Abstract
MicroRNAs (miRs) commonly regulate translation from target mRNA 3′ untranslated regions (UTRs). While effective miR-binding sites have also been identified in 5′ untranslated regions (UTRs) or open reading frames (ORFs), the mechanism(s) of miR-mediated regulation from these sites has not been defined. Here, we systematically investigate how the position of miR-binding sites influences translational regulation and characterize their mechanistic basis. We show that specific translational regulation is elicited in vitro and in vivo not only from the 3′UTR, but equally effectively from six Drosophila miR-2-binding sites in the 5′UTR or the ORF. In all cases, miR-2 triggers mRNA deadenylation and inhibits translation initiation in a cap-dependent fashion. In contrast, single or dual miR-2-binding sites in the 5′UTR or the ORF yield rather inefficient or no regulation. This work represents the first demonstration that 5′UTR and ORF miR-binding sites can function mechanistically similarly to the intensively investigated 3′UTR sites. Using single or dual binding sites, it also reveals a biological rationale for the high prevalence of miR regulatory sites in the 3′UTR.
Keywords: microRNA, translational control, 5′UTR, open reading frame, 3′UTR
INTRODUCTION
MicroRNAs (miRs) are key post-transcriptional regulators of a broad range of physiological and pathological processes in animals and plants (Bushati and Cohen 2007; Voinnet 2009). Animal miRs trigger regulation in the context of the miR-induced silencing complex (miRISC), core components of which are the Argonaute (AGO) and GW182 proteins (Peters and Meister 2007; Eulalio et al. 2009). miRs have been shown to regulate gene expression by multiple mechanisms, including deadenylation and degradation of target mRNAs, as well as inhibition of translation. In this latter case, a growing body of evidence suggests that translation is blocked at the initiation step, although alternative mechanisms have been reported as well (Filipowicz et al. 2008; Carthew and Sontheimer 2009).
The vast majority of animal miRs regulates gene expression by binding to sites located in the 3′ untranslated regions (UTRs) of their mRNA targets. This striking positional bias has led to the development of miR target search algorithms that focus on 3′UTRs and, consequently, to a further amplification of the bias for functional 3′UTR sites (Bartel 2009). However, effective miR-binding sites have also been identified in the 5′UTR or the open reading frame (ORF) of target mRNAs (e.g., Jopling et al. 2005; Duursma et al. 2008; Forman et al. 2008; Henke et al. 2008; Lal et al. 2008; Ørom et al. 2008; Tay et al. 2008; Elcheva et al. 2009; Tsai et al. 2009). Non-3′UTR miR targeting has also been studied in silico and in reporter gene assays (e.g., Kloostermann et al. 2004; Lytle et al. 2007; Stark et al. 2007). Recently, pervasive conserved miR targeting was demonstrated computationally in Drosophila ORFs; it was also suggested that the extent of physiological 5′UTR and ORF miR targeting may be greater in flies than in mammals (Schnall-Levin et al. 2010). Despite mounting evidence indicating the biological relevance of this phenomenon, the mechanisms underlying 5′UTR and ORF miR-mediated regulation are still unknown. Here we address this unresolved issue, and show that Drosophila miR-2 represses translation initiation in a cap-dependent manner and induces mRNA deadenylation with similar efficiencies from binding sites in the 5′UTR or the ORF as from standard 3′UTR positions. We also find that single or dual miR-binding sites are more effective from the 3′UTR, which may provide an explanation for the observed physiological bias for 3′UTR miR regulatory sites.
RESULTS
Effective miR-2-mediated silencing from all mRNA positions in vitro
To explore whether and how miR-mediated regulation can be confered by 5′UTR- and ORF-binding sites, we initially used the in vitro system derived from Drosophila embryos, which had previously been used to dissect the mechanisms of miR-mediated translational control via six miR-2-binding sites in the 3′UTR (Thermann and Hentze 2007; Zdanowicz et al. 2009). We generated reporter constructs based on either the firefly luciferase (FL) ORF (for translation assays) or a shortened derivative (sORF; for ribosome binding assays), bearing six copies of the reaper miR-2-binding site (wild type, wt) and 24 nuclceotides (nt) of flanking sequence alternatively in the 3′UTR, the 5′UTR, or the ORF (Figs. 1A, 2A–D). The sequence of the miR-2-binding cassette was optimized to remove all start or in-frame stop codons. As controls, we used reporters bearing mutated seed regions (mut), which are therefore not bound by miR-2. The human β-globin 5′UTR (GL, ∼60 nt) was inserted into all reporters to widen the spacing between the cap structure and the miR-2-binding sites when these are placed at the 5′ end. The resulting constructs are thus highly similar to each other and vary the miR-binding site positions with minimal changes to the overall mRNA.
