Abstract
Small regulatory RNA repertoires in biological samples are heterogeneous mixtures that may include species arising from varied biosynthetic pathways and modification events. Small RNA profiling and discovery approaches ought to capture molecules in a way that is representative of expression level. It follows that the effects of RNA modifications on representation should be minimized. The collection of high-quality, representative data, therefore, will be highly dependent on bias-free sample manipulation in advance of quantification. We examined the impact of 2′-O-methylation of the 3′-terminal nucleotide of small RNA on key enzymatic reactions of standard front-end manipulation schemes. Here we report that this common modification negatively influences the representation of these small RNA species. Deficits occurred at multiple steps as determined by gel analysis of synthetic input RNA and by quantification and sequencing of derived cDNA pools. We describe methods to minimize the effects of 2′-O-methyl modification of small RNA 3′-termini using T4 RNA ligase 2 truncated, and other optimized reaction conditions, demonstrating their use by quantifying representation of miRNAs and piRNAs in cDNA pools prepared from biological samples.
Keywords: 2′-O-methylation, RNA ligase, cDNA library, miRNA, piRNA, cloning
INTRODUCTION
Small RNAs mediate the regulation of gene expression at multiple levels that include modulation of RNA stability, translational regulation, genomic rearrangement, chromatin modification, and changes in promoter activity. Gene regulation by small, non-coding RNAs is a widely conserved phenomenon that has been described in plants, animals, and fungi and most often results in silencing. Bacterial and Archaeal species possess systems that bear striking structural or functional similarity to pathways found in the other domains of life. For instance, the CRISPR/CAS system uses small RNA guides in protein effector complexes to suppress foreign DNA (van der Oost et al. 2009; Waters and Storz 2009), and the structural homologs from prokaryotes have guided our current understanding of eukaryotic Argonaute function (Song et al. 2004; Yuan et al. 2005; Wang et al. 2009).
It is now recognized that small RNAs are the crucial targeting moieties in larger, multi-component regulatory complexes whose protein components dictate the final biological activity and consequences of targeting. Targeting is dependent on complete or partial hybridization between protein-bound guide RNAs and target molecules (for review, see Carthew and Sontheimer 2009; Malone and Hannon 2009).
Small regulatory RNAs are more diverse than originally thought. They are heterogeneous with respect to biosynthesis, size, and chemical modification. Diversity in chemical modification of regulatory small RNA classes is observed in many organisms, including Tetrahymena, Caenorhabditis elegans, Drosophila, Xenopus, and mouse (for review, see Farazi et al. 2008; Ghildiyal and Zamore 2009). 2′-O-methylation of the 3′-termini of small RNAs is a hallmark of piRNAs and endogenous siRNAs in animals, and of plant microRNAs. Differential association of 3′-end 2′-O-methylated small RNAs with distinct effector protein complexes is reported in numerous organisms (Farazi et al. 2008; Couvillion et al. 2009). Mutations in the enzymes responsible for modification have marked phenotypic consequences (Li et al. 2005), and demonstrate that 3′-end 2′-O-methylation is functionally important for small RNA stability and function (Li et al. 2005; Horwich et al. 2007; Kurth and Mochizuki 2009; Ameres et al. 2010).
5′-end modification of small RNAs is also observed. For instance, secondary siRNAs in C. elegans bear 5′-triphosphates that make them refractory to standard ligation schemes (Pak and Fire 2007; Sijen et al. 2007). Recently, the existence of a highly complex set of small 5′-7mGppp-capped RNAs was reported in the human transcriptome (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). Capped small regulatory RNAs have also been observed that correspond to the 5′-terminus of the hepatitis delta virus (Haussecker et al. 2008). Like 5′-triphosphates, m7Gppp-cap structures are unclonable by standard methods unless converted to 5′-monophosphorylated species. It is clear that small RNAs may be present in heterogeneous mixtures having modifications that are refractory to enzymatic manipulation, and could escape standard cDNA library preparation approaches.
Substantial research effort has been directed toward understanding the role of small regulatory RNAs in disease and development (for recent review, see Urbich et al. 2008; Sotiropoulou et al. 2009). Several studies have correlated small RNA expression patterns with disease prognosis, origin, and differentiation, providing an impetus for the development of specific small RNA biomarker panels (Sotiropoulou et al. 2009). The adaptation of small RNA-based biomarker panels to clinical use will require that accurate and reproducible methods be developed for their quantification.
Quantification of small RNA expression has closely followed the development of nucleic acid detection technologies. Northern blot hybridization is a robust, yet relatively low-throughput and insensitive approach that depends on prior knowledge of target sequence and labeled nucleic acid probes. Extending the throughput of hybridization techniques, microarray-based approaches allow for the simultaneous detection of thousands of potential targets. Microarrays rely on either the direct labeling or reverse-transcription/labeling of input RNA followed by hybridization and detection. Direct labeling for microarrays is commonly achieved by modification of RNA 3′-ends, often using T4 RNA ligase 1 (T4 Rnl1). Quantitative PCR greatly increases the dynamic range of array-based detection techniques, but does so at the expense of throughput. This technique relies on reverse transcription of small RNAs, and often 3′-end modification with the ligation of an adapter containing a priming site, or the addition of homoribopolymer tails.
The application of high-throughput sequencing approaches to the discovery and quantification of small RNAs has vastly increased the volume of data accrued and expanded our understanding of small RNA regulatory pathways. New small RNAs can be discovered, and digital counting techniques can be used to quantify levels of small RNAs (Ghildiyal and Zamore 2009). All schemes to prepare small RNA for high-throughput sequencing currently require the enzymatic modification of small RNAs to add priming sites to 5′- and 3′-ends. The adapter-ligated small RNAs are reverse-transcribed, amplified, and used as starting material for sequencing reactions.
This study aims to examine the potential impact of known small RNA modifications on the representation of modified RNAs in discovery, detection, and quantification experiments. Comparing commercially available enzymes, we analyze the effect of 2′-O-methylation at the 3′-terminus of small RNA tailing using poly(A) and poly(U) polymerases, and adapter ligations using T4 Rnl1 and T4 RNA ligase 2 truncated [T4 Rnl2tr; also known as T4 Rnl2 (1–249)]. Furthermore, we examined the effects of 2′-O-methylation on reverse transcription across 3′-RNA ligation junctions. To circumvent RNA 5′-modifications, we take the 5′-ligation independent approach of Pak and Fire (2007) as a starting point from which we examine the efficiency of commercial DNA and RNA ligases to promote the attachment of DNA adapters to reverse transcripts.
We present data illustrating shortcomings of conventional enzymatic methodology for manipulation of small RNAs that are modified. Specifically, we show that the common 2′-O-methyl modification of small RNA 3′-ends can result in their under-representation in discovery and quantification experiments. Through optimization of reaction parameters, we demonstrate that this 3′-end bias can be minimized, and present reaction conditions optimized for unbiased representation of 5′-modified small RNAs in cDNA pools.
RESULTS
Poly(A) tailing is sensitive to 2′-O-methylation of the terminal nucleotide
A common approach to adding a known sequence to the 3′-ends of unknown RNAs uses Poly(A) Polymerase (PAP) to add homoribopolymer tails—usually poly(A), but sometimes poly(C). After homopolymer tailing, hybridization with the complement to the tail is often used for reverse transcription. We compared the efficiency with which Escherichia coli PAP adds A-tails to unmodified small RNAs or to 2′-O-methyl-modified RNAs. Measuring the proportion of input RNA converted to higher-molecular-weight species in polyadenylation assays (Fig. 1), we observed a marked decrease in the conversion of 2′-O-methylated small RNAs as compared to unmodified. Overall, 2′-O-methylated small RNAs were converted to poly(A)+ <20% as efficiently as 2′-OH small RNAs.
FIGURE 1.
Poly(A) polymerase and poly(U) polymerase tailing bias against 2′-O-methylated small RNA 3′-ends. (A) Gel analysis of small RNA polyadenylation. Products of poly(A) polymerase (PAP), or poly(U) polymerase (PUP) polyadenylation reactions were resolved by denaturing PAGE and visualized by SYBR Gold staining. Tailing reactions contained 21-nt RNAs that were unmodified (2′-OH) or 2′-O-methylated (2′-O-Me) as indicated. (B) Quantification of 2′-O-methyl 3′-end nucleotide bias. Small RNA polyadenylation reactions using PAP or PUP were performed on unmodified or 2′-O-methylated small RNAs that terminated with A, C, G, or U. The extent of polyadenylation was determined by densitometry. Plotted points represent the mean ± standard error of the mean (SEM); n = 3 experimental replicates.
Exploring further, we examined the 3′-terminal base preference of PAP. Oligonucleotides were of identical sequence except the 3′-terminal nucleotide, which was varied to include 2′-O-methylated and unmethylated versions of all four ribonucleotides. When these synthetic RNAs were polyadenylated, we observed a general preference of A = G > C > U with all four unmodified nucleotides being adenylated within twofold efficiency of each other. The 2′-O-methyl 3′-termini were adenylated nearly 10-fold less efficiently than their unmodified counterparts with meG > meA = meU > meC (Fig. 1 B).
