in vitro assay was developed to identify inhibitors of the Leishmania tarentolae editing reaction from HTS of chemical libraries (Fig. 1A; Liang and Connell 2009). The reporter for the assay contains a streptavidin-binding aptamer that is activated by a conformational change in response to editing. Immobilization of the edited RNA at the bottom of a streptavidin-coated microtiter plate followed by electrical stimulation generates an ECL signal from a ruthenium complex linked to the reporter. Since non-edited reporter RNA does not become immobilized at the well bottom, it does not generate significant ECL.
FIGURE 1.
Inhibitors of a trypanosomatid editing reaction. (A) The RNA reporter for the HTS contains a streptavidin aptamer (yellow) that is only active after the insertion of three U's (red) by the in vitro editing reaction. Upon immobilization of the RNA at the bottom of a streptavidin-coated microtiter plate and electrical stimulation, an ECL signal is generated from a ruthenium complex attached to the reporter. (B) Confirmation of the inhibitory activity using a radiolabeled reporter that is based on the cytochrome b mRNA sequence. The gRNA used for the assay has two guiding nucleotides. Compounds were dissolved in DMSO and used at a final concentration of 10 μM. Reactions contained 0.01% TX-100, and products were analyzed after electrophoresis on a denaturing gel. No editing activity was detected in the presence of any of the inhibitors (n = 3). (C) Confirmation of the inhibitory activity of compounds obtained from the HTS using a radiolabeled SELEX-optimized reporter with two guiding nucleotides. Reactions contained 0.01% TX-100, and products were analyzed after electrophoresis on a denaturing gel. The percent inhibition for correct editing (+2) in the presence of each compound is relative to the DMSO-only control (n = 3).
The new assay was used to screen a library containing 1280 pharmacologically active compounds for inhibitors of the RNA editing reaction (see Materials and Methods). The library included a large cross-section of natural modulators for diverse biological reactions, which made it feasible to identify potential modulators of the editing reaction from a small screen. The compounds of the library were dissolved in DMSO and were at a final concentration of 10 μM during the screen. The library screen was conducted within 384-well plates, and the z′-value for each plate during the initial screen ranged from 0.5 to 0.6; the z-value is a statistical parameter that takes into consideration both the variance and the signal-to-noise ratio of an assay, and a value ≥0.5 has empirically been found to be suitable for use in HTS (Zhang et al. 1999). Twenty-eight hits were obtained from the screen, where a hit was defined as a compound that resulted in >30% inhibition of the positive control signal within each of two replicates.
Secondary screens to identify false positives
False positives are common with HTS, and compounds that interfere with the assay are one potential source. Four compounds in this category were readily apparent because the resultant ECL signal was significantly less (<70%) than the signal from the background binding of the pre-edited reporter within the negative control reactions (data not shown). A feature shared by these four compounds is that they are highly negatively charged, containing three to six sulfate or carboxyl groups. A fifth identified inhibitor, ruthenium red, was also excluded from further characterization because it contains three coordinated ruthenium ions and likely interferes with the activation of the ruthenium complex on the reporter.
Another major source of false positives can result from promiscuous inhibitors that form aggregates with proteins, and the large size of the editing complex may make it especially vulnerable to this artifact. Since nonspecific aggregates have been demonstrated in other systems to be sensitive to the inclusion of 0.01% TX-100 (McGovern et al. 2002), an editing assay that is independent of the ECL-linked reporter was used in the presence of detergent as a secondary screen (Fig. 1B). This secondary screen exploited a radiolabeled reporter that is based on the cytochrome b mRNA (Oppegard et al. 2000; Oppegard and Connell 2002). The gRNA for the cytochrome b mRNA acts in trans, like the majority of natural gRNAs. Editing extract was pre-incubated with the reporter RNA and the selected compounds or DMSO for 15 min prior to initiating the editing reaction through the addition of UTP. After resolution of the edited product by gel electrophoresis, the indicated compounds were identified that inhibited 100% of the editing when used at 10 μM in the presence of 0.01% TX-100 (Fig. 1B). An additional detergent-resistant inhibitor, 6-hydroxy-DL-DOPA, is highly susceptible to oxidation, and whereas older solutions reproducibly inhibited the editing reaction, fresh solutions did not (data not shown). As a result, this inhibitor was not further characterized.
