Abstract
The choice of synonymous codons used to encode a polypeptide contributes to substantial differences in translation efficiency between genes. However, both the magnitude and the mechanisms of codon-mediated effects are unknown, as neither the effects of individual codons nor the parameters that modulate codon-mediated regulation are understood, particularly in eukaryotes. To explore this problem in Saccharomyces cerevisiae, we performed the first systematic analysis of codon effects on expression. We find that the arginine codon CGA is strongly inhibitory, resulting in progressively and sharply reduced expression with increased CGA codon dosage. CGA-mediated inhibition of expression is primarily due to wobble decoding of CGA, since it is nearly completely suppressed by coexpression of an exact match anticodon-mutated tRNAArg(UCG), and is associated with generation of a smaller RNA fragment, likely due to endonucleolytic cleavage at a stalled ribosome. Moreover, CGA codon pairs are more effective inhibitors of expression than individual CGA codons. These results directly implicate decoding by the ribosome and interactions at neighboring sites within the ribosome as mediators of codon-specific translation efficiency.
Keywords: yeast, translation, genetic code, ribosome, codons, tRNA
INTRODUCTION
Differences in translation efficiency contribute substantially to the large variation in protein expression, with translation of individual mRNAs ranging over at least three orders of magnitude in the yeast Saccharomyces cerevisiae (Arava et al. 2003; de Godoy et al. 2008; Ingolia et al. 2009). While differential translation efficiency is the result of many factors, substantial evidence from both bacteria and eukaryotes indicates that the amount of polypeptide produced per mRNA is influenced by the particular choice of synonymous codons, codons that specify insertion of the same amino acid (Ghaemmaghami et al. 2003; Brockmann et al. 2007).
In organisms such as Escherichia coli, S. cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster, a subset of synonymous codons, generally those decoded by the most abundant tRNAs without wobble decoding (Ikemura 1982), are “preferred codons” for efficient translation and are used more frequently than their synonyms (dos Reis et al. 2004; Parmley and Hurst 2007). Preferred codons are used nearly exclusively in many highly expressed genes, sharply differentiating these genes from the majority of other genes (Grantham et al. 1980; Bennetzen and Hall 1982; Duret and Mouchiroud 1999). Indeed, codon usage and protein abundance correlate genome-wide in S. cerevisiae and E. coli (Ghaemmaghami et al. 2003; Tuller et al. 2007; Ishihama et al. 2008; Tuller et al. 2010b). Remarkably, despite changes in the actual identity of the preferred codons, the selection for preferred codons in many gene families is maintained across species (Man and Pilpel 2007). In addition, expression of genes in E. coli and in S. cerevisiae is frequently improved by recoding genes with preferred codons (Gustafsson et al. 2004; Burgess-Brown et al. 2008; Keppler-Ross et al. 2008; Quartley et al. 2009; Welch et al. 2009), or by overproducing rare tRNAs in E. coli (Burgess-Brown et al. 2008).
Nonetheless, the degree to which codons affect expression in E. coli has been the topic of some debate, with two recent papers arriving at very different conclusions about the quantitative importance of codon effects. Kudla et al. (2009) concluded that codon bias had little significant effect on protein levels of GFP variants, while Welch et al. (2009) found that variation in expression of two genes was strongly correlated with codon biases for 10 amino acids. However, it is likely that the coupling between transcription and translation complicates the analysis of codon effects in bacteria, since some codon changes are known to cause premature rho-mediated transcription termination (Deana et al. 1998). This difference in the molecular basis for codon effects between bacteria and eukaryotes is an argument to examine the role of codon usage in a eukaryote.
Assessing the contribution of codons to gene expression in eukaryotes still has at least two difficulties. First, synonymous codon choice influences several aspects of translation, including the accuracy of amino acid insertion (Kramer and Farabaugh 2007), reading frame maintenance (Farabaugh et al. 2006), and folding of the nascent polypeptide (Kimchi-Sarfaty et al. 2007), each of which may differentially affect the stability or activity of the resulting protein. Second, synonymous codon choice affects the mRNA sequence, thereby affecting mRNA structure, which itself directly affects translation (Kudla et al. 2009). The mRNA sequence also influences the binding of proteins and RNAs that regulate splicing, decay, and translation efficiency (Doma and Parker 2006; Isken and Maquat 2007).
Although the preferred codons are well characterized, neither the identity nor the properties of codons or codon combinations that cause reduced expression in eukaryotes are known. Codons that impair expression in yeast are difficult to decipher because the 37 unpreferred codons have different properties, with some decoded by rare tRNAs, others requiring wobble decoding, and still others are simply underused. Moreover, it remains controversial whether the mediator of reduced expression is individual codons, adjacent codons (codon pairs), or clusters of rare codons (Smith and Yarus 1989; Irwin et al. 1995; Kane 1995; Moura et al. 2005; Buchan et al. 2006; Coleman et al. 2008). Knowledge of the identity and properties of codon-initiated inhibitory signals is required to understand the underlying mechanisms for this regulation.
Here we describe the first systematic study of codon effects on expression. We assessed the effect of each of 59 codons in the yeast S. cerevisiae by monitoring firefly luciferase activity of constructs with repeats of individual codons. We identify the Arg codon CGA as strongly inhibitory, an effect due almost entirely to I•A wobble decoding of the CGA codon. We find that CGA codons give rise to a stable mRNA fragment containing sequences downstream from the CGA codons, from which we infer that these codons likely cause arrest of the ribosome. Moreover, we find that adjacent CGA codons are much more potent inhibitors of expression than separated CGA codons. Thus, we conclude that translation efficiency is directly modulated by decoding interactions within the ribosome.