Poly(U) polymerase (PUP) from Schizosaccharomyces pombe is an essential gene that functions in regulated RNA stability (for review, see Rissland and Norbury 2008). We tested the relative efficiency with which PUP adenylated unmodified or 2′-O-Me small RNAs. We observed a similar overall bias against the extension of 2′-O-Me small RNAs, albeit to a smaller magnitude than we observed with PAP. Examining the 3′-terminal base preference, we observed the general preference A = G > C > U with less than twofold differences between the unmodified nucleotides. For 2′-O-methylated termini, meA was adenylated ∼1.5-fold to fourfold less efficiently than A, while meG = meU > meC; each being adenylated fourfold to 10-fold less efficiently than unmethylated termini.
We noted that for the oligonucleotides we tested, those terminating with U (Fig. 1A) were never 100% converted to adenylated species. This was in contrast to similar oligonucleotides that terminated with A, C, or G, for which we observed complete polyadenylation (Fig. 1B). This might be explained by minor differences in template utilization, and it is possible that the addition of more enzyme might improve the conversion of these oligonucleotides to adenylated species (Sano and Feix 1976).
T4 RNA ligase 1 exhibits decreased ligation efficiency using 2′-O-methylated acceptor substrates with pre-adenylated donors
An alternate approach to homopolymer addition is the attachment of known sequence to the 3′-ends of small RNAs of unknown sequence by ligation of adapter oligonucleotides using RNA ligase. Enterobacteriophage T4 encodes two distinct RNA ligases—T4 Rnl1 and T4 Rnl2. Both ligases are members of the DNA ligase/mRNA capping, catalytic domain superfamily of enzymes. T4 Rnl1 and T4 Rnl2 join RNA 5′-PO4 ends to 3′-OH ends using the same three-step mechanism. In the first step, the ligase reacts with ATP to form a covalent enzyme–AMP intermediate and pyrophosphate. In the second step, the AMP is transferred to the 5′-PO4 to form an adenylated RNA intermediate (AppRNA). In the final reaction, attack by an RNA 3′-OH on the AppRNA promotes phosphodiester bond formation, releasing AMP (for review, see Shuman and Lima 2004).
T4 Rnl1 and T4 Rnl2 are capable of using DNA ligation donors (5′-PO4 ends). T4 Rnl1, but not T4 Rnl2, can use a DNA acceptor (3′-OH) (Sugino et al. 1977; Hinton et al. 1978; McCoy and Gumport 1980), while T4 Rnl2 requires 2 ribonucleotide residues on the acceptor side of the ligation junction (Nandakumar and Shuman 2004; Nandakumar et al. 2004). The rates at which ligation is catalyzed for all of the possible combinations of DNA and RNA acceptor donor pairs have not been exhaustively compared, but those that have been measured appear to vary greatly, and for both ligases, the preferred substrates are RNAs (Bullard and Bowater 2006).
T4 Rnl1 and T4 Rnl2 are divergent in primary amino acid sequence outside of their core nucleotidyl transferase domains. While the C-terminal domain of T4 Rnl1 functions in its biological role of tRNA repair, the biological role of T4 Rnl2 is unknown (for review, see Pascal 2008). The C-terminal domain of T4 Rnl2 is implicated in step 2 of the ligation reaction, and its deletion creates the requirement for pre-adenylated donor molecules (Ho et al. 2004). Importantly, ligation reactions can be performed with full-length T4 Rnl1 and with T4 Rnl2 in the absence of ATP if the donor molecule (RNA or DNA) is pre-adenylated. Lau and colleagues, in some of the earliest small RNA cloning work, used T4 Rnl1 and pre-adenylated DNA adapters to reduce self-ligated RNA circles and concatemer ligation side products in library creation since input RNA 5′-ends could not serve as ligation donors (Lau et al. 2001). Because it is deficient in step 2 of the ligation reaction (Ho et al. 2004), T4 Rnl2tr also has the advantage of producing fewer circular RNA ligation products, and its use has been reported for this purpose (Aravin and Tuschl 2005).
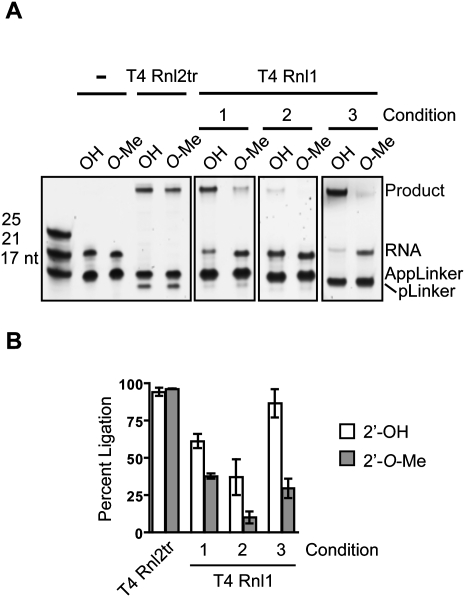
The 2′-OH group on the 3′-terminus of the ligation acceptor plays an important role in the ligation reaction (Nandakumar and Shuman 2004). We were interested in the efficiency with which T4 Rnl1 attaches a pre-adenylated adapter to single-stranded small RNAs that were unmodified, or 2′-O-Me at the 3′-end. We tested conditions corresponding to published protocols (Gu et al. 2009) and conditions similar to those recommended by a commercial supplier. We compared these to the relative efficiency of ligation of identical oligonucleotides using an optimized protocol (see below) that uses T4 Rnl2tr. We ligated a pre-adenylated 17-nt DNA oligo (AppLinker in Fig. 2) to the 3′-end of chemically synthesized 21-nt RNAs. The RNAs terminated with a uridine residue that was either unmodified or 2′-O-methylated. In all cases, we used a twofold molar excess of adenylated DNA adapter and monitored conversion of the small RNA input to ligated form.
FIGURE 2.
RNA 3′-end attachment. (A) Comparison of optimized T4 Rnl2tr ligation to published ligation conditions. Synthetic ssRNA oligonucleotides with either 2′-hydroxyl (OH) or 2′-O-methyl (O-Me) 3′-ends were ligated to pre-adenylated DNA adapter (AppLinker) using T4 Rnl2tr or T4 Rnl1 under different ligation conditions (conditions 1, 2, 3; detailed in Materials and Methods). Ligation products were resolved and visualized by SYBR Gold staining. (B) Quantification of ligation efficiency. Percent ligation refers to the amount of input RNA converted to ligated species as measured by densitometry. Data points represent the mean ± SEM; n = 3 experimental replicates.
We found that using a commercial supplier's recommended conditions, with either 25% (w/v) PEG 8000 (condition 1) or 10% (v/v) DMSO (condition 2) as ligation enhancer, 61.3% ± 4.94% and 37.0% ± 12.0% of 2′-OH RNA was converted to ligated species, respectively. Ligation efficiency decreased when the 3′-nucleotide of the input RNA was 2′-O-methylated; decreasing to 37.7% ± 1.66% and 10.2% ± 3.91%. Using ligation conditions reported by the Mello lab (condition 3), we observed 86.7% ± 9.44% ligation of unmodified RNAs. Ligation of 2′-O-methyl RNAs under the same conditions was 29.5% ± 2.67%. In contrast, under our conditions using T4 Rnl2tr and 25% (w/v) PEG 8000 (detailed in Materials and Methods), we achieved 94.2% ± 2.67% ligation of unmodified RNA and 96.3% ± 0.17% of the 2′-O-methyl modified RNA. We routinely noted a faster migrating species on SYBR Gold-stained gels of separated ligation reactions that coincided with long incubation times and higher concentrations of ligase. High concentrations of T4 Rnl2tr produced less of the faster migrating band than high concentrations of T4 Rnl1. We attributed these species to deadenylated DNA adapters (pLinker) since they co-migrated with 5′-phosphorylated, 3′-amine-blocked oligonucleotides of identical sequence to our pre-adenylated adapter, and their formation is inhibited by including ATP in the reaction buffer (Supplemental Fig. S1).
Considered together, we interpreted these data to indicate that buffer conditions, ligation enhancers, time, and temperature of ligation can have a marked effect on overall ligation efficiency of small RNAs with adenylated DNA adapters using T4 Rnl1. Modulating these conditions has differential effects on the efficiency with which 2′-O-Me small RNAs are ligated.
We extended our analysis to examine the effect of 3′-end nucleotide identity on the joining of adenylated DNA adapters to small RNA. Using input small RNAs identical to those used in our polyadenylation experiments (Fig. 1), we ligated adenylated DNA adapters with either T4 Rnl1 using condition 3 above or T4 Rnl2tr, and compared the efficiency of ligation of small RNAs terminating in unmodified or 2′-O-methyl-modified A, C, G, or U. Using optimized conditions with T4 Rnl2tr, we obtained 100% ligation regardless of 3′-nucleotide identity or methylation at the 2′-position (Fig. 3). We did not observe joining adenylated DNA adapters to unmodified RNA 3′-ends using T4 Rnl1 to be disfavored for any base under these conditions. However, we consistently observed decreased ligation efficiency when RNA 3′-ends were 2′-O-methylated. Furthermore, we observed a nucleotide-specific bias against ligation to an RNA acceptor with a 2′-O-methylated 3′-end with C = G > A > U. Compared to the ∼100% ligation efficiency of unmethylated RNAs, ligation of 2′-O-methyl RNAs terminating with A, C, G, or U was reduced to 63.0% ± 3.9%, 78.8% ± 3.2%, 72.0% ± 2.6%, and 36.0% ± 1.3%, respectively.
FIGURE 3.