The effect of the detergent-resistant inhibitors was also tested on the in vitro editing of a radiolabeled SELEX-optimized reporter that has a gRNA in cis. The optimized reporter was previously demonstrated to have many of the attributes associated with natural mRNA substrates; these include a dependence on guiding nucleotides, similar sequence and secondary constraints, and the co-purification of the editing activities. The advantage of this reporter is that it is one of the most efficient substrates described for the complete in vitro reaction, and the reaction has been demonstrated to increase linearly with the quantity of added extract (Pai et al. 2003). All six of the tested compounds inhibited between 86% and 98% of the editing activity (Fig. 1C), and since inhibition was observed with three independent reporters (Fig. 1A–C), it is unlikely caused by sequence-dependent interactions with the RNA. To prevent U-additions at the 3′ end, the reporter RNA was circularized with T4 RNA ligase prior to the assay (Brown et al. 1999). Faint bands, consistent with the size expected of the linear reaction intermediates, are visible in some reactions (labeled linear in Fig. 1C). However, the significance of these products is difficult to interpret since they can also result from unrelated minor degradation of the circular RNA (Pai et al. 2003).
Specificity of the selected inhibitors
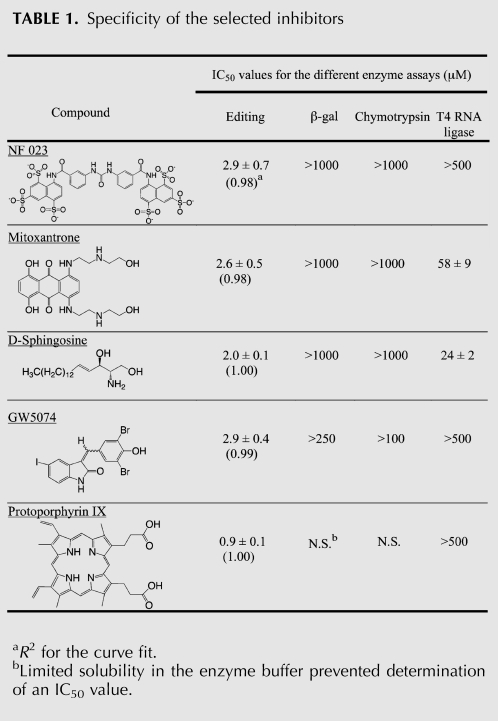
The selected detergent-resistant inhibitors include GW5074, mitoxantrone, NF 023, protoporphyrin IX, and D-sphingosine (Fig. 1B,C). These compounds inhibit the editing reaction with IC50 values between 1 and 3 μM (Table 1). NF 023 is an analog of suramin, which is a drug of undefined mechanism that has been used to treat Trypanosoma infections (for review, see Voogd et al. 1993). When tested in the editing assay, 10 μM suramin was found to inhibit the editing activity (Fig. 1B,C). Although the inhibition of the editing reaction by NF 023 and suramin is suggestive of a possible therapeutic mechanism, suramin inhibits a large number of enzymes, possibly because of its high charge density. In fact, suramin was one of the four compounds identified from the initial library screen but not subsequently characterized because of interference with the ECL assay (data not shown). GW5074 is an inhibitor of RAF-1 (Lackey et al. 2000), which is part of the MAPK/ERK signal transduction pathway, and mitoxantrone is a DNA intercalator that has also been demonstrated to interact with a bulged loop structure within a tau mRNA variant (Zheng et al. 2009). Sphingosine and protoporphyrin IX are both natural products with the former being an 18-carbon amino alcohol and the latter being the final porphyrin precursor in the synthesis of heme.