RESULTS
In a systematic analysis, most codons exert only slight effects on expression
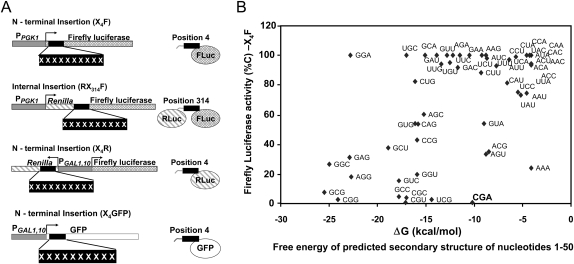
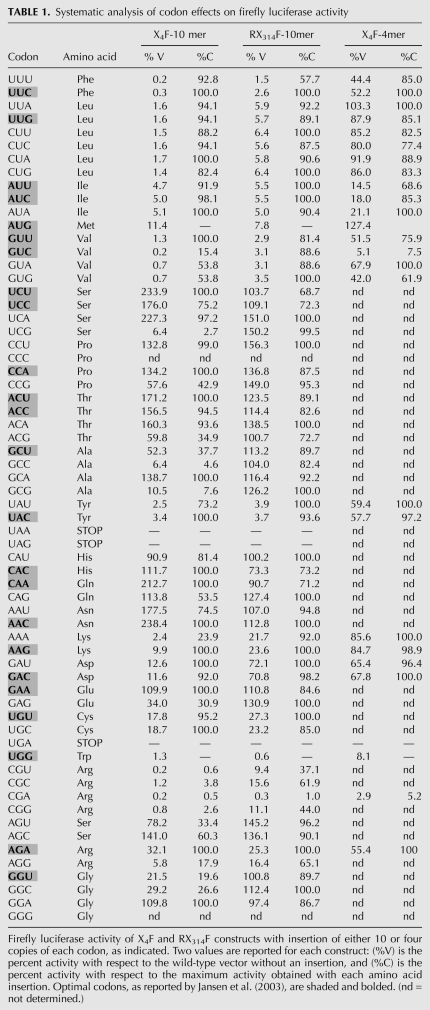
To assay the effects of sequences containing codon repeats, we inserted them into common flanking sequences, either near the N-terminus at amino acid 4 of the firefly luciferase gene (Grentzmann et al. 1998; Keeling et al. 2004) (referred to as X4F), or at amino acid 314 between the fused Renilla and firefly luciferase genes (RX314F) (Fig. 1A). We examined the effect of repeats of 10 identical codons in both positions, for 59 codons (except [CCC]10 and [GGG]10 for technical reasons). We also examined the effect of repeats of four identical codons in the N-terminal position, for 23 codons, specifying nine amino acids that were found to be inhibitory with all specifying codons in the 10-mers. Relative luciferase activity of yeast expressing each of these constructs was assayed and reported, both with respect to expression from a vector with no insert (%V), and with respect to the identical polypeptide encoded by the synonymous codon that gave maximal activity (%C) (Table 1).
FIGURE 1.
Systematic screen of the effects of synonymous codon repeats on firefly luciferase expression. (A) Schematic of reporter constructs used in these studies: Codon repeats (X) were inserted upstream of a firefly luciferase reporter gene either at position 4 of the coding region (X4F), at position 314 in the Renilla-firefly fusion construct (RX314F) or at position 4 of the coding region of Renilla luciferase (X4R) or superfolder GFP (X4G) (Pedelacq et al. 2006). X4F and RX314F were used for a systematic screen of the effects of 59 codons on expression. (B) Inhibitory effects of several codon repeats, but not CGA repeats, in X4F are likely due to formation of strong RNA secondary structures. Luciferase activity for each 10-mer codon repeat at the N-terminus (%C) is plotted as a function of the free energy of the predicted RNA secondary structure from 1 to 50. Free energy of folding (nucleotides 1–50 of X4F-reporter constructs containing 10 repeats of a particular codon for 59 of the 61 sense codons) (kcal/mol) was calculated using RNAstructure (Mathews et al. 1999) version 4.6 (http://rna.urmc.rochester.edu/RNAstructure.html). Similar results were obtained from calculations of free energy of folding from −4 to +37.
TABLE 1.
Systematic analysis of codon effects on firefly luciferase activity
We draw three conclusions from the analysis of the 57 codons with one or more synonymous codons. First, most synonymous codons have little effect on expression in these reporters. Most constructs yielded >66%C activity, including 36 and 51 of 57 codon repeats with 10 amino acids in X4F and in RX314F, respectively, and 20 of 23 codon repeats with four amino acids in X4F. As expected, most of the codons identified as preferred codons in S. cerevisiae (Jansen et al. 2003) yielded high expression (19 of 22 in X4F and 22 of 22 in RX314F) (see Table 1).
Second, the inhibitory effects of many codon repeats in X4F are likely due to formation of secondary structures in the mRNA near the AUG, and thus may not reflect codon effects. For 15 codon repeats, we find that poor expression of X4F correlates with predicted thermodynamic stability in the first 50 bases (folding free energy less than −16 kcal/mol) (Fig. 1B). Since strong secondary structures near the 5′ end are inhibitory in E. coli (Kudla et al. 2009) and are selected against in both E.coli and S. cerevisiae (Tuller et al. 2010b), we infer that secondary structure may explain some or all of the inhibitory effects of these codon repeats in the N-terminal location, and, thus, these codons were not further analyzed. Similarly, the effects of Lys AAA (24%C) are likely due to the polyA sequence, which is known to inhibit expression (Dimitrova et al. 2009).
Third, Arg CGA was strongly inhibitory both in the N-terminal and internal positions (Fig. 2A; Table 1). In the N-terminal position, four other sequences likely encode inhibitory codons (Fig. 1B; Table 1,), including Thr ACG, Val GUA, and possibly Ser UCG codons, which are decoded by essential single (or two copy) tRNAs (Johansson et al. 2008) and Ser AGU, which, like Arg CGA, is decoded by wobble decoding. In the internal position, four other Arg codons, as well as Phe UUU, yielded <66%C (Fig. 2A; Table 1).
FIGURE 2.
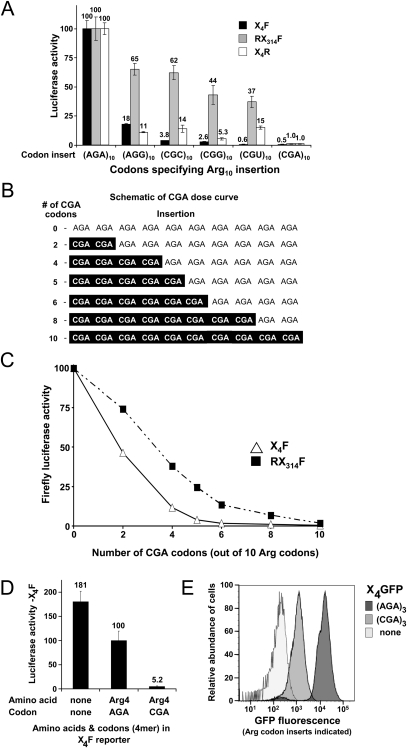
CGA codon repeats inhibit expression in a dose-dependent manner. (A) Luciferase activity of three different reporter constructs (X4F, RX314F, and X4R), containing (Arg)10 insertions specified by each of the six Arg codons. (B) The relationship between CGA codon dose and luciferase activity was examined with (Arg)10 inserts containing increasing CGA content, as illustrated in the schematic. (C) Firefly luciferase activity as a function of CGA codon dose in X4F (Δ) and RX314F (▪)—reporter constructs. (D) Firefly luciferase activity of X4F-reporter constructs containing either no insertion, or insertion of four Arg residues, specified with the indicated CGA or AGA codons. Values were normalized to the X4F-(AGA)4 reporter. (E) Expression of integrated GFP constructs with three Arg residues inserted at amino acid 4 specified by AGA or CGA codons. GFP expression was assessed based on the distribution of yeast cells exhibiting different intensities of fluorescence emission at 530 nm.
CGA codon repeats severely inhibit expression in a dose-dependent manner
The Arg CGA codon stood out as a candidate for a strongly inhibitory codon, because of both the magnitude and generality of its effects on expression. The CGA codon repeats were strong inhibitors of expression in both positions, with 10 codon repeats resulting in 0.5%C and 1%C firefly luciferase activity, respectively (Fig. 2A). CGA repeats also exerted strong inhibitory effects upstream of Renilla luciferase (Fig. 2A). Although all of the unpreferred Arg codon repeats were inhibitory, none affected all three reporters to nearly the same extent as CGA.