RNA 3′-end adapter ligation bias against 2′-O-methylated small RNA 3′-ends. Synthetic ssRNA oligonucleotides with either 2′-hydroxyl (OH) or 2′-O-methyl (O-Me) 3′-ends and different 3′-terminal nucleotides (A, C, G, or U) were ligated to a pre-adenylated DNA adapter (AppLinker) using either T4 Rnl2tr or T4 Rnl1. Ligation products were resolved and visualized by SYBR Gold staining. Percent ligation refers to the relative amount of input RNA converted to ligated species as measured by densitometry. Data points represent the mean ± SEM; n = 3 experimental replicates.
We take these results to indicate that T4 Rnl1 joining a pre-adenylated DNA adapter to an RNA displays a nucleotide bias for RNA 3′-end acceptors that is evident when the 2′-position is methylated. The bias is not apparent when RNA 3′-ends are unmodified. T4 Rnl2, under optimized conditions, does not exhibit bias based on 3′-end 2′-methylation or base identity.
Optimized conditions for 3′-ligation using T4 RNA ligase 2 truncated
Since we observed that ligation of pre-adenylated DNA adapters to small RNAs with T4 Rnl2tr had significant advantages over the same ligations using T4 Rnl1, we optimized the conditions for small RNA 3′-ligation using T4 Rnl2tr.
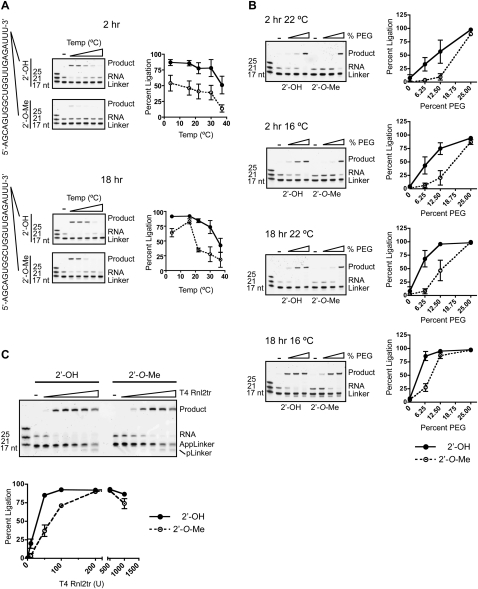
We first analyzed the efficiency of small RNA ligation to adenylated DNA adapters at various temperatures using buffer conditions as described in Materials and Methods with 12.5% (w/v) PEG 8000 in 2 h and overnight reactions. We monitored the conversion to higher-molecular-weight species of input RNA in ligation reactions containing twofold molar excess of adenylated DNA adapter. In 2-h reactions we observed that a decreasing proportion of small RNA was ligated as temperatures were increased from 4°C to 37°C (Fig. 4A). We noted a striking decrease in the ligation efficiency of 2′-O-methyl RNA as compared to unmodified RNAs after 2 h, even at lower temperatures. Extending the incubation to 18 h lessened this marked discrepancy. Efficiency of 2′-OH or 2′-O-Me RNA ligation to adenylated DNA adapters was approximately equivalent when incubated overnight below 16°C.
FIGURE 4.
Optimization of RNA 3′-end adapter ligation. (A) Temperature optimization. Synthetic ssRNA oligonucleotides with either 2′-hydroxyl (OH) or 2′-O-methyl (O-Me) 3′-ends were ligated to pre-adenylated DNA adapters (Linker) at different temperatures for either 2 or 18 h with 200 units of T4 Rnl2tr or without enzyme (−; input control). Ligation products were resolved and visualized by SYBR Gold staining. Ligation efficiency at varying temperatures is graphically represented as the mean ± SEM of four independent experiments. (B) Polyethylene glycol (PEG) as a ligation enhancer. Ligations were performed in the presence of varying concentrations of polyethylene glycol 8000 (PEG). Final concentrations in the reaction were 6.25%, 12.5%, and 25% (w/v). Ligation reactions were incubated for either 2 h or 18 h at 22°C or 16°C as indicated using 200 units of T4 Rnl2tr. (−) Indicates the absence of ligase. Ligation efficiency at varying concentrations of PEG 8000 is graphically represented as the mean ± SEM of three independent experiments. (C) Enzyme concentration. Ligations were performed using increasing amounts truncated T4 Rnl2tr (0, 10, 50, 100, 200, 500, 1000 units) in a reaction buffer containing 25% PEG 8000 (w/v) for 2 h at room temperature. Ligation efficiency using increasing amounts of enzyme are graphically represented as the mean ± SEM of three independent experiments.
PEG is a well-known enhancer of intermolecular ligation and is thought to effectively increase the concentration of donor and acceptor ends by macromolecular crowding (Harrison and Zimmerman 1984). We examined the impact of increasing the concentration of PEG 8000 in ligation reactions containing unmodified or 2′-O-methylated RNA and adenylated DNA adapter. Reactions contained 200 units of enzyme and proceeded for either 2 h or 18 h at 22°C or 16°C (Fig. 4B). In general, increasing the concentration of PEG increased the efficiency of ligation of both modified and unmodified RNAs. Ligation efficiency approached 100% with 25% (w/v) PEG 8000 for all times, temperatures, and regardless of 2′-modification. Differences in ligation efficiency between unmodified and 2′-O-methyl-modified small RNAs became apparent when ligation reactions contained decreased concentrations of PEG 8000 or were incubated for reduced time (less than overnight) or at elevated temperature.
We then examined the effect of ligase concentration on ligation of unmodified of 2′-O-methylated small RNA to adenylated DNA adapters. Reactions were incubated for 2 h at room temperature as described in Materials and Methods. By monitoring the conversion of input RNA to ligated species under conditions of excess adapter, we observed, in general, that increasing the concentration of T4 Rnl2tr increased the efficiency of small RNA ligation. As shown in Figure 4C, ligation was maximal for unmodified small RNAs using 100 units of enzyme. For 2′-O-methyl-modified RNAs, extent of ligation was maximal at 200 units of enzyme.
Considered together with results guiding RNA ligase choice in the previous section, these data guide the following recommendation for efficient and unbiased 3′-adapter ligation of small RNAs in cDNA library construction. Ligations should use T4 Rnl2tr in reactions containing a two- to fivefold molar excess of adenylated DNA adapter with 25% (w/v) PEG 8000 and be incubated overnight at 16°C.
Reverse transcription through 2′-O-methyl ligation junctions is impaired
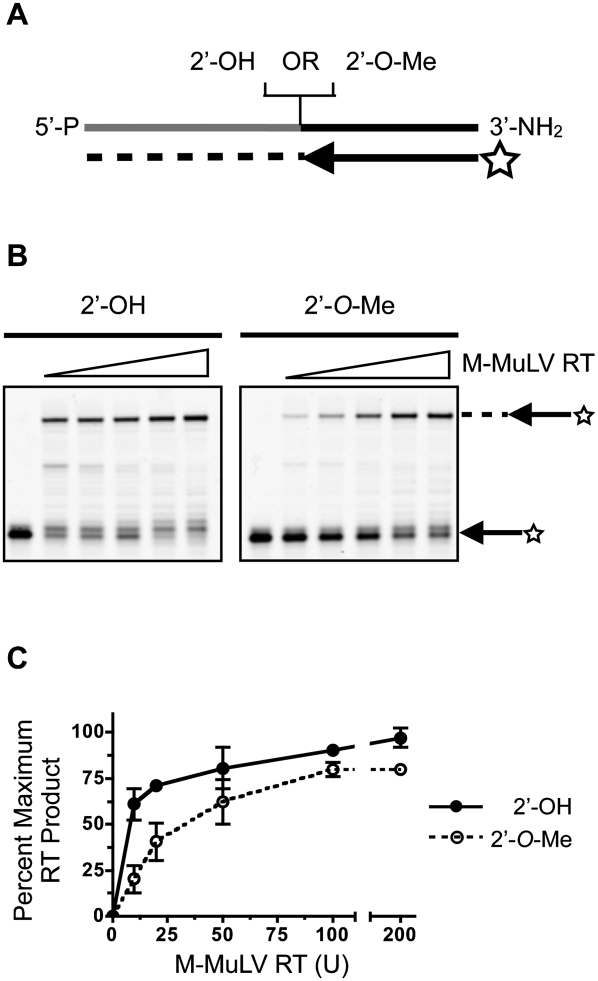
Reverse transcriptase (RT) is known to be sensitive to 2′-O-methyl residues in RNA templates. In fact, this sensitivity can be used to map methylation sites on RNAs. 2′-O-methylation mapping uses AMV or M-MuLV RT under conditions of low dNTP concentration to detect methylation sites by truncation of primer extension products (Maden et al. 1995). We asked whether, under standard RT conditions, reverse transcription over ligation junctions such as those produced in small RNA 3′-ligation reactions would be impaired for 3′-terminal 2′-O-methyl small RNAs.