TABLE 1.
Specificity of the selected inhibitors
The detergent-resistant inhibitors were assayed for their effect on chymotrypsin, β-galactosidase, and T4 RNA ligase, as a means to further assess the promiscuity of the inhibition (Table 1). T4 RNA ligase was chosen as one of the negative controls because it was earlier demonstrated to interact with inhibitors identified from in silico docking of compounds to the crystal structure of REL-1 (Amaro et al. 2008), one of the RNA ligases of the editing complex. A linearized version of the SELEX-optimized editing reporter was used as a substrate for the T4 RNA ligase reaction, which provided a means to test whether the selected inhibitors interact with the RNA rather than the editing complex. The IC50 measurements in some cases were limited by the poor solubility of the compounds in the enzyme buffers, but all five compounds showed significant specificity for inhibition of the editing reaction (Table 1).
Effect of protoporphyrin IX derivatives and precursors on editing
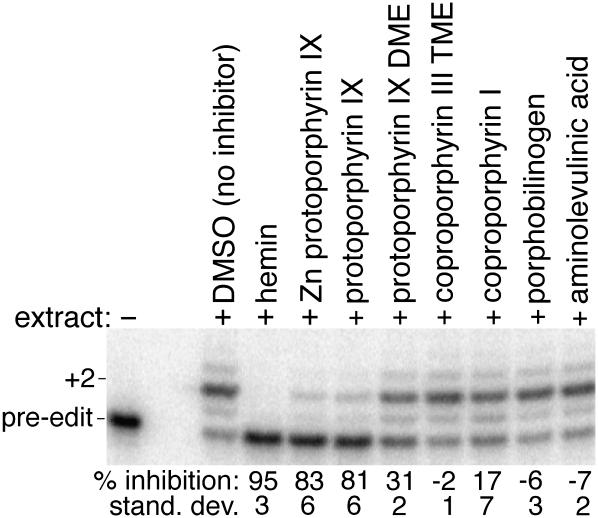
Two of the identified inhibitors, sphingosine and protoporphyrin IX, are normally present within the trypanosomatids (Sah et al. 2002; Zhang et al. 2007), and as a result, there is the potential for both to modulate in vivo editing. Protoporphyrin IX is of especial interest because the trypanosomatids are auxotrophic for it, and the environmental concentration will vary dramatically during their complex life cycles. To initially characterize the inhibitory properties of protoporphyrin IX, several derivatives and precursors were assayed for their effect on editing. The dimethyl ester of protoporphyrin IX is a poor inhibitor, but the zinc(II) coordinated porphyrin and hemin, an iron(III) coordinated porphyrin, are effective (Fig. 2). Aside from a small but reproducible stimulation of in vitro editing by the precursors, aminolevulinic acid and porphobilinogen, the other assayed compounds had little or no remarkable effect, which further emphasizes the specificity of the inhibition.
FIGURE 2.
Inhibition of in vitro editing by several derivatives and precursors of protoporphyrin IX using the radiolabeled SELEX-optimized reporter. The percent inhibition for the formation of correctly edited product (+2) is relative to the DMSO-only control (n = 4). Minus signs denote reaction stimulation rather than inhibition. (DME) Dimethyl ester; (TME) trimethyl ester.
Although it is a good inhibitor of the in vitro editing reaction (Table 1), protoporphyrin IX does not significantly inhibit in vivo editing of the L. tarentolae mitochondrial mRNAs (data not shown). However, rather than a pharmacodynamic limitation, this could result from the protoporphyrin IX not being distributed into the mitochondria, where editing occurs; the mechanisms for heme and protoporphyrin IX transport into both the cell and mitochondria are poorly defined and also likely to be highly regulated to prevent toxicity.