The effects of CGA codons on expression are unlikely to be due to RNA secondary structure for two reasons. First, the predicted free energy of the CGA repeat (−10.2 kcal/mol) is inconsistent with an inhibitory RNA secondary structure (Fig. 1B). Second, CGA repeats in the other reading frames did not cause similar effects. The Asp GAC and Thr ACG codons, which are permutations of CGA, form repeats of the same sequence, and thus, likely form structures similar to (CGA)10, but neither codon repeat inhibits expression nearly as much as CGA, either in terms of absolute, or polypeptide-adjusted, levels of luciferase (Table 1).
We also found that very few CGA codons are required for severe inhibition of expression. First, to determine the extent to which CGA codons affect expression, we examined the expression of firefly luciferase as a function of CGA codon dosage. In each case, we made identical Arg10-containing luciferase proteins, with the Arg residues coded by increasing numbers of CGA codons (Fig. 2B). In this context, two CGA codons in the N-terminal position caused a decrease in firefly luciferase activity to 46%C, while four CGA codons caused a decrease to 12%C (Fig. 2C). Within the first 14 amino acids, the position of the codons did not matter, as the constructs (CGA)5–(AGA)5 and (AGA)5–(CGA)5 yielded similar levels of luciferase activity (see Fig. 7B below). Furthermore, CGA codons were also strongly inhibitory in the internal location (RX314F), although slightly more CGA codons were required to exert similar levels of inhibition in this position compared to the N-terminal location (Fig. 2C). Second, the inhibitory effects of CGA were apparent with only three or four Arg residues. Inhibition by CGA codons (5.2%C) is observed with only four Arg residues upstream of firefly luciferase (Fig. 2D; Table 1), and three arginines upstream of GFP (6.7%C) (Fig. 2E). Thus, inhibition by CGA is effective with very few codons and does not require an extended arginine repeat.
FIGURE 7.
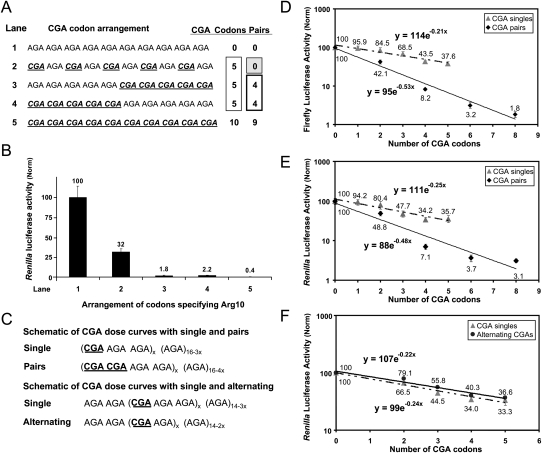
CGA codon pairs are more inhibitory than single CGA codons. (A,B) The effects of the arrangement of five CGA codons within an (Arg)10 insert on Renilla luciferase activity were compared to each other and to Renilla luciferase activity from constructs containing (Arg)10 insertions specified by (AGA)10 or (CGA)10. (A) The arrangements of Arg CGA codons with the number of CGA codons and codon pairs indicated and (B) the resulting luciferase activity with lanes corresponding to constructs in A. Reported values were corrected to an independently transcribed firefly luciferase gene, and then normalized to (AGA)10. (C) Schematic of the arrangement of CGA codons in test constructs designed to examine the effects of single CGA codons, pairs of CGA codons, and alternating CGA codons. (D) Relative luciferase activity of firefly luciferase reporter constructs containing (Arg)16 insertions with increasing CGA content arranged either as single codons or codon pairs. Reported values were corrected to an independently transcribed Renilla luciferase gene, and then normalized to (AGA)16. (E) Relative luciferase activity of Renilla luciferase reporter constructs containing (Arg)16 insertions with increasing CGA content arranged either as single codons or codon pairs. Reported values were corrected to an independently transcribed firefly luciferase gene, and then normalized to (AGA)16. (F) Relative luciferase activity of Renilla luciferase reporter constructs containing (Arg)16 insertions with increasing CGA content. CGA codons are clustered either as single codons separated by two AGA codons or as alternating codons (CGA–AGA). Reported values were corrected to an independently transcribed firefly luciferase gene, and then normalized to (AGA)16.
Inhibition by CGA is due to I•A wobble decoding
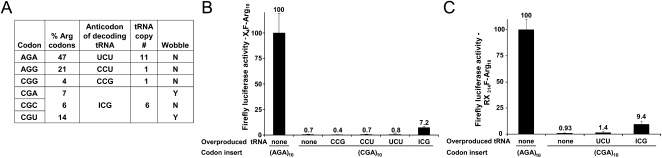
To determine explicitly if the CGA codon inhibits expression, we examined the effect of overproducing the corresponding tRNA species on expression of (CGA)10 constructs. CGA codons are decoded by tRNAArg(ICG), which is encoded by six tRNAArg genes (Fig. 3A) bearing an ACG anticodon that is subsequently converted to ICG (Grosjean et al. 2010). Because the number of gene copies of tRNAs vary from one to 17, and the amounts of various tRNA species correspond closely to the number of genes that specify each variant (Tuller et al. 2010a), these six copies should yield sufficient amounts of tRNA to decode CGA. Nonetheless, overproduction of tRNAArg(ICG), but not any other Arg isoacceptor, resulted in an ∼10-fold increase in X4F–(CGA)10 luciferase activity compared to the vector control (Fig. 3B), and similarly suppressed the inhibitory effect of the internal CGA codon repeat (Fig. 3C). This increase in expression to ∼7% of X4F–(AGA)10 luciferase activity (Fig. 3B) was as much as would be expected from the likely increase in the concentration of tRNAArg(ICG) due to the tRNA gene on the multicopy plasmid, since the multicopy plasmid added ∼25 copies of the tRNAArg(ICG) gene (Schneider and Guarente 1991) to the six genomic copies, resulting in an approximately fivefold increase in gene copy number. Thus, we conclude that some of the CGA-mediated reduced expression is due to a defect in translation of CGA codons.
FIGURE 3.
Suppression of CGA-mediated expression defects by its cognate tRNA. (A) The Arg codons are compared with respect to use in the genome, gene copy number of the decoding tRNAArg species and the requirement for wobble decoding. (B) Effect of overproduction of the indicated tRNAArg species from high copy plasmids on firefly luciferase activity of X4F-reporter constructs containing (CGA)10 insertions. Values were compared to activity of X4F-(AGA)10 luciferase in a strain with an empty vector. (C) Effect of overproduction of tRNAArg(ICG) from a high copy plasmid on firefly luciferase activity of RX314F-reporter constructs containing (CGA)10 insertions at amino acid 314. Values were normalized to (AGA)10 overexpressing a vector control.