We hybridized a chimeric RNA/DNA similar to a small RNA ligation product to a 5′-fluorescently labeled DNA RT primer as shown schematically in Figure 5A, and reverse-transcribed the chimera using 50 U of wild-type M-MuLV under standard conditions. Extension products were resolved and quantified to examine the effect of a single 2′-O-methyl residue at the ligation junction. Comparing primer extension on templates that did not contain a 2′-O-methyl residue to templates that did, we observed that primer extension by M-MuLV was impaired to a small degree (Fig. 5B,C). We examined the impairment in greater detail, titrating the concentration of the enzyme in primer extension reactions on unmodified templates or templates that contained a 2′-O-methyl residue at the ligation junction. We observed that impairment of reverse transcription through the 2′-O-methyl junction was more pronounced under conditions of low enzyme concentration (Fig. 5B) and that increasing the enzyme concentration to 200 U of M-MuLV reduced the difference in primer extension over the two junctions (Fig. 5C). Increasing the concentration of dNTPs or MgCl2 did not improve the efficiency of reverse transcription over 2′-O-methyl junctions (Supplemental Figs. S2, S3).
FIGURE 5.
Impairment of reverse transcription across 2′-O-methyl residues in small RNA ligation products. (A) Schematic representation of reverse transcription reactions. 5′-End IRDye 700 labeled reverse transcription primers were hybridized to a chimeric single-stranded RNA/DNA oligo that mimics 3′-ligated small RNAs. The RNA residue adjacent to the 5′-most DNA residue was either 2′-O-methyl, or 2′-hydroxyl as indicated. (Gray) The RNA part of the oligo; (black) the DNA; (star) IRDye 700; (dashed line) cDNA. (B) M-MuLV Reverse transcriptase titration. Reverse transcription was performed using increasing amounts of M-MuLV-RT (0, 10, 20, 50, 100, and 200 units). Primer extension products were resolved by denaturing PAGE and visualized by IR fluorescence imaging. (−) No enzyme. (C) Reverse transcription efficiency. Reverse transcription efficiency of 2′-OH and 2′-O-Me templates was quantified by densitometry of scanned gels. Percent maximum yield refers to the proportion of extension products normalized for the molar excess of primer over template. The data shown represent the mean ± SEM of two independent experiments.
Moving the 3′-terminus of the RT primer 3 nt 3′ of the ligation junction with respect to the template strand had no appreciable effect on the production of full-length transcripts. Instead, we observed an accumulation of RT primers that were extended 3 nt, which coincided with the position of the methylated nucleotide in the reverse transcription template (Supplemental Figs. S2–S4).
Comparing wild-type M-MuLV to AMV and RNase H− variants of M-MuLV RT showed no appreciable advantage to any of the enzymes with regard to the production of full-length reverse transcripts from templates containing 2′-O-methyl junctions versus unmodified junctions (Supplemental Fig. S4). However, differences in the accumulation of truncated reverse transcripts that corresponded to 3-nt extensions noticeably differed. AMV RT did not produce appreciable amounts of truncated reverse transcripts that were prominent in wild-type and variant M-MuLV RT reactions. The increased recommended reaction temperature of the thermostable RNase H− M-MuLV RT reaction had no discernable benefit for extension through 2′-O-methylated ligation junctions.
Together, we interpret these data to indicate that in addition to 3′-RNA ligation, reverse transcription of ligation products is another potential source of bias against 3′-terminally, 2′-O-methyl-modified RNA species. Modification of reverse transcription conditions, by using either AMV RT or increased concentrations of M-MuLV RT, equalizes primer extension efficiency across 2′-O-methylated residues.
Ligation efficiency of RNA and DNA ligases for single-stranded adapter ligation to reverse transcription products
To facilitate the capture of mixtures of RNAs with heterogeneous 5′-ends, Pak and Fire (2007) recently described a library creation protocol that ignores 5′-modifications. In their approach, a second adapter is attached to the 3′-end of the cDNA instead of to the 5′-end of the small RNA.
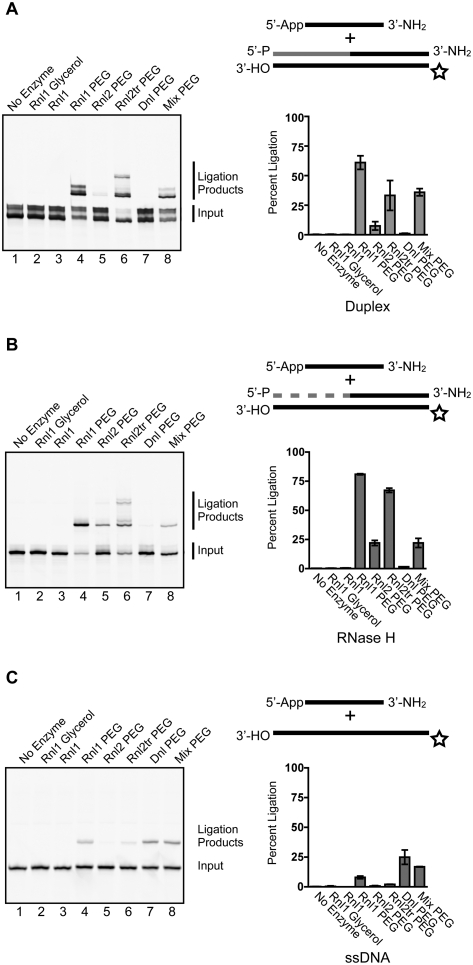
Ligation of an adenylated DNA adapter to cDNA 3′-ends in double-stranded molecules composed of chimeric RNA/DNA ligation product hybridized to a nascent reverse transcript (Fig. 6A) is an “off-label” use for commercially available ligases. We tested the activity of T4 DNA ligase, T4 Rnl1, T4 Rnl2, and T4 Rnl2tr to promote the ligation of these molecules under various reaction conditions. The donor for the ligation reaction was a 24-nt pre-adenylated DNA adapter, blocked at the 3′-end with an amine group, and the acceptors were synthetic oligonucleotides that mimic reverse transcription products of 3′-ligated small RNAs (Fig. 6A). The cDNA strand mimic (acceptor) was labeled at the 5′-terminus with IRDye 700. Duplex oligonucleotides were treated with Exonuclease I to remove single-stranded oligos from the hybridization mixture, and ligations were performed with a twofold excess of adapter to acceptor. We monitored the conversion of the labeled cDNA strand to higher-molecular-weight species by denaturing gel electrophoresis.
FIGURE 6.
Ligation of adenylated adapters to cDNA 3′-ends. Pre-adenylated DNA oligonucleotides were ligated to synthetic double-stranded, partially double-stranded, or single-stranded oligonucleotides that mimic reaction products from reverse transcription of 3′-end ligated small RNAs. In the schematic representations of ligation inputs shown: (black lines) DNA; (gray lines) RNA; (star) IRDye 700. (A) Ligation of pre-adenylated DNA adapters to double-stranded reverse transcription products. Ligation products were separated by denaturing PAGE and visualized by IR fluorescence scanning. Ligation efficiency was determined as described in the Materials and Methods and is presented as the mean ± SEM of three independent experiments. Incubation and buffer conditions are detailed in the Materials and Methods section. (B) Ligation of pre-adenylated adapters to RNase H-treated reverse transcription products. The efficiency of ligation of pre-adenylated DNA adapters to RNase H-treated substrates is represented graphically as the mean ± SEM of three independent experiments. Ligase, buffer composition, and incubation conditions correspond to those in panel A. (C) Ligation of pre-adenylated adapters to single-stranded DNA oligonucleotides. The efficiency of ligation of pre-adenylated DNA adapters to synthetic single-stranded DNA oligonucleotides is represented graphically as the mean ± SEM of three independent experiments. Ligase, buffer composition, and incubation conditions correspond to those in panel A.
We observed that all of the ligases tested were able to catalyze the reaction, albeit to vastly different extents (Fig. 6). T4 Rnl1 and T4 Rnl2tr had the highest activity in these ligations, and activity was stimulated by adding 25% PEG 8000. Figure 6A illustrates the relative activity of T4 Rnl1, Rnl2, Rnl2tr, and T4 DNA ligase under various conditions with a fully double-stranded substrate. We considered any IRDye 700 signal that migrated more slowly than the input to be a ligation product for quantification purposes.
Ligation efficiencies for T4 Rnl1 using published conditions (Lau et al. 2001; Pak and Fire 2007) and NEB T4 Rnl1 buffer without ATP were 0.35% ± 0.18%, and 0.32% ± 0.07%, respectively (Fig. 6A, lanes 2,3). Adding 25% PEG 8000 to the reaction increased ligation efficiency to 61.03% ± 5.8% (Fig. 6A, lane 4). T4 RNA ligase 2 with 25% PEG 8000 ligated 7.55% ± 3.6% of this substrate (Fig. 6A, lane 5). T4 Rnl2tr ligated 33.27% ± 12.6% of the substrate and produced higher-molecular-weight species, possibly resulting from ligation of the 5′-end of the RNA with the adjacent 3′-end of the cDNA (Fig. 6A, lane 6). T4 DNA ligase was ineffective at joining pre-adenylated adapters to this double-strand structure (Fig. 6A, lane 7), ligating 1.0% ± 0.4% of the molecules. Mixing T4 Rnl1 and T4 DNA ligase did not improve ligation efficiency (35.95% ± 3.0%) (Fig. 6A, lane 8).