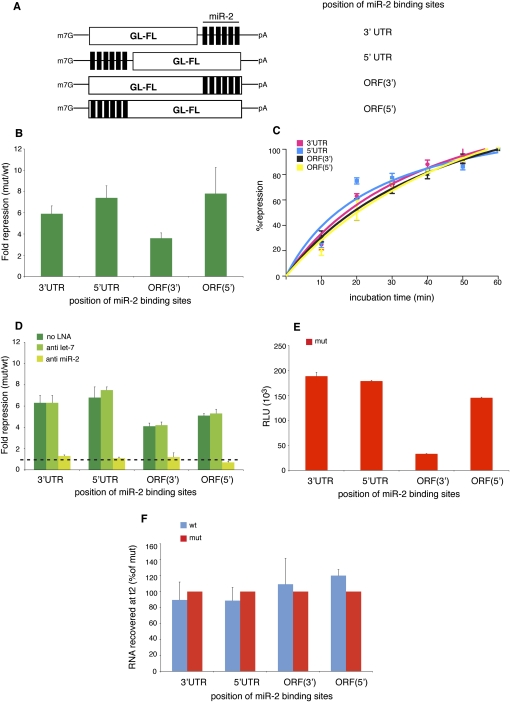
FIGURE 1.
miR-2 represses translation from 5′UTR- and ORF-binding sites in vitro. (A) Schematic representation of the reporters used in this study. The firefly luciferase open reading frame is indicated by a white box (GL-FL); six binding sites for miR-2 are shown as black bars. (B) miR-2 represses translation from both UTRs and from ORF-binding sites in the Drosophila cell-free extracts. (C) Kinetic analysis of translational repression. Samples are analyzed at 10-min intervals during the repression assay. One-hundred percent repression for each construct corresponds to the values indicated in B. (D) The repression observed requires miR-2, since it is specifically relieved by anti miR-2 LNAs. (E) Luciferase activity from the different mut constructs is indicated to show the effect of the position of the miR-2-binding sites on enzymatic activity. (F) The RT–qPCR analyses show no significant destabilization of the reporter mRNAs at the end (t2) of the repression assay. Shown are averages and standard deviations of five (B,D) or three (C,F) independent experiments. (E) One experiment performed in duplicate from B.
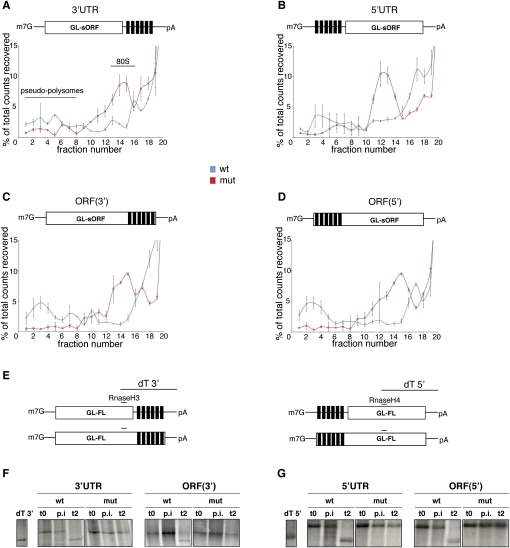
FIGURE 2.
miR-2 controls translation initiation and triggers deadenylation from both UTR- and ORF-binding sites. (A–D) The radiolabeled GL-sORF reporters were incubated in the Drosophila cell-free extracts in the presence of cycloheximide. At the end of the reaction, they were resolved through 15%–45% sucrose gradients, fractionated, and analyzed by scintillation counting. On all wt reporters, inhibition of 80S complex assembly (fractions 10–15) and the formation of pseudopolysomes (fractions 1–8) can be detected. Shown are averages and standard deviations from three independent experiments. (E) Schematic representation of the oligonucleotides used in the deadenylation assays. The dT 3′ and dT 5′ fragments were obtained by annealing the reporter mRNAs to both an RnaseH and a dT oligo and subsequent digestion with RNase H. Therefore, they serve as size markers for the identification of deadenylated reporter mRNAs. Both the wt reporters bearing the miR-2-binding sites at the 3′ end (F) and at the 5′ end (G) are deadenylated at the end of the repression assay (t2), while mut RNAs maintain full poly(A) tail lengths. No change in the adenylation status is observed after the preincubation (p.i.) step. t0 represents input samples at the beginning of the repression assay. The data shown are representative of three independent experiments.