Since suppression by the overproduced tRNAArg(ICG) was incomplete, we inferred that either a portion of the CGA effects is not codon-dependent, or that decoding of the CGA codon by its cognate tRNAArg(ICG) is inherently inefficient. Decoding of the CGA codon might be inefficient either due to inherently poor tRNA function of tRNAArg(ICG) or due to the I•A wobble decoding necessary to decode the CGA codon. To address these issues, we constructed two tRNAs that should exactly base pair (an anticodon-match) to the CGA codon, by mutating the anticodon of the major tRNAArg(UCU) species from UCU to UCG, and of tRNAArg(ICG) from ACG to UCG.
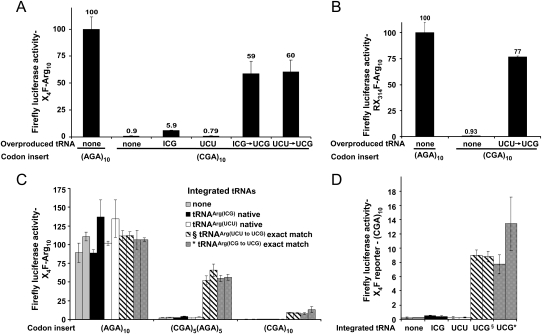
The introduction of an anticodon-matched isoaccepting tRNA almost completely suppressed the expression defect caused by the 10 CGA codons. Overproduction of either the mutant tRNAArg(UCU to UCG) or the mutant tRNAArg(ICG to UCG) resulted in a 60-fold increase in luciferase activity, to nearly the levels of X4F–(AGA)10 (Fig. 4A). Similar suppression was observed for the reporter with CGA codons at amino acid 314 (Fig. 4B). This result demonstrates that almost all effects of the (CGA)10 sequence are due to the use of this codon in translation, suggests that CGA decoding is severely deficient, and implies that I•A wobble decoding is a prominent factor in CGA effects on translation efficiency.
FIGURE 4.
Suppression of CGA-mediated expression defects by an exact base-pairing anticodon mutated tRNAArg(UCG). (A) Effect of indicated native and anticodon-mutated tRNAArg species, expressed on high copy plasmids, on firefly luciferase activity of X4F-reporter constructs containing (CGA)10 insertions. The parent tRNA of each anticodon-mutated tRNAArg is indicated by the first anticodon shown below the figure. Values were compared to activity of X4F-(AGA)10 luciferase in a strain with an empty vector. (B) Effect of an anticodon-mutated tRNAArg species, expressed on a high copy plasmid, on firefly luciferase activity of a RX314F-reporter construct containing a (CGA)10 insertion. (C) Suppression of the inhibitory effects of CGA codons by integrated copies of the mutant tRNAArg(UCG) species that decode CGA without wobble. Luciferase activity of X4F-reporter constructs containing (Arg)10 insertions, specified by (AGA)10, [(CGA)5(AGA)5], or (CGA)10, in yeast strains bearing an integrated copy of either of two native tRNAArg species, two nonnatural exact match tRNAArg species, or a vector control as indicated. Two individual integrant strains were analyzed in each case. Values were normalized to the average of the activity of X4F-(AGA)10 luciferase in the two integrant strains expressing a vector control. Error bars represent the standard deviation from three independent transformants, each assayed in triplicate. (D) A closer examination of expression of X4F-reporter constructs containing (CGA)10 insertions in indicated strains with an expanded scale of the y-axis. Values are normalized as in C.
To evaluate the importance of tRNA abundance in decoding CGA with an exact match tRNA, we examined suppression of CGA-mediated inhibition of expression by a single copy of the anticodon mutated tRNA genes integrated into the yeast chromosome (Fig. 4C,D). Remarkably, we found that an integrated copy of either tRNAArg(ICG to UCG) or tRNAArg(UCU to UCG) strongly suppressed the expression defect caused by CGA codons, resulting in a 40- to 50-fold improvement in expression of the (CGA)10 construct (Fig. 4D). Similarly, expression of a (CGA)5(AGA)5 luciferase construct improved ∼20-fold in strains expressing these exact match single copy tRNAs (Fig. 4C). The simplest interpretation of this strong suppression is that the exact match tRNAArg(UCG) decodes a disproportionate fraction of CGA codons relative to that decoded by the more abundant tRNAArg(ICG). Thus, the decoding properties of the exact match tRNAArg(UCG), not its abundance, are responsible for efficient expression.
CGA codons do not result in reduced mRNA amounts, but do yield a shortened RNA
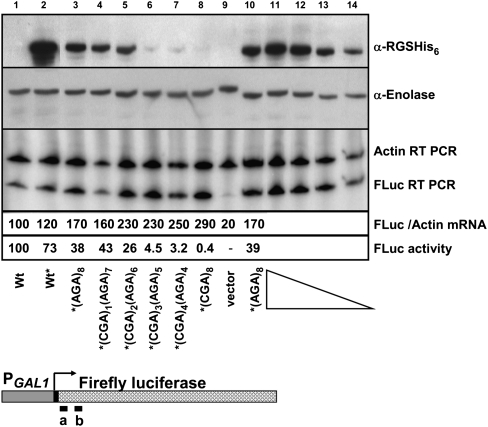
Defects in translation are often closely coupled to mRNA degradation (Frischmeyer et al. 2002; van Hoof et al. 2002; Inada and Aiba 2005; Doma and Parker 2006; Isken and Maquat 2007) and sometimes also affect protein folding and activity (Kimchi-Sarfaty et al. 2007). To directly assess mRNA and protein levels, we introduced an RGS-(His)6 epitope tag upstream of an (Arg)8 insertion, coded by (AGA)8 or increasing increments of CGA up to (CGA)8. The introduction of progressively more CGA codons resulted in reduced firefly luciferase activity (Fig. 5) and in a parallel loss of protein (Fig. 5). Firefly luciferase protein was weakly detectable from strains bearing the (CGA)3(AGA)5 and (CGA)4(AGA)4 constructs, which produced 8%C–12%C luciferase activity, and was undetectable from strains bearing the (CGA)8 constructs. However, we did not observe concurrent loss of firefly luciferase mRNA from any of the (CGA)-containing constructs, relative to that from constructs with (AGA)8, using an RT-PCR assay with the indicated probes (Fig. 5).
FIGURE 5.
CGA-mediated inhibition affects firefly luciferase activity and protein levels, but does not result in reduced luciferase mRNA. Analysis of firefly luciferase protein, mRNA, and activity in X4F-reporter constructs containing RGS-(His)6-(Arg)8 insertions, from yeast strains bearing the indicated constructs (lanes 1–10). Firefly luciferase protein was detected with antibody to RGS-(His)6, with antibody to enolase serving as an internal control. In lanes 11–14, the top two panels contain dilutions of crude extracts expressing the X4F- RGS-(His)6 (AGA)8 reporter construct (10, 7.5, 5.0, 2.5 μg). Firefly luciferase mRNA was measured by RT-PCR analysis using primers homologous to bases 21–46 and 148–173, as indicated on the diagram below the panel; sequences are given in Supplemental Table S3; measurements of Actin mRNA served as an internal control. In the third panel, lanes 11-14 contain RT-PCR reactions in which dilutions of DNase-treated RNA from X4F-RGS-(His)6 (CGA)8-containing sample (0.6, 0.3, 0.15, 0.075 μg) were used in the RT reactions, with equal volumes of the RT reaction in each PCR reaction. Reverse transcriptase reactions in lanes 1–10 contain 0.3 μg total RNA input. Firefly luciferase mRNA amounts were first normalized to the levels of Actin mRNA and then quantified relative to the wild-type X4F-reporter construct with no insertion; the FLuc mRNA levels in the wild-type strain were arbitrarily set to 100. The presence of RGS-(His)6 is indicated by an asterisk (*). Firefly luciferase activity of X4F-reporter constructs containing RGS-(His)6-(Arg)8 insertions was normalized to the wild-type X4F-reporter construct with no insertion.
