Abstract
Effective small interfering RNA (siRNA)–mediated therapeutics require the siRNA to be delivered into the cellular RNA-induced silencing complex (RISC). Quantitative information of this essential delivery step is currently inferred from the efficacy of gene silencing and siRNA uptake in the tissue. Here we report an approach to directly quantify siRNA in the RISC in rodents and monkey. This is achieved by specific immunoprecipitation of the RISC from tissue lysates and quantification of small RNAs in the immunoprecipitates by stem–loop PCR. The method, expected to be independent of delivery vehicle and target, is label-free, and the throughput is acceptable for preclinical animal studies. We characterized a lipid-formulated siRNA by integrating these approaches and obtained a quantitative perspective on siRNA tissue accumulation, RISC loading, and gene silencing. The described methodologies have utility for the study of silencing mechanism, the development of siRNA therapeutics, and clinical trial design.
Keywords: siRNA, Argonaute 2, RISC, in vivo delivery
INTRODUCTION
Within a decade, RNA interference (RNAi) has advanced from an inspiring discovery to a promising therapeutic approach. On the discovery front, RNAi is recognized as a fundamental process whereby a small effector RNA induces the site-specific cleavage of a mRNA containing a complementary sequence, leading to the degradation of the mRNA and subsequent down-regulation of target gene expression (Carthew and Sontheimer 2009; Ghildiyal and Zamore 2009; Jinek and Doudna 2009). The small effector RNA originates from a duplex RNA named small interfering RNA (siRNA), which is 21–23 nucleotides (nt) long and can be introduced exogenously. In a regulated assembly event, a siRNA duplex is loaded into RNA-induced silencing complex (RISC), during which one strand (passenger strand) is degraded, while the other strand (guide strand) becomes the effector small RNA remaining in the RISC (Ghildiyal and Zamore 2009). In mammals, Argonaute (Ago) 2, which belongs to the Ago subfamily of Ago proteins (Carmell et al. 2002), is the protein within the RISC that binds to the guide strand and catalyzes the cleavage of the target mRNA (Liu et al. 2004; Meister et al. 2004). Other members of the Ago subfamily, including Ago1, Ago3, and Ago4, also bind to siRNA but have no target cleavage activity (Wang et al. 2009; Yoda et al. 2010). In most mammalian cells, however, no endogenous siRNAs have been identified, and the primary small RNA species that Ago proteins bind to is microRNA (miRNA). miRNAs are also derived from double-stranded RNA precursors and predominantly function as post-transcriptional regulators of endogenous genes (Bartel 2009; Carthew and Sontheimer 2009). It is proposed that in mammalian cells, exogenous siRNA utilizes the miRNA machinery and guides cleavage of mRNA by associating with Ago2 (Farazi et al. 2008).
In therapeutic development, RNAi has achieved preclinical validation in several disease models and has advanced into clinical trials (Castanotto and Rossi 2009; Whitehead et al. 2009). This progress, however, is still overshadowed by many challenges, and the greatest is siRNA delivery (Sepp-Lorenzino and Ruddy 2008; Tseng et al. 2009). From a pharmacodynamic perspective, the essential destination of therapeutic siRNA is the cellular Ago2 in targeted cells. Successful siRNA delivery requires the administered siRNA to be internalized by the desired cells, to escape from endosomes if endocytosed, and to be loaded into Ago2. Inefficiency in any of these steps will result in reduced gene silencing. In order to overcome the delivery barriers, several strategies have been developed, including formulating siRNA with lipids or synthetic polymers (Whitehead et al. 2009). Because the potency of the delivery vehicles depends on their composition and formulation process, it is often necessary to screen multiple vehicles for the most effective variant.
To date, in vivo delivery of siRNA has been assessed via the following means (Mescalchin et al. 2007): (1) measuring reduction of target mRNA/protein; (2) quantification of siRNA uptake in a tissue; and (3) imaging, which monitors the localization of labeled siRNA. Reduction in the target mRNA/protein level provides a critical measure of siRNA pharmacodynamics (Svensson et al. 2008) but provides a relatively indirect measure of cellular delivery and may be complicated by unidentified target sequence polymorphism and off-target effects not related to RNAi (Kleinman et al. 2008). In addition, because reduction of target expression can reach a steady state at elevated doses, it is often necessary to perform resource-intensive dose titrations to differentiate siRNAs or vehicles. The tissue quantification method presents data on classical tissue pharmacokinetics. It, however, fails to examine steps beyond organ uptake such as cellular internalization and endosome escape, which are important for siRNA delivery. Because of this, the total siRNA in tissues does not always correlate with efficacy (Abrams et al. 2010). Imaging analysis may yield relevant information on subcellular siRNA delivery, but it is not always precise in quantification and it cannot distinguish between intact siRNA and the free labels released from siRNA by metabolism (Morin et al. 2009).
To improve the evaluation of siRNA delivery in vivo, we tailored an approach that quantifies the small RNAs residing in the RISC from the tissues of rodents and monkey. Application of our approach to preclinical animal studies yielded quantitative insights into the in vivo efficiency of siRNA delivery and the mechanism of RNA silencing.