Upon incubation of the capped and polyadenylated GL-FL-reporter mRNAs in the Drosophila cell-free system, we observe that miR-2 triggers effective repression of the GL-FL wt reporters from the 5′UTR and both ORF sites, as well as from the 3′UTR-binding sites (Fig. 1B). This effect is confirmed by kinetic analyses (Fig. 1C) and is specific because it is relieved by sequestration with a complementary locked nucleic acid (LNA) anti-miR but not an unrelated anti-miR (Fig. 1D). We also notice that the luciferase activities from the mut reporters are not greatly affected by the position of the miR-2-binding sites, indicating that the effects on translational repression of the wt reporters are likely to be comparable between the different constructs (Fig. 1E). The lower (approximately sixfold) luciferase counts from the ORF(3′) construct are likely due to a negative effect of C-terminal fusions on the catalytic activity of luciferase (Waud et al. 1996; see below). To check whether the stability of the reporter mRNAs is affected by miR-2, we performed RT–qPCR analyses of RNA recovered at the end of the translation reactions (t2). As previously shown for the 3′UTR reporters (Thermann and Hentze 2007), all GL-FL wt reporters are as stable as their mut counterparts, indicating that miR-2 does not alter mRNA stability and that repression occurs at the level of translation (Fig. 1F). We conclude that Drosophila miR-2 can principally inhibit translation from the 5′UTR or the ORF as efficiently as from the 3′UTR.
miR-2 inhibits translation initiation and triggers deadenylation from all sites
miR-2 binding to 3′UTR sites induces deadenylation and repression of translation initiation (Thermann and Hentze 2007; Zdanowicz et al. 2009). We next wanted to explore whether this is also the case for the non-3′UTR-binding sites, or whether miR-2 acts in a different way from either of the alternative sites. We performed sucrose gradient analyses of translation initiation complex formation in reactions with cycloheximide to monitor 80S ribosomal complex formation. As shown for miR-2 regulation via 3′UTR-binding sites (Thermann and Hentze 2007), we detect the formation of pseudopolysomes and a reduction of 80S complex formation on all wt reporters, indicative of a miR-2-mediated block of translation initiation (Fig. 2A–D).
We next analyzed the adenylation status of the reporter mRNAs using the described RNase H cleavage protocol (Zdanowicz et al. 2009) with the oligonucleotides depicted in Figure 2E. These analyses reveal effective and specific deadenylation of all wt reporter mRNAs regardless of the location of the miR-2-binding sites (Fig. 2F,G).
Another defining feature of “classical” miR regulation is that it relies on the interaction between the AGO1 (but not AGO2) and GW182 proteins in the context of the miRISC (Eulalio et al. 2008); this requirement is maintained in our Drosophila embryo cell-free system (Till et al. 2007). To investigate whether repression from non-3′UTR sites also requires this interaction, we made use of a dominant-negative derivative of Drosophila GW182 [GW182(1-592)]. This polypeptide contains the AGO1-binding domain but lacks the so-called silencing domain, and is thought to act by sequestering AGO1 into inactive complexes (Eulalio et al. 2008). The addition of this protein strongly and specifically derepresses all reporter mRNAs, indicating that full repression requires the interaction between AGO1 and GW182 in all cases (Fig. 3A).
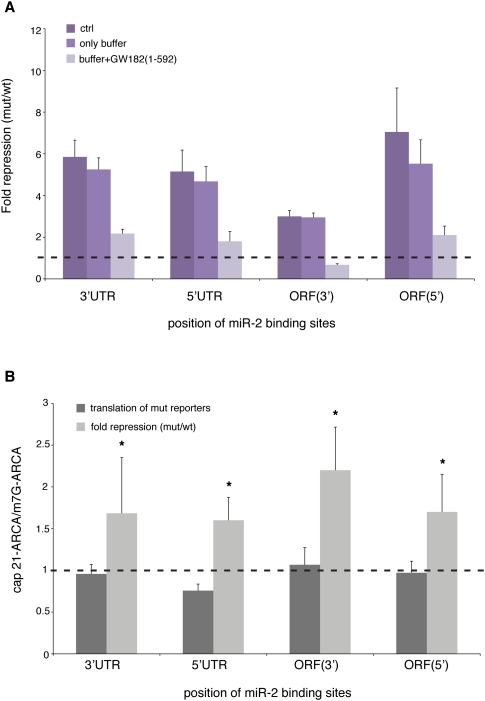
FIGURE 3.
Effective repression of all of the reporters requires the interaction between AGO1 and GW182 and targets cap-dependent translation. (A) In vitro translation reactions were incubated with either a dominant-negative version of the Drosophila GW182 protein [GW182(1-592)] or with protein buffer. The addition of 200 nM dominant-negative protein significantly derepresses all wt reporters, while no effect is observed with buffer alone. The graph shows averages and standard deviations from three independent experiments. (B) GL-FL-reporter mRNAs bearing either the m7G-ARCA cap or the 21-ARCA cap analog (Zdanowicz et al. 2009) were synthesized in vitro and incubated in the Drosophila embryo extract. The luciferase counts of the mut constructs (dark-gray bars) and the fold repression of each wt-mut pair (light-gray bars) bearing cap 21-ARCA were compared with their m7G-ARCA-capped counterparts. In all cases, cap 21-ARCA displays no effect on general translation, while augmenting miR-mediated repression. The graph shows averages and standard deviations from six independent experiments. Statistical significance was evaluated through an unpaired two-tailed t-test: (*) P < 0.05.