We asked whether treatment of double-stranded ligation acceptor with RNase H before ligation would increase the efficiency of the reaction (Fig. 6B). We observed a generally similar activity profile for the ligases on this acceptor substrate. T4 Rnl1 in the absence of PEG had negligible activity (0.38% ± 0.1% and 0.35% ± 0.2% for Rnl1 with glycerol and Rnl1, respectively). Including 25% PEG 8000 increased the efficiency of T4 Rnl1 to 73.4% ± 7.6%. T4 Rnl2 in the presence of PEG ligated 25.0% ± 3.3% of the substrate molecules, and T4 Rnl2tr ligated 58.2% ± 9.0% of the molecules. T4 DNA ligase was inefficient at promoting ligation, joining 1.1% ± 0.5% of the substrate. Mixture of T4 DNA ligase with T4 Rnl1 ligated 16.8% ± 5.7% of input.
Finally, we tested the ligation activity of this panel of ligases on single-stranded acceptor DNA molecules (Fig. 6C). On the whole, all of the ligases that we tested under these conditions ligated single-stranded substrates more poorly. Without PEG, T4 Rnl1 had minimal activity. The addition of PEG increased ligation to 7.95% ± 1.3%. T4 Rnl2, and T4 Rnl2tr joined 0.69% ± 0.3% and 2.06% ± 0.1%, respectively. T4 DNA ligase was the most efficient, ligating 24.9% ± 6.1% of the substrates, while a mixture of T4 DNA ligase and T4 Rnl1 ligated 16.84% ± 0.2% of input molecules.
We noted that the input (Fig. 6A, lane 1) and the ligation products (Fig. 6A, lanes 4,7,8) appeared as heterogeneous bands. We attribute this heterogeneity to partial reannealing of the duplexes, since we did not observe them when ligation inputs were first treated with RNase H (Fig. 6B) or were single-stranded (Fig. 6C). In ligations that used T4 Rnl2tr with fully double-stranded, or RNase H-digested double-stranded cDNA (Fig. 6A,B, lane 6), we routinely noted the formation of ligation products that migrated with larger apparent size. Although these species contain at least one IR Dye 700 label, their precise nature is not known.
We conclude from these data that T4 Rnl1 in the presence of 25% PEG 8000 ligates single-stranded pre-adenylated adapters to RT products with the highest efficiency, and fewest side-ligation products. Furthermore, we recommend ligation when the RT product is in a double-stranded structure. Digestion of the ligation acceptor substrate with RNase H would disrupt the A-form helix that arises in a DNA/RNA hybrid, possibly to yield sequence-dependent, heterogeneous, single-stranded structures. We suggest that performing the ligation reaction using the double-stranded hybrid may maintain the A-form helical structure regardless of primary sequence and provide more consistent ligations.
High-efficiency ligation using T4 Rnl2tr reduces 2′-O-methyl capture bias in cDNA pools
Our observations detailed in the previous sections led us to hypothesize that 2′-O-methylated small RNA species would be under-represented in cDNA pools made using strategies that use T4 Rnl1 in the first step. We tested this hypothesis by ligating adenylated adapters to size-fractionated small RNA from mouse testes, which is known to contain mixtures of miRNAs and piRNAs. We performed the first ligation using T4 Rnl1 or T4 Rnl2tr. Reverse transcription of ligated RNA used 5′-extended primers that provided a priming site for qPCR. We quantified the amount of cDNA corresponding to piRNAs and miRNAs known to be expressed in mouse testes using specific forward primers and a common reverse primer (Fig. 7A).
FIGURE 7.
Under-representation of 2′-O-methylated small RNAs in cDNA pools. (A) Schematic representation of ligated small RNA qPCR. Size-fractionated small RNA from mouse testis was ligated to a pre-adenylated DNA adapter. The RT primer contained a 5′-overhang region that provides a PCR priming site. QPCR was performed for specific small RNA targets in the pools using unique forward primers with a common reverse primer. (B) Relative quantification of ligated small RNA. Normalized amounts of small RNAs in cDNA pools were compared in cDNA pools prepared using T4 Rnl1 and T4 Rnl2tr. Signals corresponding to small RNAs in T4 Rnl2tr pools are shown as means ± SEM of three or more experimental replicates relative to signal from T4 Rnl1 cDNA pools. (C) Schematic representation of cloning workflow. Equimolar mixtures of unmodified or 2′-O-methylated 21-nt RNAs were ligated to a pre-adenylated DNA adapter. Input RNAs were distinguishable by reversal of the nucleotides at positions 10 and 11 (bold). Samples were further processed in parallel and subjected to sequence analysis. (D) Quantification of sequenced small RNAs. Identities of recovered clones corresponding to input small RNAs were scored in a blinded fashion. Proportions of cDNAs arising from unmodified or 2′-O-methylated RNA sequenced were compared to an expected recovery ratio of 1:1 using the exact binomial test. P-values from these tests are shown below (n = 3 experimental replicates, 248 ligation junctions sequenced).
We normalized the levels of mouse piRNAs and microRNAs to levels of microRNA 125b within each pool and expressed the levels detected with T4 Rnl2tr relative to those detected in cDNA pools prepared with T4 Rnl1. We confirmed the equivalent efficiency of ligating an adenylated adapter to miR-125b using T4 Rnl1 and T4 Rnl2tr in gel-based assays (Supplemental Fig. S5). piR-100127, piR-140075, piR-1L, piR-126274, and piR-142227 were recovered 3.77 ± 0.43–, 4.01 ± 0.94–, 2.88 ± 0.31–, 3.08 ± 0.45–, and 3.00 ± 0.38–fold more, respectively, in cDNA pools prepared with T4 Rn2tr (Fig. 7B). The observed level of enrichment for these piRNAs was significantly different from 1—the mean predicted by the null hypothesis (p < 0.05). In contrast, enrichment of miR-16, miR-143, and miR-34c in T4 Rnl2tr-prepared cDNA pools was 0.79 ± 0.3–, 1.7 ± 0.5–, and 1.58 ± 0.2–fold, respectively, not significantly different from the hypothesized mean of 1.
We confirmed our qPCR results by Northern blot analysis of ligated small RNAs (Supplemental Fig. S6). Ligation reactions that used mouse testis small RNA and pre-adenylated DNA adapters with either T4 Rnl1 or T4 Rnl2tr were separated by denaturing PAGE and hybridized with small RNA-specific probes. We observed equivalent hybridization signal for ligated mmu-mir-125b when small RNAs were ligated with T4 Rnl1 or T4 Rnl2tr. The entire hybridization signal for the small RNAs migrated with higher apparent molecular weight than no ligase control reactions when T4 Rnl2tr was used. However, the higher-molecular-weight hybridization signal was markedly decreased for the piRNAs, but not mmu-mir125b, in where T4 Rnl1 was used to attach the pre-adenylated DNA adapter. This decrease corresponded to an increase in hybridization signal that comigrated with the signal from no ligase control reactions.
These results supported our hypothesis that 2′-O-methylated small RNAs in biological samples are under-represented in cDNA pools prepared with T4 Rnl1 as compared to those prepared with T4 Rnl2tr.
To further test the hypothesis that 2′-O-methyl-modified small RNAs are under-represented in cDNA libraries, we performed a cloning experiment in which synthetic small RNA, either 2′-O-methyl modified or not at their 3′-termini, were starting material in a cDNA library construction approach guided by our findings detailed above. We began with equimolar mixtures of synthetic RNAs that had identical sequence composition but were distinguishable by swapping the bases at positions 10 and 11 (Fig. 7C). Small RNA pools were ligated to pre-adenylated DNA adapters using T4 Rnl1, or T4 Rnl2tr using optimized conditions. Ligation products were cloned and subjected to Sanger sequencing as detailed in the Materials and Methods.
We sequenced a total of 248 ligation junctions from three experimental replicates. One hundred twenty-seven junctions ligated with T4 Rnl1, and 121 ligated with T4 Rnl2tr. Beginning with an equimolar mixture of modified and unmodified small RNAs, we observed that 30% of clones from T4 Rnl1-prepared libraries corresponded to 2′-O-methylated input RNA. Performing the initial ligation with T4 Rnl2tr increased the proportion of recovered 2′-O-methyl-modified RNA to 43% (Fig. 7D). The ratio of 2′-O-methyl RNA to 2′-OH RNA ligation junctions differed significantly from the expected ratio (p = 6.97 × 10−6) in the T4 Rnl1 libraries, whereas the deviation from the ratio was not significantly different for T4 Rnl2tr libraries (p = 0.145). We did not observe significant differences in ratios between experimental replicates within a group. Reflecting this observation, P-values of the heterogeneity G-values between experimental replicates using the replicated G-test of goodness-of-fit were 0.52 and 0.56 for T4 Rnl1 and for T4 Rnl2tr libraries, respectively.
We interpret these results to indicate that 2′-O-methyl-modified RNAs are under-represented in small RNA library preparation using T4 Rnl1. This bias is minimized when T4 Rnl2tr is used under optimized conditions.
We did not observe evidence of untemplated nucleotide addition to cDNA 3′-ends in our sequencing data. Others have reported and used the untemplated addition of C's, which are used for producing double-stranded cDNA libraries by adding 3′-GGG-containing primers for second-strand synthesis. The template switching approach uses modified reaction conditions or mutant M-MuLV RT (Cloonan et al. 2008). Untemplated nucleotides introduce a degree of uncertainty as to the precise identities of small RNA 5′-ends that must be taken into account for discovery and mapping.