Inhibitors acting at an early stage of editing
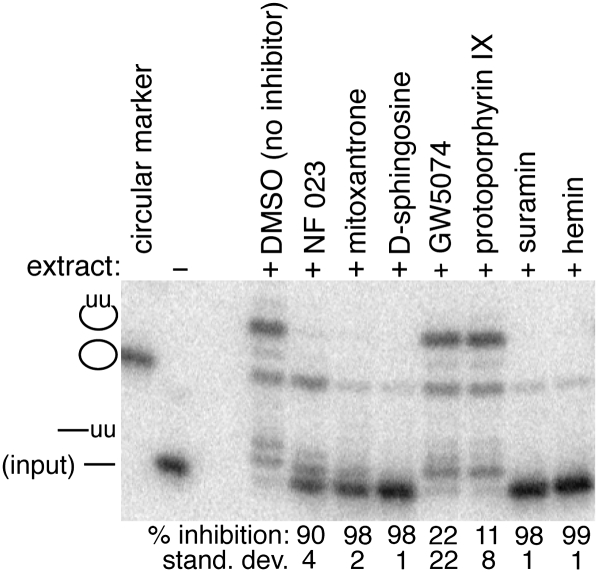
Endonuclease cleavage normally initiates the editing reaction by creating a 3′ end at which the appropriate uridylate deletions or insertions occur, and the reaction is completed by subsequent religation of the mRNA fragments (Blum et al. 1990; Kable et al. 1996; Seiwert et al. 1996). To identify the step of the editing reaction at which the selected inhibitors act, an RNA was synthesized corresponding to the product formed by endonuclease cleavage at the editing site. Since the SELEX-optimized reporter used for the complete in vitro editing reaction is circular to prevent U-additions at the 3′ end (Pai et al. 2003), the intermediate resulting from endonuclease cleavage at the editing site is the full-length linear molecule (Fig. 3, input). When the linear intermediate is treated with editing extract in the absence of inhibitor, the different steps of the reaction are clearly evident (Fig. 3, +DMSO): Two guided U's are added to the 3′ end of the linearized molecule, and the ligase activity of the complex re-forms the circular RNA with the two gRNA-directed U-insertions at the editing site. This is consistent both with an earlier assay exploiting a cleaved substrate (Igo et al. 2002) and with the currently accepted reaction mechanism.
FIGURE 3.
The inhibitors affect different aspects of the editing reaction. The radiolabeled RNA reporter for these reactions corresponds to the intermediate formed by endonuclease cleavage at an editing site. Since the reporter for the full-reaction is normally circular in order to prevent U-additions to the 3′ end, the intermediate resulting from endonuclease cleavage at the editing site is the indicated full-length linear molecule (input). In the absence of inhibitor, two guided U's are added to the linear molecule, and ligated circular product containing two U-insertions is formed. The percent inhibition for the formation of the edited circular product is relative to the DMSO-only control (n = 3). GW5074 and protoporphyrin IX only weakly inhibit the editing of the post-cleavage substrate as compared to the corresponding full reaction indicated in Figure 1C.
Although several of the inhibitors appear to affect multiple aspects of the RNA editing reaction, two are relatively specific for early events. NF 023, suramin, mitoxantrone, and D-sphingosine inhibit the formation of the edited product with the post-cleavage substrate to the same extent as in the full reaction (cf. Figs. 1C and 3). This suggests that these inhibitors are either acting at a step following the endonuclease cleavage or alternatively having global effects on the editing complex. The latter possibility is more likely since the large accumulation of linear molecules predicted from inhibition of a post-cleavage step during the complete reaction was not observed (Fig. 1C). A fraction of the input linear RNA appears to be ligated to circles without U-insertions, and this reaction is partially inhibited by mitoxantrone, D-sphingosine, and suramin; the inhibition of the ligase activity is also consistent with global impairment of the complex by these inhibitors. In contrast, GW5074 and protoporphyrin IX only weakly inhibit the formation of correctly edited +2U circular product in the post-cleavage reaction (cf. Figs. 1C with 3), and the ligation of the non-modified input RNA is unaffected. This suggests that GW5074 and protoporphyrin IX are predominantly acting either at or before the endonuclease cleavage. However, hemin, the Fe (III) coordinated porphyrin, is an effective inhibitor of the post-cleavage reaction. Since hemin is also a twofold better inhibitor of the complete reaction than protoporphyrin IX (Fig. 2; data not shown), it may be making additional contacts that are impacting the complex.