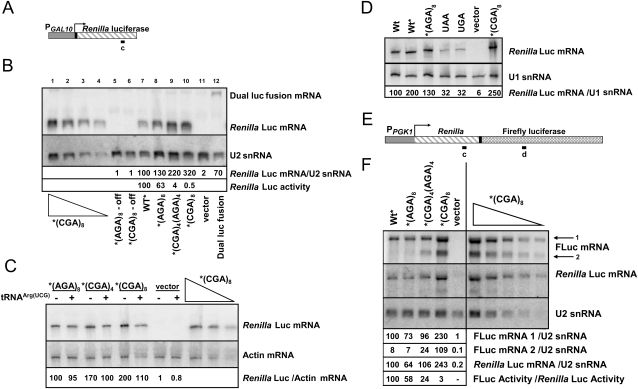
To ascertain whether mRNA stability of transcripts bearing CGA codons is a general phenomenon and to examine the RNA species more carefully, we also examined Renilla and firefly luciferase mRNAs by Northern analysis of RNA from strains bearing the RGS-(His)6-(Arg)8 insertions at either amino acid 4 of Renilla luciferase (X4R) or at amino acid 314 in the Renilla-firefly luciferase fusion (RX314F). In both cases, we observed that the amount of full-length luciferase RNA from the (CGA)8 construct is similar to (or slightly greater than) that from the corresponding (AGA)8 construct (Fig. 6B,F). Neither the amount nor the approximate size of Renilla luciferase mRNA from the X4R reporter was reduced in RNA from strains bearing either the (CGA)4(AGA)4 or the (CGA)8 constructs, relative to that of the (AGA)8 or no-insert constructs (Fig. 6A,B). In fact, the CGA-containing mRNAs were approximately twofold more abundant (Fig. 6B), a phenomenon that was not observed if tRNAArg(ICG to UCG) was coexpressed (Fig. 6C). As expected, the amounts of Renilla luciferase protein were reduced by CGA codons, and restored by coexpression of the exact match tRNAArg(UCG) (data not shown). Likewise, the large RNA from the RX314F fusion that hybridized to both the Renilla luciferase and firefly luciferase probes was approximately twofold more abundant in strains bearing constructs with (CGA)8, compared to strains bearing constructs with (AGA)8, [(CGA)4(AGA)4], or no insert (Fig. 6F). Furthermore, we note that Renilla luciferase mRNA is itself suitable for RNA degradation, since introduction of either a UAA or a UGA stop codon at amino acid 4, which would be expected to elicit nonsense-mediated decay in susceptible genes (Isken and Maquat 2007), results in reduction of Renilla luciferase mRNA to ∼32% of the no-insert control (Fig. 6D). Thus, we conclude that CGA-mediated inhibition of expression is not generally accompanied by mRNA degradation.
FIGURE 6.
The effects of CGA codons at amino acids 4 and 314 on full-length mRNA, examined by Northern analysis. (A) Diagram of the X4R Renilla luciferase reporter indicating the position of the oligonucleotide probe used in panels B–D. Probe c is located at base pairs 682–705 in the 935 base-pair Renilla luciferase coding sequence. (B) Analysis of the effects of insertion of CGA codons at amino acid 4 upstream of Renilla luciferase on Renilla luciferase mRNA and activity. Renilla luciferase mRNA from X4R reporter constructs containing RGS-(His)6-(Arg)8 was examined by Northern blots with the probe c indicated in panel A; U2 snRNA served as an internal control. Renilla luciferase mRNA levels were first normalized to levels of U2 snRNA and then compared to the wild-type Renilla luciferase mRNA containing RGS-(His)6. In the lower panel, luciferase activity of Renilla luciferase reporter constructs containing RGS-(His)6-(Arg)8 insertions are shown. (Off, expression under repressing conditions.) Lanes 1–4 contain different amounts of input total RNA from the X4R- RGS-(His)6 (CGA)8 bearing strain (20, 10, 5, 2.5 μg), while lanes 5–12 contain RNA (20 μg) from yeast strains bearing the indicated constructs. (C) Decoding of CGA codons with an anticodon-mutated tRNAArg species suppresses the CGA-mediated increase in Renilla luciferase mRNA. Northern blot analysis of Renilla luciferase mRNA from Renilla luciferase reporter constructs containing RGS-(His)6-(Arg)8, using Actin as an internal control. Strains expressed either a nonnatural exact match tRNA or a vector control. Luciferase mRNA amounts were normalized to the levels of Actin mRNA and then to RGS-(His)6 (AGA)8; In the titration indicated with the triangle, mRNA input from the strain X4R- RGS-(His)6 (CGA)8 in a vector control was varied (20, 10, 5 μg). (D) Insertion of stop codons at amino acid 4 of Renilla luciferase resulted in a reduction in stable Renilla luciferase mRNA. Northern blot analysis of Renilla luciferase mRNA from indicated Renilla luciferase reporter constructs. Renilla luciferase mRNA was first normalized to levels of U1 snRNA and then to RLuc (wt) mRNA. (E) A diagram of the RX314F reporter indicating the relative position of the probes used in panel F. Probe c is located at base pairs 682–705 in Renilla luciferase (935 base pairs), while probe d is located at base pairs 1564–1593 in the RX314F reporter (2631 base pairs); this corresponds to base pairs 601–630 in firefly luciferase (1668 base pairs). Probe sequences are shown in Supplemental Table S3. (F) Analysis of the effects of insertion of CGA codons at amino acid 314 in the RX314F reporter on Renilla-firefly luciferase mRNA. RNA from strains bearing RX314F constructs with RGS-(His)6, or RGS-(His)6-(Arg)8 encoded with either (AGA)8, [(CGA)4(AGA)4], or (CGA)8 as indicated was examined by Northern blots with the probes indicated in panel E. U2 snRNA served as an internal control.
In addition to the full-length RLuc-FLuc fusion RNA, we also detected a smaller RNA species from strains expressing either the (CGA)8 or the [(CGA)4(AGA)4] RX314F reporters (Fig. 6E,F). If the ribosome stops or stalls at CGA codons, the mRNA might undergo cleavage, since many stalls during translation trigger endonucleolytic cleavage near the site at which the ribosome stalls (Doma and Parker 2006). The smaller RNA (labeled FLuc mRNA 2 in Fig. 6F), had the properties expected for an RNA cleaved at or near the CGA codons. This smaller RNA was detected with the firefly luciferase probe, but not the Renilla luciferase probe, and was found only in strains bearing the (CGA)8 or [(CGA)4(AGA)4] constructs (Fig. 6F). Moreover, there is significantly more small RNA from strains with the (CGA)8 reporters than in the [(CGA)4(AGA)4] reporters. We did not observe a unique band corresponding to the RNA fragment from strains with constructs in which the CGA codons are near the N-terminus, most likely because the expected difference in size between the full-length and truncated RNAs is small (39–60 bases out of >800 bases) (Fig. 6B). Chen et al. (2010) have reported similar results: they observed an RNA cleavage product from the PGK1 gene bearing four CGA codons. Thus, we consider it highly likely that this shorter mRNA with homology to firefly luciferase is the result of a ribosome stall at or near the CGA codons.