To complete our mechanistic analyses, we wanted to probe miR regulation of mRNAs bearing different cap structure analogs. This approach recently uncovered a specific hallmark of miR regulation via the 3′UTR, identifying two chemical analogs of the cap structure (cap 16 and cap 21) that augment miR regulation. Moreover, these experiments also suggested that translational repression from the 3′UTR may occur via a mechanism related to that of the 4E-binding proteins (4E-BPs) (Zdanowicz et al. 2009). We therefore in vitro synthesized GL-FL-reporter mRNAs bearing either the physiological cap structure (m7G anti-reverse cap analog, ARCA) or the cap analog 21-ARCA, which contains a hexaphosphate instead of a triphosphate linker (Zdanowicz et al. 2009). Reproducing our earlier results for 3′UTR miR-2 sites (Zdanowicz et al. 2009), the incorporation of cap 21-ARCA did not affect general translation of reporter mRNAs, whereas it augmented miR repression from the 3′UTR (Fig. 3B). This feature also characterizes repression from the other miR positions (Fig. 3B), implicating cap-mediated translation initiation as the primary target for repression also for 5′UTR and ORF miR-binding sites.
We conclude that miR-2-mediated translational control from 3′UTR sites and non-3′UTR sites displays striking mechanistic similarities with regard to (1) controlling translation initiation, (2) leading to mRNA deadenylation, (3) requiring the interaction between AGO1 and GW182, and (4) targeting cap-dependent translation, possibly via a mechanism related to the 4E-BPs.
Analyzing miR-2-mediated silencing in vivo
To evaluate the significance of our findings in vivo, we tested corresponding reporter constructs in Drosophila S2 cells. First, we transfected the capped and polyadenylated GL-FL-reporter mRNAs. Confirming our findings in the Drosophila embryo extracts (Fig. 1B,D), we observe that the endogenous miR-2 specifically represses the expression of the wt reporters bearing binding sites in the 3′UTR, the 5′UTR, or the 5′ end of the ORF (Fig. 4A,B). Whereas luciferase activities encoded by the 3′UTR, 5′UTR, and ORF(5′) reporters are similar (data not shown), the measured luciferase activities from the reporters bearing binding sites at the 3′ end of the ORF (which corresponds to the C terminus of the encoded protein) are not significantly above background, and these constructs had to be excluded from further in vivo analyses. The mRNA levels of these ORF(3′) constructs were found to be simlar to the other reporters (data not shown), indicating that the lack of luciferase activity is caused by misfolding or degradation of the protein product. C-terminal fusions of firefly luciferase have indeed been reported to cause misfolding and degradation of such fusion proteins in vivo (Waud et al. 1996).
FIGURE 4.
miR-2 represses translation from 5′UTR- and ORF-binding sites in vivo. (A) In vitro transcribed GL-FL-reporter mRNAs are repressed by endogenous miR-2 following transfection into Drosophila S2 cells. Luciferase activity of the ORF(3′) constructs is not significantly over background, possibly due to misfolding and degradation of luciferase C-terminal fusion proteins in S2 cells. (B) The repression is mediated by miR-2, since it is specifically relieved by cotransfection of 100 nM anti-miR-2 LNAs. (C) miR-2 represses luciferase expression from transfected reporter plasmids. (D) This repression is also specifically relieved upon cotransfection of 100 nM anti-miR-2 LNAs. (E) Total RNA was extracted from S2 cells transfected with the reporter plasmids and subjected to RT–qPCR analyses. The wt reporter mRNAs are equally stable throughout the experiment. mRNA levels could also be measured for the ORF(3′) constructs, as expected. Shown are averages and standard deviations from five independent experiments. Statistical significance was evaluated through an unpaired two-tailed t-test: (*) P < 0.05.
To overcome limitations of the RNA transfection strategy (i.e., the absence of a nuclear history of the transfected mRNA) and technical difficulties to accurately determine mRNA levels (Barreau et al. 2006), we also performed plasmid transfections. As seen before with the RNA transfections, we observe miR-2-specific repression of the three wt reporter constructs that can be analyzed in vivo (Fig. 4C,D). Interestingly, in plasmid transfection experiments, the repression of the 5′UTR and ORF(5′) constructs is about half of the repression via the 3′UTR. Although we do not know the reason for this difference, it is tempting to speculate that aspects of the more physiological mRNP biogenesis via nuclear transcription and export introduce a bias in favor of 3′UTR sites. We then performed RT–qPCR analyses on total RNA from the S2 cells transfected with the reporter plasmids. Confirming the results obtained in vitro (Fig. 1F), we detected no significant changes in reporter mRNA levels (Fig. 4E), indicating that miR-2-mediated regulation is exerted at the translational level also in vivo. We conclude that, both in vivo and in vitro, miR-2 can silence translation also from 5′UTR and ORF-binding sites.