DISCUSSION
Experimental approaches for discovery, detection, and quantification of small RNAs should aim to be as representative as possible. This study examined the impact of 2′-O-methylation—a common small RNA modification—on the representation of small RNAs in cDNA libraries for detection, quantification, and discovery. We assessed the efficiency and 2′-O-methyl bias of homoribopolymer tailing, DNA adapter ligation, and reverse transcription, optimizing reactions where necessary. We further optimized adenylated DNA linker ligation to first-strand cDNA to enable 5′-ligation-independent small RNA library creation.
Our results confirm earlier observations of inefficient tailing of 2′-O-methyl RNAs using PAP (Ebhardt et al. 2005; Yang 2006) and indicate that PUP displays a similar bias. In addition, we observed a small bias against adenylation of 2′-OH RNA containing U as the 3′-base using both PAP and PUP. We conclude that polyadenylation of small RNAs for library creation will result in libraries that largely under-represent small RNAs that terminate in 2′-O-methyl nucleotides and will be biased against 3′-terminal U RNAs. For quantification experiments of small RNAs known to be unmethylated, 3′-polyadenylation may suffice, but will under-represent small RNAs that terminate in U. Clearly, the use of polyadenylation for small RNA discovery is not ideal since entire classes of small RNAs may be ignored.
We approached our analysis of 3′-adapter ligation efficiency hoping to improve established protocols. Our analyses revealed that the impairment of adenylated DNA adapter attachment to 2′-O-methyl small RNA using T4 Rnl1 is additionally influenced by reaction conditions and by base composition. These findings expand and update an earlier report that ligation of an RNA adapter to 2′-O-methyl RNA 3′-ends using T4 Rnl1 is impaired (Ebhardt et al. 2005). Our comparisons showed that published methods of ligating pre-adenylated DNA adapters to the ends of small RNAs with T4 Rnl1 varied greatly with respect to the efficiency of ligation to both 2′-OH and 2′-O-methyl RNAs, indicating that sequencing-based measures of quantification of cDNA library members should be interpreted with caution.
We optimized conditions to capture 100% of small RNAs regardless of 2′-O-methyl modification or 3′-terminal base identity using T4 Rnl2tr. It is possible that conditions for Rnl1-mediated ligation could be found that would similarly minimize attachment efficiency, but the advantage afforded by Rnl2tr in terms of reduced undesired ligation side products is clear.
How does a 3′-end 2′-O-methyl group inhibit the ligation activity of the T4 RNA ligases? It is clear the 2′-position of the ligation acceptor plays an important role in phosphodiester bond formation between the 3′-OH and the adenylated 5′-acceptor for T4 Rnl2 since replacement of the 2′-OH with 2′-H slows nick sealing by a factor of at least 25 for full-length T4 Rnl2 on a double-stranded substrate (Nandakumar and Shuman 2005). However, the 2′-position of the 3′-terminal residue is unimportant for nick recognition, and ligation is clearly possible when the 3′-end of a nicked substrate is 2′-O-methylated (Nandakumar and Shuman 2004; this study). Postulated roles for the 2′-OH on the 3′-nucleotide include coordination of the AMP phosphates on the ligation donor, assistance in activation of the adjacent 3′-OH for attack on the 5′-P at the ligation junction, or indirect influence on the conformation of the furanose sugar pucker on the terminal residue (Nandakumar and Shuman 2004). Because our data show that extended incubations using increased enzyme concentration and ligation enhancers equalize the efficiency with which 2′-O-methyl RNAs are ligated, we would argue that replacement of the 2′-OH with 2′-O-methyl slows phosphodiester bond formation in single-stranded ligations.
Pausing of reverse transcriptase opposite modified ribonucleotides is known and has been exploited for mapping sites of 2′-O-methylation on ribosomal RNA. Consistent with previous studies of rRNA (Maden et al. 1995), our data indicated that reverse transcription over modified ligation junctions is impaired—even when dNTPs are not limiting. This is similar to some, but not all, sites of 2′-O-methylation in animal rRNA (Maden et al. 1995). Thus, 2′-O-methylation of small RNA 3′-ends may impair representation in cDNA libraries by also reducing reverse transcription. It remains to be seen whether other modifications such as pseudouridylation, or base modifications are also present, and as of yet unrecognized, in small RNA repertoires because of under-representation in cDNA libraries.
Recent reports have suggested that quantification of small RNAs by high-throughput sequencing or by microarray can appear variable depending on the methods used (Bissels et al. 2009; Linsen et al. 2009). Because these studies have used synthetic input small RNAs and known concentrations, differences in detection are likely attributable to complicated sequence preferences of the enzymes used for labeling and ligation—a phenomenon observed almost 30 years ago (Romaniuk et al. 1982). These sequence preferences appear not to be limited to the termini of small RNAs, as single nucleotide differences in the middle of otherwise identical oligonucleotides resulted in significantly different recovery (Linsen et al. 2009). Our recommendations for small RNA library preparation include the use of molecular crowding agents and extended incubations that result in the efficient ligation of suboptimal ligation substrates that bear 3′-terminal 2′-O-Me groups. It is plausible that these conditions may also improve the capture of difficult-to-ligate sequences. Further studies will address whether the improvements that we report here also aid in the more complete and equal representation of all sequence variants in sequencing libraries and labeled molecular pools.
If small RNA 5′-modifications are known and can be converted to 5′-monophosphates, current 5′-ligation-dependent approaches will capture them, at least to some degree. Recent reports of naturally occurring small-molecule 5′-RNA conjugates such as CoA derivatives and NAD (Chen et al. 2009; Kowtoniuk et al. 2009) indicate that RNAs may be modified in unexpected ways that could result in their invisibility to current high-throughput sequencing library preparation techniques. This underscores the need for cDNA library construction that is insensitive to 5′-end modifications and the development of molecular biology tools to detect, analyze, and manipulate a range of RNA modifications.
Our findings are theoretically applicable beyond the scope of small RNAs and may enable further discoveries in regulatory RNA discovery and analysis, especially when RNA ends are not ideal for sequence attachment.
MATERIALS AND METHODS
Oligonucleotide substrates, adapters, and primers
Oligonucleotides were synthesized at Integrated DNA Technologies. To produce adenylated DNA adapters by enzymatic adenylation, 5′-phosphorylated adapters were annealed to complementary deoxyoligonucleotides to form 3′-overhang structures by incubating the mixture for 15 min at 95°C, transferring it for 15 min to 37°C, and allowing it to cool to room temperature over a 60-min period. Ten nanomoles of the annealed oligonucleotide mixture was incubated with 100,000 units of T4 DNA ligase (2000 units/μL; New England Biolabs) in 1× quick ligation reaction buffer for 18 h at room temperature in a final volume of 250 μL.
3′-small RNA Adapter: 5′-P-GCTCGTATGCCGTCTTCTGCTTG-NH2-3′
3′-small RNA Adapter Template: 5′-CAAGCAGAAGACGGCATACGAGCAACCGATTTCAGATGGTGCTA-3′
3′-cDNA Adapter: 5′-P-CAACCGTCGGATCGTCGAGCTGTAGAACTCTG-NH2-3′
3′-cDNA Adapter Template: 5′-CAGAGTTCTACAGCTCGACGATCCGACGGTTGAACCGATTTCAGATGGTGCTA-3′
Adenylated species were gel-purified before use.
Gel analyses
All denaturing polyacrylamide TBE-Urea gels were prepared using SequaGel reagents (National Diagnostics) following the manufacturer's instructions. Samples were prepared in 2× RNA denaturing loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, bromophenol blue, and xylene cyanol) and denatured for 5 min at 90°C and then placed on ice prior to loading. Gels were stained with SYBR Gold Nucleic Acid Gel Stain (Life Technologies) for 15 min and visualized on a Typhoon 9400 Variable Mode Imager (GE Healthcare Biosciences Corp.). For IRDye labeled samples, gels were scanned using the Odyssey Infrared Imaging System (LI-COR, Inc.).
RNA 3′-polyadenylation and polyuridylation
Ten picomoles of synthetic ssRNA oligonucleotides with either 2′-hydroxyl or 2′-O-methyl 3′-ends and different 3′ terminal nucleotides:
rA-OH: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrA-3′
rA-O-Me: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUmA-3′
rC-OH: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrC-3′
rC-O-Me: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUmC-3′
rG-OH: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrG-3′
rG-O-Me: 5′-PrArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUmG-3′
rU-OH: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrU-3′
rU-O-Me: 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUmU-3′
were polyadenylated using E. coli Poly(A) Polymerase (New England Biolabs Inc.), or Poly(U) Polymerase (New England Biolabs Inc.) using their respective reaction buffers for 30 or 60 min at 37°C. Reactions were terminated by the addition of 2× loading buffer (95% [v/v] formamide, 18 mM EDTA, 0.025% [w/v] SDS), and products were separated by gel electrophoresis. The extent of RNA adenylated was calculated by quantifying the percent of unextended input RNA remaining, normalizing to the internal control DNA oligo, and subtracting this number from 100%.