Adjacent CGA codons are more effective inhibitors than separated CGA codons
Although the existence of codon pair bias is well documented, its significance in gene expression has not been explicitly examined (Smith and Yarus 1989; Irwin et al. 1995; Kane 1995; Moura et al. 2005; Buchan et al. 2006; Coleman et al. 2008). To elucidate the importance of codon interactions in modulating expression, we compared the inhibitory effects of different arrangements of five CGA and five AGA codons (Fig. 7A,B). Since there was ∼15-fold more luciferase from strains in which the construct lacks any adjacent CGA codons [(CGA–AGA)5] compared to those with five adjacent CGA codons [(AGA5CGA5) or (CGA5AGA5)], codon arrangement plays an important role in CGA-mediated inhibition of expression.
To quantitatively assess the contribution of individual CGA codons, adjacent codon pairs and alternating codons, we increased the Arg repeat to 16. This number allowed us to insert spacers of (AGA)2 between various CGA-containing test sequences, such that CGA codons in adjacent test sequences could not reside within a ribosome simultaneously, and to insert a sufficient number of test sequences to establish the relationship between expression and CGA dosage by systematic variation (Fig. 7C).
For all three arrangements of CGA codon test sequences, there was an exponential dependence of the inhibition of expression on the number of CGA codons (Fig. 7D–F), indicating that each test sequence acts independently. For each series, we derived the exponential coefficient of translation inhibition, called kTI. In each case, this coefficient had a very similar value for CGA codons upstream of firefly or Renilla luciferase (Fig. 7D–F; Table 2).
TABLE 2.
Exponential coefficient of translation inhibition, kTI values, for CGA in different contexts
Strikingly, CGA–CGA codon pairs were far more effective inhibitors of expression than isolated CGA codons or alternating CGA–AGA codon combinations. Whereas the kTIS (for single isolated codons) was −0.21 and −0.25 for the two reporters, the kTIP (for codon pairs) was −0.53 and −0.48 for CGA codon pairs, for the two reporters, respectively (Table 2). Thus, there appears to be a synergistic interaction between codons that occupy the ribosome simultaneously in adjacent sites. This synergy does not occur with CGA codons that might occupy the separated A and E sites, since the kTIA for alternating (CGA–AGA) codons was −0.22, nearly identical to that of isolated CGA codons (Fig. 7F). We note, however, that even individual CGA codons were themselves strongly inhibitory, since 10 isolated CGA codons would be expected to yield ∼14%C (y = 114e−0.21x, where x = 10) (Fig. 7D).
DISCUSSION
The major result that emerges from this study is that decoding of the CGA codon in yeast interferes with an intrinsic aspect of the translation elongation reaction. Three features of the CGA translational inhibitory signal are notable. First, the critical defect in decoding CGA appears to be I•A wobble decoding, and not tRNA abundance, since a single copy of an exact base-pairing mutant tRNAArg(UCG) efficiently suppressed the expression defects caused by the CGA codon, whereas an approximate sixfold excess of tRNAArg(ICG) only mildly suppressed the expression defect. This means that wobble decoding modulates the efficiency of elongation, a primary catalytic function of the ribosome. The inhibition of expression reported here is clearly caused by translation of the CGA codon, and not by poly-arginine, since the expression effect is codon-dependent, is suppressible by the anticodon-mutated tRNAArg(UCG), and does not require long stretches of arginine. Second, although the loss of expression caused by defects in decoding CGA codons is not mediated by mRNA degradation, CGA codons do result in formation of a novel RNA, most likely due to endonucleolytic cleavage near a stalled ribosome. This means that translation of sense codons can induce a strong and efficient halt to translation. Third, adjacent CGA codons are far more potent inhibitors of expression than separated CGA codons, a phenomenon most easily ascribed to interactions within the ribosome. Thus, profound differences in translation efficiency can stem directly from decoding interactions in the ribosome.
Observations from our work and others provide two clues to the mechanism by which CGA decoding impairs translation efficiency. First, the rate of acceptance of the tRNAArg(ICG) into an A site occupied with a CGA codon may be slow, since overproduction of the native tRNAArg(ICG) partially suppressed the CGA-mediated expression defect, indicating that charged tRNAArg(ICG) is limiting for this codon. Acceptance into the A site may be slow because of the altered geometry of the ICG anticodon base-paired with a CGA codon in the ribosome (Murphy and Ramakrishnan 2004). Second, CGA codons are likely to exert their effects in two successive steps during translation, since adjacent CGA codons have a synergistic effect on expression. This might mean that ribosomes have a memory of the last decoding reaction in the A site, an idea for which there is no precedent, or that the I•A wobble interaction is also recognized as defective in the P site. Zaher and Green (2009) recently identified a quality control pathway in E. coli in which a mismatch in the P site provokes an increased rate of misincorporation into the A site, and ultimately results in premature release. A similar pathway in yeast that probes the codon–anticodon interaction in the P site would account for the increased potency of adjacent CGA codons.
Many translation defects are closely coupled to mRNA degradation. For example, the presence of a premature stop codon results in nonsense-mediated decay (Isken and Maquat 2007), an RNA structure that blocks ribosome progress results in no-go decay (Doma and Parker 2006), and the absence of a stop codon or translation into the 3′ UTR results in nonstop decay (Frischmeyer et al. 2002; van Hoof et al. 2002; Inada and Aiba 2005). It is therefore not surprising that CGA-mediated inhibition of expression is associated with cleavage of the mRNA at or near the CGA codons, although it is surprising that the resulting 3′ fragment appears to be stable, since there is a large amount of it. We infer that ribosomes arrest at CGA codons, since CGA codons in two contexts result in a new RNA with properties expected of an endonucleolytic cleavage product near the CGA codons (our data; as well as Chen et al. 2010). The ribosome may remain bound to the 5′ end of the fragment either because the arginine residues in the exit tunnel hold it there (Dimitrova et al. 2009), or because the ribosome with both the A and P sites occupied with tRNAArg(ICG) is arrested but left in a translation competent mode. Tuller et al. (2010a) noted that clusters of poorly adapted codons are enriched within the first 50 codons, and proposed that such clusters function to stall ribosomes early in translation to prevent collisions between elongating ribosomes. If rare codons do play a role in mediating ribosome density, then mechanisms may have evolved to prevent mRNA decay in response to codon-mediated ribosome arrest.