miR-2-mediated regulation in vitro and in vivo from single or dual binding sites
Most cellular mRNAs that are regulated by miRs have single or dual miR-binding sites; as a consequence, the extent to which the expression of these mRNAs is affected by miRs rarely exceeds a factor of two (Grimson et al. 2007; Selbach et al. 2008; Bartel 2009). The correlation between the number of 3′UTR miR-binding sites and the degree of repression is recapitulated for miR-2-mediated regulation in the Drosophila cell-free system, and significant (twofold or less) repression is observed from dual or single 3′UTR miR-2 sites in vitro (Thermann and Hentze 2007). To assess the function of single or dual 5′UTR and ORF miR-binding sites, we first analyzed appropriate GL-FL-reporter constructs in vitro.
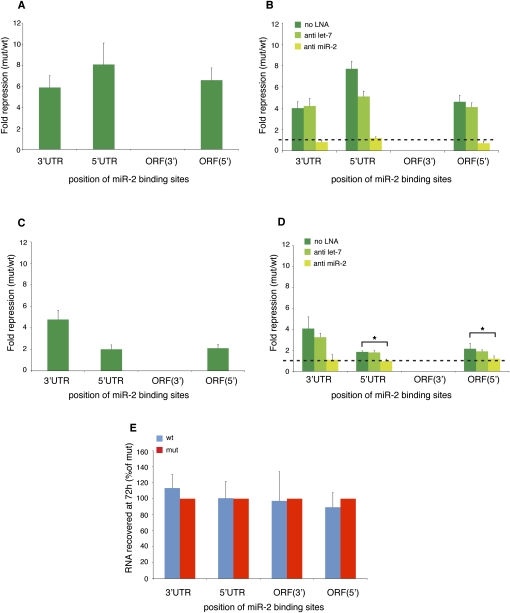
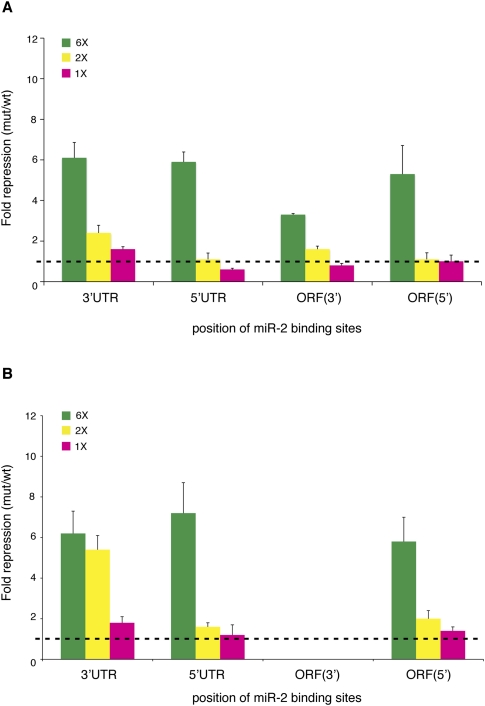
Upon incubation in the Drosophila cell-free system, we observe that two binding sites mediate just over twofold repression from the 3′UTR, whereas repression is less efficient from the 3′ end of the ORF and completely ineffective from the 5′UTR and the 5′ end of the ORF (Fig. 5A). Regulation from a single binding site in vitro is only observed from the 3′UTR (Fig. 5A). We next transfected the reporter mRNAs into S2 cells. Two binding sites trigger robust repression when located in the 3′UTR, but are much less effective in the 5′UTR and at the 5′ end of the ORF (Fig. 5B). A single binding site is ineffective from the 5′UTR, while it mediates weak repression from the 3′UTR and the ORF (Fig. 5B). Taken together, these findings show that single or dual miR-binding sites in the 5′UTR or the ORF are generally less functional than in the 3′UTR or even completely nonfunctional.
FIGURE 5.
miR-2-mediated regulation from single or dual binding sites in vitro and in vivo. (A) The GL-FL-reporter mRNAs containing one, two, or six miR-2-binding sites were incubated in the Drosophila cell-free extract. A single binding site is only effective from the 3′UTR. Two binding sites yield over twofold repression from the 3′UTR and mediate ∼1.5-fold repression from the 3′ end of the ORF, but are ineffective from the 5′UTR and the 5′ end of the ORF. (B) The GL-FL-reporter mRNAs containing one, two, or six miR-2-binding sites were transfected into Drosophila S2 cells. A single binding site triggers modest repression from the 3′UTR and the 5′ end of the ORF. Two binding sites trigger robust repression from the 3′UTR, but are only moderately effective from the 5′UTR and the 5′ end of the ORF. Shown are averages and standard deviations of five independent experiments.