Conclusion
Even though screens of much larger chemical libraries are required to identify novel therapeutics, several important inhibitors were obtained from the screen of the 1280-compound library described here. The library includes a large cross-section of natural modulators for diverse biological reactions, which made it feasible to identify potential modulators of the editing reaction from a small screen. Since both protoporphyrin IX and sphingosine are present within the trypanosomatids, their identification as inhibitors of in vitro editing is suggestive of the possibility that they or their derivatives could interact with the editing complex within the cell. Further studies, though, are clearly required to determine whether these two compounds modulate in vivo editing. Both GW5074 and protoporphyrin IX were demonstrated to significantly inhibit the complete editing reaction, but not the reaction with the post-cleavage substrate. This suggests that these compounds primarily affect either the endonuclease or a step preceding it. Since neither of these early editing events is well characterized, the two inhibitors will be useful as biochemical probes to identify the interacting component, which will potentially establish a functional role for some of the editing complex proteins. The study also demonstrates that HTS of larger chemical libraries is not only possible but also likely to be highly productive.
MATERIALS AND METHODS
Screening of a chemical library for editing inhibitors
The screen of the Sigma LOPAC 1280 library was performed in streptavidin-coated, standard-capacity 384-well ECL plates (Meso Scale Discovery). Plates were pre-blocked with Superblock buffer (Thermo Scientific) and washed three times prior to use with 0.05% Tween 20 containing 50 mM NaCl and 25 mM Tris (pH 8.0, 27°C) and once with binding buffer (4 mM MgCl2, 50 mM KCl, and 5 mM Tris at pH 8.0). Plate washing was performed with a Perkin Elmer Flexdrop. The library compounds were at a concentration of 10 mM in DMSO, and an Echo liquid handler (Labcyte) was used to transfer 12 nL of each compound to designated wells.
The assay reactions contained 100 fmol of reporter RNA and 60 units of an L. tarentolae editing extract that were prepared as previously described (Liang and Connell 2009); one unit of editing activity is defined as the amount that results in 1 fmol of correctly edited product in 1 h using our standard reaction conditions. The pre-edited reporter RNA was initially denatured for 5 min at 65°C in 2.4 volumes of denaturing buffer containing 0.2 mM EDTA and 25 mM Tris (pH 8.0, 27°C). The RNA was mixed with three volumes containing 4 mM ATP, 4 mM dithiothreitol, 20 μg/mL leupeptin, 12 mM MgCl2, and 2 mg/mL Pefabloc SC, and then incubated for 10 min at room temperature. The editing extract, 3.6 volumes at 16.7 units/μL, was added to the RNA solution, and 9 μL of the mixture delivered into each well. The RNA-editing extract mixture was incubated with the previously delivered library compounds at room temperature for 15 min prior to initiating editing through the addition of a 3-μL UTP solution (4 mM UTP, 8 mM MgCl2). Liquid dispensing of 9 μL and 3 μL was performed by a Multidrop Combi (Thermo Scientific). The plates were then incubated for 4 h at 27°C followed by washing four times with wash buffer (150 mM KCl, 4 mM MgCl2, and 5 mM Tris at pH 8.0). To avoid differences in dissociation caused by read buffer delivery, a Biomek FX Laboratory Automation Workstation (Beckman Coulter) was used to simultaneously add 35 μL of read buffer T plus surfactant (Meso Scale Discovery) to each well. The ECL signals were recorded using a SECTOR Imager 6000 Reader (Meso Scale Discovery).