Comparison of small RNA ligation conditions
Synthetic ssRNA oligonucleotides (21-mer; 5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrU-3′) with either 2′-hydroxyl or 2′-O-methyl 3′-ends were ligated to the Universal miRNA Cloning Linker (AppLinker; New England Biolabs Inc.)—5′- rAppCTGTAGGCACCATCAAT-NH2-3′—using 200 units of truncated T4 RNA ligase 2 in 1× reaction buffer (50 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT at pH 7.5, 25°C) containing 25% (w/v) PEG for 18 h at 16°C or T4 Rnl1 under different ligation conditions. Condition 1 was 1 unit of T4 Rnl1 (Epicentre Biotechnology) in 1× reaction buffer without ATP (33 mM Tris-acetate at pH 7.8, 66 mM potassium acetate, 10 mM magnesium acetate, and 0.5 mM DTT) containing 25% (w/v) PEG. Condition 2 was similar to condition 1 except that 10% (v/v) DMSO was substituted for PEG. Conditions 1 and 2 included incubation for 2 h at room temperature, mimicking ligation conditions recommended by Integrated DNA Technologies. Condition 3 used 30 units of T4 Rnl1 (New England Biolabs Inc.) in 1× reaction buffer (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 10 mM DTT, 60 μg/mL BSA) containing 10% DMSO for 2 h at room temperature, followed by transfer for 18 h to 4°C. These reaction conditions were similar to those recommended by the Mello Lab (University of Massachusetts Medical School) (Gu et al. 2009). Reactions were stopped with 2× RNA denaturing loading buffer and analyzed by gel electrophoresis. Ligation efficiency between reactions was compared by dividing the pixel score of ligated input by the pixel score of the total input in the same lane. Deadenylation reactions were performed for 2 h in the absence of RNA acceptors with T4 Rnl2tr as described above or under condition 3 for Rnl1.
RNA 3′-end ligation optimization
Ligation reactions contained 10 pmol of synthetic ssRNA oligonucleotides (21-mer) either with 2′-hydroxyl or 2′-O-methyl 3′-ends, 20 pmol of pre-adenylated DNA adapter (Universal miRNA Cloning Linker; New England Biolabs Inc.) in 1× reaction buffer (50 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT at pH 7.5, 25°C) in 20-μL final reaction volumes. We varied the amount of T4 Rnl2tr (New England Biolabs Inc.), ligation time, ligation temperature, and concentration of Polyethylene Glycol 8000 as indicated in the figures. Reactions were stopped with denaturing loading buffer and analyzed by gel electrophoresis.
3′-terminal nucleotide bias analysis
Synthetic ssRNA oligonucleotides with either 2′-hydroxyl or 2′-O-methyl 3′-ends and different 3′ terminal nucleotides were ligated onto the Universal miRNA Cloning Linker (New England Biolabs Inc.) using either 200 units of T4 Rnl2tr in 1× buffer reaction buffer (50 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT at pH 7.5, 25°C) containing 25% (w/v) PEG for 18 h at 16°C or 30 units of T4 Rnl1 in 1× reaction buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 60 μg/mL BSA at pH 7.5, 25°C) containing 10% DMSO for 2 h at room temperature and then for 18 h at 4°C. The reactions were stopped with 2× RNA denaturing loading buffer and analyzed by gel electrophoresis.
Reverse transcription
Reverse transcription of a synthetic RNA-DNA (5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrUCTGTAGGCACCATCAAT-3′) was performed as follows: 10 pmol of template and 20 pmol of 5′-end IRDye 700 labeled reverse transcription primers 5′-IRDye 700-ATTGATGGTGCCTACAG-3′, or 5′-IRDye 700-ATTGATGGTGCCTA-3′ in a final volume of 10 μL were denatured for 5 min at 90°C and placed on ice. Ten microliters of 2× reverse transcription mix (2× M-MuLV-Reverse Transcriptase reaction buffer, 0–200 units of M-MuLV-Reverse Transcriptase enzyme [New England Biolabs Inc.], or 10 units of AMV Reverse Transcriptase [New England Biolabs Inc.], 1 mM dNTPs [New England Biolabs Inc.], and 40 units of Murine RNase Inhibitor [New England Biolabs Inc.]) were added to the template mix. Samples were incubated for 30 min at 42°C. The reactions were quenched with 2× RNA denaturing loading buffer and separated by denaturing PAGE. For dNTP titration reactions, 1 mM free Mg2+ was maintained by increasing the concentration of MgCl2 proportionally with the increased dNTPs. Reverse transcription comparing wild-type M-MuLV, AMV, SuperScript II, and SuperScript III used 200 U of M-MuLV or 10 U of AMV (New England Biolabs Inc.) as described above, or SuperScript II or III (Life Technologies) according to the manufacturer's suggestions with incubation temperatures of 42°C or 50°C, respectively.
cDNA 3′-end adapter ligation
RT product mimics were obtained by annealing an equimolar mixture of a synthetic RNA–DNA (5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrUCTGTAGGCACCATCAAT-NH2-3′) with its reverse complement infrared dye-labeled ssDNA (5′-IRDye 700-ATTGATGGTGCCTACAGAAATCTCAACCAGCCACTGCT-3′). The mixture was denatured for 5 min at 95°C, then transferred for 1 h to 37°C, followed by incubation for 1 h at 22°C, and cooled on ice. The RT product mimic was treated with Exonuclease I (New England Biolabs Inc.) to remove single-stranded oligos as follows: 750 pmol of duplex mixture was treated with 100 units of Exonuclease I for 30 min at 37°C and then phenol/chloroform extracted and ethanol-precipitated. In some cases, the Exonuclease I–treated duplex (250 pmol) was further digested with 5 units of RNase H (New England Biolabs Inc.).
In all ligation reactions, 10 pmol of cDNA mimic was ligated to 20 pmol of the Universal miRNA Cloning Linker (New England Biolabs Inc.). The reaction in Figure 6A–C, lane 1 contained no ligase. Published conditions (Pak and Fire 2007) were reproduced and run in lane 2 (T4 Rnl1 glycerol) by using 20 units of T4 Rnl1 in 50 mM HEPES (pH 8.3), 10 mM MgCl2, 3.3 mM DTT, 10 μg/mL BSA, and 8.3% (v/v) glycerol; the reaction was incubated for 2 h at 22°C. Lane 3 (Rnl1) contained 30 U of T4 Rnl1 in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 60 μg/mL BSA incubated for 2 h at 22°C, followed by incubation for 18 h at 4°C. Lane 4 (Rnl1 PEG) was run under similar conditions to lane 3, but including 25% (w/v) PEG 8000. Lane 5 (Rnl2 PEG) contained T4 RNA ligase 2 and the same buffer and incubation conditions as lane 4. Lane 6 (Rnl2tr PEG) contained 200 units of T4 Rnl2tr in 50 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT (pH 7.5, 25°C) with 25% (w/v) PEG 8000 incubated for 18 h at 16°C. Lane 7 (Dnl PEG) contained 2000 units of T4 DNA ligase in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 60 μg/mL BSA, and 25% (w/v) PEG 8000 incubated for 2 h at 22°C, then transferred for 18 h to 4°C. Lane 8 (Mix PEG) contained 30 units of T4 Rnl1 and 2000 units of T4 Dnl under the same buffer and incubation conditions as lane 5.
Ligation was assessed by integrating the IRDye 700 signal that migrated more slowly than the input ligation substrate, and percent ligation is expressed as the amount of slower migrating material divided by the total signal in the lane.
Quantitation of 3′-ligated small RNAs in cDNA pools
RNA ligation
Size-fractionated small RNAs from mouse testes (Bioo) were dephosphorylated using 1 unit of Alkaline Phosphatase, Calf Intestinal (New England Biolabs Inc.) per picomole of input RNA, incubating for 1 h at 37°C. Following extraction with phenol:chloroform:isoamyl alcohol, 25:24:1 and ethanol precipitation, RNA (5–15 pmol) was ligated to an adenylated DNA adapter with the sequence 5′-AppTCGTATGCCGTCTTCTGCTTGT-NH2-3′ using T4 Rnl1 or T4 Rnl2tr at a 2:1 molar ratio of adapter to small RNA. T4 Rnl1 was added to reactions at a ratio of 4 units per picomole of junctions in 1× ligation buffer without ATP (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 10 mM DTT, 60 μg/mL BSA) containing 10% DMSO and 40 units of Murine RNase Inhibitor (New England Biolabs Inc.) in a 10-μL final reaction volume for 2 h at room temperature, then transferred for 18 h to 4°C. T4 Rnl2tr was added at a ratio of 40 units per picomole of RNA in 1× reaction buffer (50 mM Tris-HCl, 2 mM MgCl2, 1 mM DTT at pH 7.5, 25°C) containing 25% (w/v) PEG 8000 and 40 units of Murine RNase Inhibitor (New England Biolabs Inc.) in a 20-μL final reaction volume for 18 h at 16°C.
The ligation products were separated on a 15% acrylamide TBE-urea gel (20 × 20 cm, 0.8-mm thickness, 1-cm-wide well) and run sufficiently far to resolve ligated (28–56 nt) from unligated (17–35 nt) species as judged by co-electrophoresis of size standards (microRNA marker and Low Range ssRNA Ladder; New England Biolabs Inc.). Ligation products were excised and purified before reverse transcription.
First-strand cDNA synthesis
Ligated RNAs were reverse-transcribed with AMV Reverse Transcriptase (New England Biolabs Inc.) and a 5× molar excess of the reverse transcription primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGGCAAGCAGAAGACGGCATACGA-3′). Samples were incubated for 30 min at 42°C. The reverse transcript was digested with 40 units of Exonuclease I (New England Biolabs Inc.) for 30 min at 37°C to degrade excess RT primers. Samples were extracted with phenol:chloroform:isoamyl alcohol, 25:24:1, and ethanol-precipitated.