DISCUSSION
Although the vast majority of identified endogenous targets for miRs bear their miRISC interaction site(s) in the 3′UTR, physiologically relevant examples of miR regulation from binding sites in the 5′UTR or the ORF have been described (Jopling et al. 2005; Duursma et al. 2008; Forman et al. 2008; Henke et al. 2008; Lal et al. 2008; Ørom et al. 2008; Tay et al. 2008; Elcheva et al. 2009; Tsai et al. 2009). Our work provides, to our knowledge, the first biochemical mechanistic analyses of miR regulation from positions 5′ of the 3′UTR. We show that miRs can principally regulate translation initiation, in a cap-dependent manner, from all (tested) sites of an mRNA, and that the establishment of repression is accompanied by miR-2-induced deadenylation (Figs. 2A–G, 3B), implying that the deadenylation machinery is recruited to the mRNA irrespectively of the position of the miRISC.
For miR-binding sites in the 5′UTR (and the ORF), it is conceivable that the miRISC acts by a dual mechanism: by interfering with translation initiation, and by causing steric hindrance to scanning/translating ribosomes that escape a cap-dependent initiation block (Zdanowicz et al. 2009). Several regulators binding to the 5′UTR have, indeed, already been shown to act sterically (Stripecke et al. 1994; Gebauer and Hentze 2004). For miR regulation from the 5′UTR, this possibility has been raised (Lee et al. 2009), and it will be interesting to study its potential contribution to repression. We note that repression from all sites, including the 5′UTR, is relieved equally strongly by the dominant-negative GW182(1-592) polypeptide (Fig. 3A). If we assume that these inhibited miRISC complexes remain bound to the target mRNAs by virtue of the miR–mRNA interaction, this result suggests that at least these miRISCs do not act primarily by simple steric hindrance.
miRs targeting the 5′UTRs have been reported to trigger both positive (Jopling et al. 2005; Ørom et al. 2008; Tsai et al. 2009) and negative (Lytle et al. 2007; Grey et al. 2010; this study) responses. Although these different regulatory effects are not yet mechanistically explained, it is tempting to speculate that they may be affected by specific structural features of the miR–mRNA interaction (e.g., seed-mediated recognition vs. noncanonical miR target sites) (Ørom et al. 2008), by the miRISC-mediated recruitment of auxiliary repressor/activator complexes, or even by the nature of the targeted RNA itself (e.g., the HCV genome) (Jopling et al. 2005). The availability of in vitro systems may, in the future, help to shed light on this unresolved issue.
We also validated the key findings of this study by in vivo transfection analyses. In line with previous findings (Lytle et al. 2007), these experiments uncover significant differences in quantitative aspects of the experimental results, depending on the transfection method used. Whereas we observe similar repression upon reporter mRNA transfection, regulation from the 5′UTR and the 5′ end of the ORF is weaker than from the 3′UTR when plasmid DNA is transfected (Fig. 4, cf. A and C). Although these results do not challenge the major mechanistic conclusion, they may illuminate physiological aspects of miR regulation, especially regarding single or dual binding sites. Furthermore, they may provide mechanistic clues regarding the relationship between the miRISC, the mRNP, and the translation (initiation) process. We envisage a scenario in which the 5′UTR and ORF miRISCs are challenged by ribosomes that have escaped the initiation block. The ribosomal challenge that the miRISC has to face is proportional, on the one hand, to the efficiency of the translation initiation process (and therefore the percentage of ribosomes that manage to escape the miRISC block) and, on the other hand, to ribosomal processivity (Bartel 2009). In our case, these parameters may differ between the Drosophila embryo in vitro system, transfection experiments with in vitro transcribed mRNAs or with mRNAs that are transcribed in vivo. Especially in the latter case, the nuclear history of the mRNA is expected to endow it with a different mRNP.
Previous reporter studies in mammals and zebrafish have used a fixed number of miR-binding sites (either two or four) to evaluate regulation from the 5′UTR or the ORF. This could possibly explain the discordant conclusions reached regarding the effectiveness of 5′UTR or ORF miR targeting (Kloostermann et al. 2004; Lytle et al. 2007; Gu et al. 2009). Therefore, we also systematically compared miR-mediated regulation by different numbers of miR-2-binding sites in the 3′UTR, the 5′UTR, or the ORF. We observe that single or dual miR-binding sites are less efficient from the 5′UTR and ORF than from the 3′UTR (Fig. 5A,B). Consistent with recent findings in mammalian cells (Gu et al. 2009), one or two miRISCs are expected to impose a weaker initiation block than six, and are therefore more likely to be displaced by “escaping ribosomes,” hence hampering repression.