The z′-value for each plate was calculated from 16 wells of both positive and negative control reactions located within the two outside columns and using the following equation (Zhang et al. 1999):
where μc+ and μc− are the mean values for the positive and negative control assay signals, respectively, and σc+ and σc− are the corresponding standard deviations for these values. The positive control reactions consisted of the complete editing reaction in the presence of DMSO but with no drug. Since there were not any known specific inhibitors of the complete editing reaction prior to this study, the negative control reaction consisted of the complete reaction minus the critical UTP cofactor.
Enzyme assays
To confirm the effect of the selected compounds on the editing reaction, 0.4 pmol of the radiolabeled A-1 RNA editing substrate (Pai et al. 2003) was denatured for 5 min at 65°C in 9 μL of denaturing buffer containing 0.2 mM EDTA and 25 mM Tris (pH 8.0, 27°C). The RNA was then mixed with 12.5 μL of a solution containing 4 mM ATP, 4 mM dithiothreitol, 20 μg/mL leupeptin, 12 mM MgCl2, and 2 mg/mL Pefabloc SC. It was incubated for 10 min at room temperature prior to the addition of 15 μL of 16.7 units/μL editing extract. The editing extract also contained 0.034% TX-100 where indicated. Either 1 μL of DMSO or the indicated quantity of inhibitor in DMSO was added to the RNA-extract mixture and incubated for 15 min at room temperature. Editing was initiated through the addition of 12.5 μL of a UTP solution (4 mM UTP, 8 mM MgCl2) and then incubated for 1 h at 27°C. Reactions were terminated as previously described (Oppegard et al. 2003), and the edited RNA resolved from non-edited by denaturing gel electrophoresis. The fraction of correctly edited (+2 U) RNA was quantified by PhosphorImage analysis, and the percent activity for each compound was determined relative to the DMSO control.
The post-cleavage editing assay was performed similar to the complete reaction with the exception that the ATP was added after the 15-min incubation of inhibitor and RNA-extract mixture. This limited RNA ligase activity prior to inhibition of the complex. The RNA reporter for the reactions was a T7 RNA polymerase transcript from the following template: (5′-UCCCACAAATAAAGCAACCAACAAGAACAAGTTAAACG CCGCGGAGGATTGAAACTTAACATTAAATTCCTATAGTGAGTCGTATTA-3′). The two 5′-nucleotides contained a C2′-methoxyl group (Kao et al. 1999), and the T7 RNA polymerase reaction was performed in the presence of GMP as previously described (Milligan and Uhlenbeck 1989).
The chymotrypsin and β-galactosidase assays were performed under previously described conditions (McGovern et al. 2002). The assays contained either 10 nM chymotrypsin or 1.5 nM β-galactosidase that had been pre-treated for 15 min at room temperature with DMSO or the selected compounds in DMSO; compounds were used at a final concentration of 1–1000 μM. The chymotrypsin assays were initiated with 200 μM of the succinyl-ala-ala-pro-phe-p-nitroanilide substrate and the β-galactosidase with 1 mM ortho-nitrophenyl-β-galactoside. Reaction progress for both enzymes was monitored at 405 nm. The percent activity was determined by comparing the initial reaction rate in the presence of each test compound to that of the DMSO control.
The T4 RNA ligase assays contained 0.1 pmol of enzyme in a 20-μL final volume. The enzyme was pre-treated with chemical compounds as for the chymotrypsin and β-galactosidase assays. Reactions were initiated through the addition of 2 pmol of radiolabeled linear A-1 RNA editing substrate (Pai et al. 2003). The RNA was synthesized with a 5′ monophosphate, and the formation of circular product was used to monitor the ligase activity after resolution of the products by denaturing gel electrophoresis. T4 RNA ligase reaction conditions were as previously described (Brown et al. 1999).
The IC50 values were determined by plotting the percent enzymatic activity as a function of the log inhibitor concentration and fitting to the following equation:
where Y is the percent enzymatic activity relative to non-treated controls, Bottom is the value of Y at the bottom plateau, Top is the value of Y at the top plateau, and X is the log concentration. Curve fitting was performed using Kaleidagraph software.