Quantification of cDNA
cDNAs corresponding to specific small RNAs were quantified by qPCR using the DyNAmo HS SYBR Green qPCR kit (Finnzymes) and the primers listed in Table 1. Amplification reactions were performed in triplicate and contained 2 μL of diluted cDNA and 0.5 μM final concentration of primers (Table 1). Real-time quantitative PCRs were performed on DNA Engine Opticon Instruments (MJ Research). Relative quantities of small RNA targets were calculated using the delta delta CT method using mmu-miR-125b as a reference, and comparing levels of micro and piRNAs in cDNA pools prepared using T4 Rnl1 and Rnl2tr. The null hypothesis tested was that no difference exists in the relative representation of miRNAs or piRNAs tested in pools prepared using T4 Rnl2tr or T4 Rnl1. Thus, normalized relative means of micro and piRNAs detected in T4Rnl2tr prepared pools relative to T4 Rnl1 prepared were compared to a theoretical mean of 1.0 using one-sample t-tests.
TABLE 1.
Primers for small RNA qPCR
Northern blots
Mouse testicle small RNA was ligated with T4 Rnl1 or Rnl2tr as described above and ethanol-precipitated. For blots that detected mmu-miR-125b ligations used the 17-mer Universal miRNA cloning linker (New England Biolabs Inc.) because the pre-adenylated DNA adapter used for the qPCR experiments is the same size as this miRNA and obscured hybridization. For the detection of all of the piRNAs, the 21-mer pre-adenylated DNA adapter (sequence above) was used. The equivalent of 2.5–10 pmol of input small RNA per lane was separated on 15% denaturing polyacrylamide gels and electroblotted to GeneScreen Plus nylon membrane (Perkin-Elmer Life Sciences). Following UV cross-linking using the optimal cross-link setting on a Spectrolinker SL100 (Spectronics), membranes were pre-hybridized in Church Buffer (0.5 M Na2HPO4 at pH 7.2, 1 mM EDTA, 7% SDS) for 1 h at 42°C.
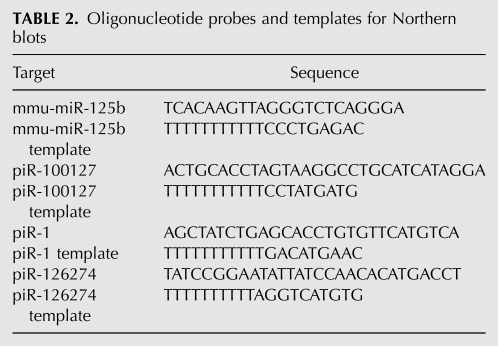
Oligonucleotide probes (Table 2) were labeled with [α-32P]dATP (Perkin-Elmer Life Sciences) by primer extension as follows: 0.5 pmol of probe and 12.5 pmol of template oligo were mixed and heated for 1 min to 95°C, annealed for 3 min at room temperature, then kept on ice. Labeling reactions contained 120 μCi of [α-32P]dATP, 5 U of Klenow Fragment (3′ → 5′ exo-), in 1× buffer 2 (New England Biolabs Inc.). Reactions were incubated for 60 min at 37°C and terminated by the addition of 40 μL of TE. Unincorporated nucleotides were removed using G-25 microspin columns (GE Healthcare).
TABLE 2.
Oligonucleotide probes and templates for Northern blots
Denatured probes were added to fresh hybridization buffer at a concentration of 1 × 106 CPM/mL and hybridized overnight at 42°C. Blots were washed three times for 15 min each with 2× SSC at 42°C, then exposed for 2–16 h to a phosphor storage screen. Images were collected using a Typhoon 9400 Variable Mode Imager. Hybridization signal was measured using QuantityOne (Bio-Rad) with local area background correction, and percent ligation was calculated by dividing higher-molecular-weight hybridization signal by the sum of the hybridization signals that corresponded to both ligated and unligated small RNAs.
Synthetic small RNA oligonucleotide mixture cloning
RNA 3′-end attachment
50 pmol of an equimolar mixture of the oligonucleotides:
mixture 1
5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUmU-3′
5′-P-rArGrCrArGrUrGrGrCrGrUrGrUrUrGrArGrArUrUrU-3′
or
mixture 2
5′-P-rArGrCrArGrUrGrGrCrUrGrGrUrUrGrArGrArUrUrU-3′
5′-P-rArGrCrArGrUrGrGrCrGrUrGrUrUrGrArGrArUrUmU-3′
were ligated to 100 pmol of the 3′-small RNA adapter using T4 Rnl1 or Rnl2tr as described above. After gel purification and reverse transcription as described above using the RT primer 5′-CAAGCAGAAGACGGCATACGAGC-3′, first-strand cDNA was treated with Exonuclease I (New England Biolabs Inc.) as described to remove excess RT primers.
cDNA 3′-end attachment
Reverse transcripts were ligated to 50 pmol of the 3′-cDNA adapter (5′-AppGATCGTCGGACTGTAGAACTCTGAAC-NH2-3′) using conditions described above (Rnl PEG). Ligation products were gel-purified and ethanol-precipitated.
PCR amplification
PCR amplification was performed using LongAmp 2× Master mix (New England Biolabs Inc.) using 25% of the 3′-ligated cDNA product with a final concentration of 1 μM Forward and Reverse USER primers [Forward USER primer: 5′-GGAGACA(dU)CAGAGTTCTACAGCTCGACGATCCGA-3′; Reverse USER primer: 5′-GGGAAAG(dU)CAAGCAGAAGACGGCATACGAGC-3′] and 25 cycles of amplification. Samples and size were separated on and purified from 4% MetaPhor (Lonza) Agarose gels, or 6% native polyacrylamide 1× TBE gels.
USER cloning
The PCR products were digested and cloned into the pNEB206A as according to the manufacturer's suggestion (USER Friendly Cloning Kit; New England Biolabs Inc.). Plasmid sequencing was performed at Agencourt Bioscience Corporation.
Data analysis
Inserts were extracted from sequencing reads using their flanking 5′- and 3′- sequences, aligned using Lasergene software, and scored by two individuals. Proportions of 2′-O-methyl RNAs recovered were compared with a theoretical 1:1 ratio using the exact binomial test of goodness-of-fit. Heterogeneity of observed ratios between experimental replicates was examined using the G-test of goodness-of-fit (McDonald 2009).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Paul Davis and Sanjay Kumar for informatics support; Weifeng Gu and the Mello lab, the Tzertzinis and McReynolds labs for helpful discussions; and Bill Jack, Elizabeth Raleigh, and Fanglei Zhuang for a critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2242610.
REFERENCES
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung J-H, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Tuschl T 2005. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett 579: 5830–5840 [DOI] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A 2009. Absolute quantification of microRNAs by using a universal reference. RNA 15: 2375–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard DR, Bowater RP 2006. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem J 398: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Methods 5: 879–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K 2009. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev 23: 2016–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Thi EP, Wang M-B, Unrau PJ 2005. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci 102: 13398–13403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B, Zimmerman SB 1984. Polymer-stimulated ligation: Enhanced ligation of oligo- and polynucleotides by T4 RNA ligase in polymer solutions. Nucleic Acids Res 12: 8235–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA 2008. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol 15: 714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton DM, Baez JA, Gumport RI 1978. T4 RNA ligase joins 2′-deoxyribonucleoside 3′,5′-bisphosphates to oligodeoxyribonucleotides. Biochemistry 17: 5091–5097 [DOI] [PubMed] [Google Scholar]
- Ho CK, Wang LK, Lima CD, Shuman S 2004. Structure and mechanism of RNA ligase. Structure 12: 327–339 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acad Sci 106: 7768–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K 2009. 2′-O-Methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SEV, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77: 22–29 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MI, Gumport RI 1980. T4 ribonucleic acid ligase joins single-strand oligo(deoxyribonucleotides). Biochemistry 19: 635–642 [DOI] [PubMed] [Google Scholar]
- McDonald JH 2009. Handbook of biological statistics. Sparky House Publishing, Baltimore, MD [Google Scholar]
- Nandakumar J, Shuman S 2004. How an RNA ligase discriminates RNA versus DNA damage. Mol Cell 16: 211–221 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S 2005. Dual mechanisms whereby a broken RNA end assists the catalysis of its repair by T4 RNA ligase 2. J Biol Chem 280: 23484–23489 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Ho CK, Lima CD, Shuman S 2004. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J Biol Chem 279: 31337–31347 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pascal JM 2008. DNA and RNA ligases: Structural variations and shared mechanisms. Curr Opin Struct Biol 18: 96–105 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ 2008. The Cid1 poly(U) polymerase. Biochim Biophys Acta 1779: 286–294 [DOI] [PubMed] [Google Scholar]
- Romaniuk E, McLaughlin LW, Neilson T, Romaniuk PJ 1982. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur J Biochem 125: 639–643 [DOI] [PubMed] [Google Scholar]
- Sano H, Feix G 1976. Terminal riboadenylate transferase from Escherichia coli. Characterization and application. Eur J Biochem 71: 577–583 [DOI] [PubMed] [Google Scholar]
- Shuman S, Lima CD 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol 14: 757–764 [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RHA 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Song J-J, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z 2009. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 15: 1443–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A, Snoper TJ, Cozzarelli NR 1977. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J Biol Chem 252: 1732–1738 [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S 2008. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 79: 581–588 [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJJ 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2006. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ 2005. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]