In summary, our results establish a mechanistic basis for miR function from the 5′UTR and the ORF. We demonstrate that the translation initation block by miR-2 can also be imposed from the 5′UTR and the ORF. Together with the finding that a higher number of binding sites is required to establish effective regulation from the 5′UTR and the ORF, our observations help to resolve the conundrum resulting from mechanistic predictions made for a (cap-dependent) block of translation initiation and the seemingly inconsistent observation that the vast majority of physiological miR regulatory sites has been found in 3′UTRs. The data may also contribute to the definition of rules for 5′UTR and ORF miR targeting, thereby facilitating the (computational) identification of new physiological miR targets.
MATERIALS AND METHODS
Plasmids
The GL-FL-reporter plasmids were obtained by insertion of the human β-globin 5′UTR (amplified with primers gl-fw, 5′-TTTTGGTACCACATTTGCTTCTG-3′; and gl-rv, 5′-TTTTGGGCCCGGTGTCTGTTTTGAGG-3′) between the KpnI and ApaI sites of the FL-mut vector (Thermann and Hentze 2007). The wt and mut miR-2-binding sites of Drosophila reaper mRNA were obtained by PCR amplification using the primers 3′box-wt-fw (5′-TTTTACTAGTGGATCCAATTTTAGTTTACTCATCAAAGCGATTGTGATATTGG-3′), 3′box-wt-rv (5′-TTTTAAGTTTTAAGATCTAAACAAAACCAATATCACAATCGCTTTGATGAGTAAAC-3′), 3′box-mut-fw (5′-TTTTACTAGTGGATCCAATTTTAGTTTACTCATCAAAGCGAGGTATTATTTGG-3′), and 3′box-mut-rv (5′-TTTTAAGCTTTTAAGATCTAAACAAAACCAAATAATACCTCGCTTTGATGAGTAAAC-3′). The cloning of one, two, or six binding sites in the 3′UTR was done as previously described (Thermann and Hentze 2007). The ORF(3′)constructs were obtained by replacing a C-terminal fragment of the luciferase ORF, to eliminate the stop codon, using the primers deltastop-fw (5′-TTTTATCGATATTGTTACAACACCCCAACATCTTCG-3′) and deltastop-rv (5′-TTTTACTAGTGGATCTCAATTTGGACTTTCCG-3′) and the restriction sites ClaI and SpeI of the GL-FL-3′UTR wt and mut reporters. The 5′UTR and ORF(5′) reporters were obtained by amplifying the binding sites with the primers ORF(5′)box-fw (5′-TTTTGGGCCCAGTCATGGCCACTAGTGGATCC-3′), 5′UTRbox-fw (5′-TTTGGGCCCTCCACTAGTGGATCC-3′), and 5′box-rv (5′-TTTTCTCGAGGCGGAAGCTTAGATCT-3′) and inserting them using the ApaI and XhoI restricion sites of a GL-FL-3′UTR vector that was previously cut with BamHI and BglII and religated so as to eliminate the miR-2-binding sites from the 3′UTR. The RL-control plasmid was previously described (Thermann and Hentze 2007). The GL-sORF-reporter plasmids were generated by replacing the firefly luciferase ORF in the GL-FL reporters with the sORF, using the primers sORF-fw (5′-TTTTCTCGAGTCATGGACTACAAAGACGACG-3′), sORF-deltastop-rv (5′-TTTTACTAGTTAACAATTTGGACTTTCCG-3′), and sORF-rv (5′-TTTTACTAGTTTATAACAATTTGGACTTTCCG-3′) and the XhoI and SpeI restriction sites. The HS-GL-FL-reporter plasmids were generated from the pCaSpeR-hs-pA vector. The GL-FL-3′UTR and -ORF(3′) fragments were cloned into the pCaSpeR-hs-pA using the KpnI and BglII restriction sites. The GL-FL-5′UTR and -ORF(5′) fragments were PCR-amplified with the primers gl-fw and fluc-rv (5′-TTTTGTTAACGGATCTATTACAATTTGGACTTTGCG-3′) and cloned via the KpnI and HpaI sites of the pCaSpeR-hs-pA vector. The HS-RL-control plasmid was previously described (Duncan et al. 2006).
In vitro transcription
The synthesis of the Gl-FL-reporter, the GL-sORF-reporter, and the RL-control mRNAs was performed as previously described (Thermann and Hentze 2007).
Preparation of Drosophila melanogaster embryo extract and translation experiments
Drosophila embryo extracts were prepared and used for translation assays as described (Thermann and Hentze 2007). The preincubation step was carried out for 1 h or 30 min at 25°C for luciferase mRNA and protein quantification and for deadenylation analyses, respectively, and the amounts of reporter mRNAs were adjusted to maintain constant RNA concentrations. Where indicated, 30 nM anti-miR LNA (dme-miR-2 or hsa-let-7 miRCURY LNA MicroRNA Knockdown Probes from Exiqon) was added to the translation reaction. For the experiment shown in Figure 3A, 200 nM GW182(1-592) protein or an equal volume of buffer was added to the translation reaction.