ACKNOWLEDGMENTS
We are grateful to Jonathan Solberg and Dr. Derek Hook of the University of Minnesota High-Throughput Facility for assistance with the chemical screen. This work was supported by U.S. Department of Defense Grant W81XWH-06-1-0794 and grants from the Minnesota Medical Foundation and University of Minnesota Graduate School.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2347310.
REFERENCES
- Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA 2008. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci 105: 17278–17283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 22: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L 1990. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198 [DOI] [PubMed] [Google Scholar]
- Brown LM, Burbach BJ, McKenzie BA, Connell GJ 1999. A cis-acting A-U sequence element induces kinetoplastid U-insertions. J Biol Chem 274: 6295–6304 [DOI] [PubMed] [Google Scholar]
- Igo RP Jr, Lawson SD, Stuart K 2002. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol Cell Biol 22: 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable ML, Seiwert SD, Heidmann S, Stuart K 1996. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rudisser S 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey K, Cory M, Davis R, Frye SV, Harris PA, Hunter RN, Jung DK, McDonald OB, McNutt RW, Peel MR, et al. 2000. The discovery of potent cRaf1 kinase inhibitors. Bioorg Med Chem Lett 10: 223–226 [DOI] [PubMed] [Google Scholar]
- Liang S, Connell GJ 2009. An electrochemiluminescent aptamer switch for a high-throughput assay of an RNA editing reaction. RNA 15: 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern SL, Caselli E, Grigorieff N, Shoichet BK 2002. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem 45: 1712–1722 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Oppegard LM, Connell GJ 2002. Direct visualization of RNA editing within a Leishmania mitochondrial extract. Int J Parasitol 32: 859–866 [DOI] [PubMed] [Google Scholar]
- Oppegard LM, Kabb AL, Connell GJ 2000. Activation of guide RNA-directed editing of a cytochrome b mRNA. J Biol Chem 275: 33911–33919 [DOI] [PubMed] [Google Scholar]
- Oppegard LM, Hillestad M, McCarthy RT, Pai RD, Connell GJ 2003. Cis-acting elements stimulating kinetoplastid guide RNA-directed editing. J Biol Chem 278: 51167–51175 [DOI] [PubMed] [Google Scholar]
- Pai RD, Oppegard LM, Connell GJ 2003. Sequence and structural requirements for optimal guide RNA-directed insertional editing within Leishmania tarentolae. RNA 9: 469–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Gygi SP, Ernst NL, Igo RP, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, et al. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol 21: 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9: 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Cruzreyes J, Piller KJ, Sollnerwebb B 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16: 4069–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah JF, Ito H, Kolli BK, Peterson DA, Sassa S, Chang K 2002. Genetic rescue of Leishmania deficiency in porphyrin biosynthesis creates mutants suitable for analysis of cellular events in uroporphyria and for photodynamic therapy. J Biol Chem 277: 14902–14909 [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP Jr, Wirtz E, Salavati R, Stuart K 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2162 [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Heidmann S, Stuart K 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84: 831–841 [DOI] [PubMed] [Google Scholar]
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R 2008. Kinetoplastids: Related protozoan pathogens, different diseases. J Clin Invest 118: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd TE, Vansterkenburg EL, Wilting J, Janssen LH 1993. Recent research on the biological activity of suramin. Pharmacol Rev 45: 177–203 [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg K 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73 [DOI] [PubMed] [Google Scholar]
- Zhang K, Pompey JM, Hsu F, Key P, Padmavathi B, Saba JD, Turk J, Beverley SM 2007. Redirection of sphingolipid metabolism toward de nova synthesis of ethanolamine in Leishmania. EMBO J 26: 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Chen Y, Donahue CP, Wolfe MS, Varani G 2009. Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone). Chem Biol 16: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]