Although codon choice has been linked for many years to the large disparities in translation efficiency (Grantham et al. 1980; Bennetzen and Hall 1982; Ikemura 1982; Duret and Mouchiroud 1999; Ghaemmaghami et al. 2003; Brockmann et al. 2007), we still do not know the extent to which codon choice directly influences translation efficiency, or the identity and properties of all of the codon-initiated signals, particularly in eukaryotes. With this systematic analysis, we found that most codon repeats have only small effects on expression, making the CGA codon conspicuous in its effects. Given that CGA codon pairs are substantially more effective inhibitors of expression than isolated CGA codons, it is likely that other codon combinations, that are more potent inhibitors of expression than the individual codon repeats, remain to be discovered.
Three lines of reasoning support the idea that codon-mediated regulation of expression is functionally important and nearly universal. First, I•A wobble decoding of CGA is conserved in the hemiascomycetes fungi, as well as in many bacteria (Curran 1995; Grosjean et al. 2010), even though it is presumed to be inefficient, as described here. Although CGA is the only codon in yeast decoded with an I•A wobble interaction (Johansson et al. 2008), it is highly unlikely that widespread retention of this decoding strategy is accidental. Since expression of the exact match tRNAArg(UCG) does not result in obvious growth defects on either minimal or rich plates at temperatures from 18°C to 37°C (data not shown), such mutations have undoubtedly occurred in nature and been discarded. Presumably the retention of this inefficient decoding has an important, but as yet undiscovered, role. Second, organisms go to great lengths to vary the decoding efficiency at different codons. All organisms use 49 (or fewer) tRNAs to decode the 61 sense codons, making wobble decoding of some codons obligatory. In many organisms, there is tremendous variation in the amounts of tRNAs that decode particular codons, as exemplified by the one to 17 copies of different tRNA genes in yeast (Grosjean et al. 2010). In addition, the sequences of tRNAs that all decode the same codon (isodecoders) vary. In humans, the 49 isoaccepting tRNA families are encoded by 450 tRNA genes, which include ∼270 isodecoders, some of which appear to be functionally distinct (Geslain and Pan 2010). Third, reduced translation efficiency of some genes may be under selective pressure. Even in human genes, clusters of codons decoded by rare codons are found more frequently than expected, are frequently conserved between human and chimp orthologs, and sometimes cluster into distinct functional groups (Neafsey and Galagan 2007; Parmley and Huynen 2009). A resolution to these issues of codon usage requires further identification of the precise codon combinations that reduce translation efficiency, and an analysis of their distribution in genomes and functional importance.
MATERIALS AND METHODS
Plasmids and strains
Both X4F-firefly luciferase (pDL306) and RX314F dual luciferase (pDL202) constructs were constructed from dual luciferase vector pDB688 (Keeling et al. 2004) and contain unique SalI and BamHI sites for codon insertions. To construct X4F (pDL306), the Renilla luciferase gene was removed by BamHI digestion followed by ligation of oligonucleotides DLO188 and DLO189 containing an ATG start codon (Supplemental Table S3). To construct RX314F (pDL202), the BamHI site upstream of the Renilla luciferase gene was mutated by PCR amplification of pDB688 with DLO99 and DLO100 (Supplemental Table S3). Vectors (JP0054 and JP0056), in which Renilla and firefly luciferase genes were expressed under PGAL1,10 control, were constructed by individual PCR amplification of the firefly and Renilla luciferase genes with additional sequences required for ligation-independent cloning into a 2μ URA3 vector BG2596, which is derived from BG2483 by insertion of sequences containing PGAL10 and the CYC1 transcription terminator (details available upon request) (Malkowski et al. 2007). Unique NheI and BamHI sites were inserted in firefly luciferase (JP0054) or Renilla luciferase (JP0056). Oligonucleotides containing codon insertions (Supplemental Table S3) were annealed and ligated into SalI-BamHI or NheI-BamHI digested vectors, which were transformed into E. coli (XL1-Blue) and confirmed by sequence analysis. Vector pEKD1024 carries the superfolder GFP (Pedelacq et al. 2006) and yeast optimized RFP (Keppler-Ross et al. 2008) under control of the bidirectional GAL1,10 promoter inserted into integrating vector JW132 (J Whipple and E Phizicky, unpubl.).
To construct 2μ plasmids containing tRNAArg species with their own promoters, tRNAs tR(CCG)L, tR(CCU)J, tR(UCU)K, and tR(ACG)D, with ∼300 base pairs of upstream and downstream sequences, were amplified by PCR from yeast genomic DNA with oligonucleotides listed in Supplemental Table S3. Purified PCR products were cloned into 2μ vector AVA0577 (Alexandrov et al. 2006) by standard ligation-independent cloning (LIC) (Aslanidis and de Jong 1990; Alexandrov et al. 2004). To construct the anticodon mutated exact base-pairing tRNAArg variants, pDL866 (tR(UCU)K) and pDL867 (tR(ACG)D) were subjected to site-directed mutagenesis with a Quickchange kit (Stratagene 200518) according to the manufacturer's instructions using oligonucleotides DLO444, DLO445, DLO508, and DLO509 (Supplemental Table S3). To integrate tRNAArg variants, tRNA genes from pDL866 (tR(UCU)K), pDL867 (tR(ACG)D), pDL869 (tR(UCU-U35G)), and pDL870 (tR(ACG-A34U)) were amplified by PCR with oligonucleotides DLO575 and DLO576, ligated into integrating vector JW132 by standard LIC cloning procedures (Alexandrov et al. 2004). A linear fragment containing the tRNAArg variants was produced by restriction digestion and transformed into yeast strain BY4741. Yeast strains, derived from BY4741 (MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), are listed in Supplemental Table S1 and plasmids used in these studies are listed in Supplemental Table S2.
Luciferase assays
Transformants were grown in SD-uracil or SC-uracil with 2% raffinose, 2% galactose media (Sherman et al. 1986) to OD600 between 0.8 and 1.0. Cells (0.8 OD-mL) were harvested by centrifugation at 4000 rpm, washed with cold ddH2O, and resuspended in 1X Passive Lysis Buffer (100 μL) (Promega), nutated for 15 min at 25°C, followed by incubation at 25°C for 20 min., then 2 μL lysate was added to 20 μL Luciferase Assay Reagent II (Dual Luciferase Reporter Assay System, Promega PAE1910). Firefly luciferase activity was measured in Relative Luminescence Units (RLUs), with a 2-sec delay and a 10-sec measurement using a Berthold FB12 luminometer. To assay Renilla luciferase activity in the same reaction, 20 μL 1X Stop and Glo (Promega) was added, and activity was measured as described above. In the systematic analysis of codon effects, activity was measured from three independent transformants, and three readings were performed on each lysate. Error bars reflect the standard deviation between three independent transformants.