RT–qPCR
Total RNA from translation reactions or from transfected cells was isolated with Trizol (Invitrogen) according to the manufacturer's protocol. Total RNA isolated from cells was treated with Turbo DNA-free kit (Ambion) and 1 μg was reverse-transcribed with random primers and SuperScript II Reverse Transcriptase (Invitrogen) following the manufacturer's protocol. RT–qPCR was then performed as described (Zdanowicz et al. 2009).
Cell culture and S2 cell transfection
Schneider 2 cells were maintained at 25°C in Schneider's Medium containing L-glutamine (Gibco) and supplemented with penicillin/streptomycin (Gibco) and 10% fetal bovine serum (Gibco). RNA transfections were performed with the TransMessenger Transfection Reagent (Qiagen) according to the manufacturer's protocol. Cells were seeded at 106 cells per well in 24-well plates 24 h before transfection. Cells were transfected with 30 fmol of each of the Gl-FL-reporter mRNAs and 6 fmol of the RL-control mRNA, plus 100 nM anti-miR LNA where indicated. Cells were lysed after 24 h in 1× Passive Lysis Buffer (Promega), and firefly and Renilla luciferase activity was determined with the Dual-Luciferase Assay System (Promega). DNA transfections were performed with the Effectene Transfection Reagent (Qiagen) according to the manufacturer's protocol. Cells were seeded at 1.5 × 106 cells per well in six-well plates 24 h before transfection. Cells were transfected with 3 fmol of each of the HS-GL-FL-reporter plasmids and 0.3 fmol of the HS-RL-control plasmid, plus 100 nM of anti-miR LNA where indicated. Cells were lysed after 72 h in 1× Passive Lysis Buffer, and firefly and Renilla luciferase activity was determined with the Dual-Luciferase Assay System.
Deadenylation assay
The deadenylation assay was carried on as previously described (Zdanowicz et al. 2009). The primers RNaseH3 (5′-CGCCCTTCTTGGCCTTTATG-3′) and RNaseH4 (5′-GCGGTCAACTATGAAGAAGTGTTCG-3′), complementary to the firefly luciferase ORF, were used as specified.
Sucrose density-gradient analyses
The analyses of 80S complex assembly were performed through 15%–45% linear sucrose density gradients in the presence of 1 mM cycloheximide as previously described (Thermann and Hentze 2007).
ACKNOWLEDGMENTS
We thank Elisa Izaurralde (Tübingen) for the GW182(1-592) expression plasmid, Agnieszka Zdanowicz for the GW182(1-592) polypeptide, Edward Darzynkiewicz (Warsaw) for the cap 21-ARCA, Jan Medenbach for the pCaSpeR-hs-pA vector, and all members of the Hentze laboratory for helpful discussions. This work was funded by a grant (HE 1442/12-1) from the Deutsche Forschungsgemeinschaft to M.W.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2384610.
REFERENCES
- Barreau C, Dutertre S, Paillard L, Osborne HB 2006. Liposome-mediated RNA transfection should be used with caution. RNA 12: 1790–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW 2006. Sex-lethal imparts a sex-specific function to UNR by recruting it to the msl-2 mRNA 3′UTR: Translational repression for dosage compensation. Genes Dev 20: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R 2008. miR-148 targets human DNMT3b protein coding region. RNA 14: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Goswami S, Noubissi FK, Spiegelman VS 2009. CRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 35: 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. GW182 interaction with Argonaute is essential for miR-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009. The GW182 protein family in animal cells: New insights into domains required for miR-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA 2008. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci 105: 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW 2004. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog 6: e100967 doi: 10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA 2009. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol 16: 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Kloostermann WP, Wienholds E, Ketting RF, Plasterk RH 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 32: 6284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullman R Jr Srikatan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. 2008. p16INK4a translation suppressed by miR-24. Plos One 3: e1864 doi: 10.1371/journal.pone.0001864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD 2009. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′UTR. Proc Natl Acad Sci 104: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G 2007. Argonaute proteins: Mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Schnall-Levin M, Zhao Y, Perrimon N, Berger B 2010. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Nat Acad Sci 107: 15751–15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradopour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripecke R, Oliveira CC, McCarthy JE, Hentze MW 1994. Proteins binding to 5′ untranslated region sites: A general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol 14: 5898–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW 2007. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447: 875–878 [DOI] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG 2007. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Tsai NP, Lin YL, Wei LN 2009. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and upregulates its protein expression. Biochem J 424: 411–418 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Waud JP, Sala-Newby GB, Matthews SB, Campbell AK 1996. Engineering the C-terminus of firefly luciferase as an indicator of covalent modification of proteins. Biochim Biophys Acta 1292: 89–98 [DOI] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darsynkiewicz E, Hentze MW 2009. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell 35: 881–888 [DOI] [PubMed] [Google Scholar]