Protein expression and RNA analysis
Yeast transformants, in which Renilla and firefly luciferase genes were under control of the GAL1,10 promoter, were grown as described (Quartley et al. 2009) except that strains were grown overnight in raffinose to an OD600 between 1.8 and 2.4, and expression was induced by addition of 3X YP media with 6% galactose (1/3 final volume), followed by growth for 6 h. Crude extract preparation and Western blot analysis were performed as described (Quartley et al. 2009). Membranes were incubated either with mouse anti-RGS-(His)6 (1:2500 dilution) or with rabbit anti-enolase (1:25,000 dilution), followed by washing as described previously, and incubation with either HRP-conjugated Goat IgG anti-mouse (1:10,000 dilution, BioRad) for anti-RGS-(His)6 or HRP-conjugated Goat IgG anti-rabbit (1:10,000 dilution, Biorad) for anti-enolase, and development with ECL Plus Western blotting detection system (GE Healthcare RPN2132).
RNA for RT-PCR was prepared from 50 OD-mL cells as described (Schmitt et al. 1990). RNA (5 μg) was treated with 1.25 U RNase free RQ DNase I (Promega M6101), followed by cDNA synthesis from DNase-treated RNA (0.3 μg) using Moloney murine leukemia virus reverse transcriptase (Invitrogen); the resulting cDNA was amplified by PCR using primers DLO527, DLO528, DLO531, and DLO532 (Supplemental Table S3).
Expression of green fluorescent protein in yeast strains bearing an integrated copy of pEKD1163, pEKD1170, or pEKD1167 was assessed after growth in YP + adenine + 2% galactose + 2% raffinose. Cells were washed in cold ddH2O, collected by centrifugation, resuspended in 1X PBS, and subjected to analysis on the FACSCantoII with excitation at 488 nM and detection of fluorescence emission using a 530/30 band pass filter.
For analysis by Northern blots, yeast transformants (150 OD-mL), grown as described above, were resuspended in 3 mL RNA isobuffer (0.5 M NaCl, 0.2 M Tris-Cl, pH 7.5, 0.01 M EDTA, 1% SDS) followed by addition of 3 mL phenol-chloroform-isoamyl alcohol (PCA) (25:24:1), equilibrated at pH 7.5, and glass beads, and lysed by vortexing 10 times for 20 sec with 1 min on ice between cycles. Additional RNA isobuffer (4.5 mL) and PCA (4.5 mL) were added, and lysates were centrifugated; the aqueous phase was re-extracted with 4.5mL PCA, nucleic acids were precipitated with ethanol, and the pellet was resuspended in TE, pH 7.5, and stored at −80°C. Total RNA (20 μg) was separated by electrophoresis either in a 1.5% formaldehyde agarose gel (Sambrook et al. 1989) or in a 6% polyacrylamide gel (19:1) containing 7 M urea, transferred to Genescreen plus membrane, and UV crosslinked to the membrane at 254 nm for 2 min. mRNA was visualized by synthetic oligonucleotide probes DLO511, DLO531, and SHO21 (Supplemental Table S3) labeled at the 5′ end with 32P using T4 polynucleotide kinase (Roche).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Eric Phizicky, Stan Fields, Gloria Culver, Roy Parker, and Mark Dumont for advice during the course of the work and for comments on the manuscript, and Joe Whipple and E.P. for the integrating vector. This work was supported by NSF grant no. MCB-0919658 awarded to E.J.G.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2411710.
REFERENCES
- Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, et al. 2004. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics 3: 934–938 [DOI] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18: 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD 1982. Codon selection in yeast. J Biol Chem 257: 3026–3031 [PubMed] [Google Scholar]
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T 2007. Posttranscriptional expression regulation: What determines translation rates? PLoS Comput Biol 3: e57 doi: 10.1371/journal.pcbi.0030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Aucott LS, Stansfield I 2006. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res 34: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O 2008. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif 59: 94–102 [DOI] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H 2010. Structure of the Dom34-Hbs1 complex and implications for its role in no-go decay. Nat Struct Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320: 1784–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF 1995. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res 23: 683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- Deana A, Ehrlich R, Reiss C 1998. Silent mutations in the Escherichia coli ompA leader peptide region strongly affect transcription and translation in vivo. Nucleic Acids Res 26: 4778–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284: 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L 2004. Solving the riddle of codon usage preferences: A test for translational selection. Nucleic Acids Res 32: 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci 96: 4482–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ, Kramer E, Vallabhaneni H, Raman A 2006. Evolution of +1 programmed frameshifting signals and frameshift-regulating tRNAs in the order Saccharomycetales. J Mol Evol 63: 545–561 [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Geslain R, Pan T 2010. Functional analysis of human tRNA isodecoders. J Mol Biol 396: 821–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Mercier R, Pave A 1980. Codon catalog usage and the genome hypothesis. Nucleic Acids Res 8: r49–r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, de Crecy-Lagard V, Marck C 2010. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett 584: 252–264 [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J 2004. Codon bias and heterologous protein expression. Trends Biotechnol 22: 346–353 [DOI] [PubMed] [Google Scholar]
- Ikemura T 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol 158: 573–597 [DOI] [PubMed] [Google Scholar]
- Inada T, Aiba H 2005. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J 24: 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin B, Heck JD, Hatfield GW 1995. Codon pair utilization biases influence translational elongation step times. J Biol Chem 270: 22801–22806 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9: 102 doi: 10.1186/1471-2164-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Jansen R, Bussemaker HJ, Gerstein M 2003. Revisiting the codon adaptation index from a whole-genome perspective: Analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res 31: 2242–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28: 3301–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JF 1995. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6: 494–500 [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Ross S, Noffz C, Dean N 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179: 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528 [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, Said M, Luft JR, Phizicky EM, DeTitta GT, et al. 2007. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc Natl Acad Sci 104: 6678–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man O, Pilpel Y 2007. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet 39: 415–421 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, Santos MA 2005. Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol 6: R28 doi: 10.1186/gb-2005-6-3-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V 2004. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11: 1251–1252 [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Galagan JE 2007. Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol Biol 7: 119 doi: 10.1186/1471-2148-7-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Hurst LD 2007. How do synonymous mutations affect fitness? Bioessays 29: 515–519 [DOI] [PubMed] [Google Scholar]
- Parmley JL, Huynen MA 2009. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet 5: e1000548 doi: 10.1371/journal.pgen.1000548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24: 79–88 [DOI] [PubMed] [Google Scholar]
- Quartley E, Alexandrov A, Mikucki M, Buckner FS, Hol WG, DeTitta GT, Phizicky EM, Grayhack EJ 2009. Heterologous expression of L. major proteins in S. cerevisiae: A test of solubility, purity, and gene recoding. J Struct Funct Genomics 10: 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. In Molecular cloning: A laboratory manual, pp. 7.43–7.48 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JC, Guarente L 1991. Vectors for expression of cloned genes in yeast: Regulation, overproduction, and underproduction. Methods Enzymol 194: 373–388 [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB 1986. In Methods in yeast genetics, pp. 145–149 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Smith D, Yarus M 1989. tRNA–tRNA interactions within cellular ribosomes. Proc Natl Acad Sci 86: 4397–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Kupiec M, Ruppin E 2007. Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput Biol 3: e248 doi: 10.1371/journal.pcbi.0030248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y 2010a. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354 [DOI] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E 2010b. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci 107: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C 2009. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE 4: e7002 doi: 10.1371/journal.pone.0007002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]