RESULTS
A biochemical method for quantification of Ago2-associated siRNA strand
Our method encompasses preparation of tissue lysates, immunoprecipitation of the RISC from the lysates, and quantification of the Ago-associated small RNAs by real-time PCR (Supplemental Fig. 1). In order to maintain the fidelity of the Ago–siRNA interaction, we flash-froze biopsy samples after harvest, performed the immunoprecipitation at 4°C, and optimized the lysis buffer. An important component of our lysis buffer is heparin, which blocks RISC assembly but does not disrupt the mature RISC (Tomari et al. 2004). To increase throughput, we utilized a semi-automated instrument for lysate preparation, carried out the immunoprecipitation in a 96-well microplate using magnetic beads, and eliminated the need for RNA purification after immunoprecipitation. In order to make the method label-free, we quantified the immunoprecipitated small RNAs by the stem–loop RT-RCR assay (Chen et al. 2005; Stratford et al. 2008; Cheng et al. 2009). The reverse transcription primer in this assay has a stem–loop structure and extends the length of the generated DNA. The longer DNA template can be subsequently amplified by conventional quantitative PCR, whereas the shorter template of the size of the small RNAs cannot. Because chemical modifications of a siRNA may have effects on the stem–loop assay (Stratford et al. 2008), we measured each siRNA strand using a standard curve derived from respective modified RNA oligonucleotides. To increase assay reproducibility, we collected tissue biopsies of comparable size, started the immunoprecipitation with lysates containing equal amounts of total protein, and normalized the siRNA strand amount with those of coimmunoprecipitated endogenous miRNAs.
To demonstrate the utility of the method, we focused on characterizing the delivery properties of an exemplar lipid nanoparticle (LNP), LNP201b (Supplemental Fig. 2), which resembles the LNP201 previously published (Abrams et al. 2010). Both LNPs contained a chemically modified siRNA (Supplemental Fig. 7A, mod2) targeting the ubiquitously expressed Ssb (Sjogren syndrome antigen B) gene (Wolin and Cedervall 2002). Because RISC formation is required for all delivered siRNAs to mediate gene silencing, we anticipate that the principle of our quantification method can be applied to other siRNA delivery vehicles and targets.
Identification of specific antibodies for rodent and monkey Ago2
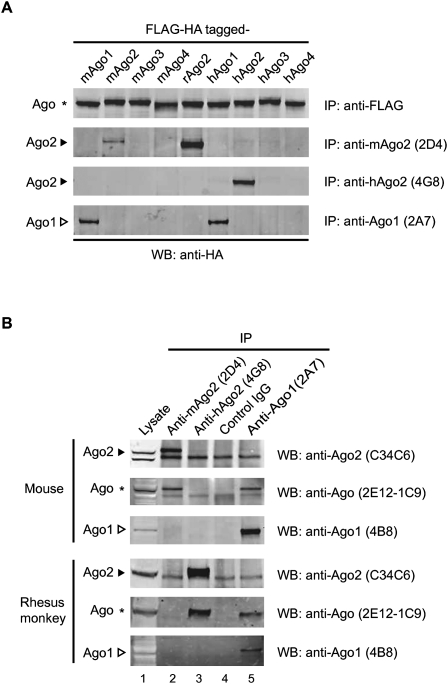
A specific antibody is the prerequisite for immunoprecipitation of Ago proteins from tissue lysates. There are multiple anti-Ago antibodies that are commercially available, but their specificity for Ago proteins from experimental animals has not been clearly defined. To expedite the characterization of antibodies, we transiently expressed a panel of Flag-HA–tagged Ago proteins, including Ago2 of mouse, rat, human, as well as three other paralogs of mouse and human, in HEK 293T cells. The overexpressed proteins were subjected to immunoprecipitation with either an anti-Flag antibody or individual test antibodies, followed by Western blotting with an anti-HA antibody. Among the antibodies we tested, an anti-mAgo2 antibody (clone 2D4) and an anti-hAgo2 antibody (clone 4G8) (Azuma-Mukai et al. 2008; Miyoshi et al. 2008) were confirmed to be specific for rodent and human Ago2, respectively (Fig. 1A). In addition, an anti-Ago1 antibody (clone 2A7) was validated to be cross-reactive with mouse and human Ago1 (Fig. 1A).
FIGURE 1.
Characterization of Ago-specific antibodies. (A) Specificity of the antibodies for immunoprecipitation of overexpressed Ago proteins. Indicated rodent and human Ago proteins with N-terminal Flag-HA tags were transiently expressed in HEK 293T cells, immunoprecipitated with the indicated antibodies, and detected in Western blot using an anti-HA antibody. (B) Immunoprecipitation effectiveness of the antibodies using either mouse or rhesus monkey total liver lysates. Lysates (30 μg) were loaded as input (5%). Antibodies used for Western blot after immunoprecipitation are indicated to the right of individual blots. The species specificity of the antibodies is represented with the abbreviation (m, mouse; h, human/monkey) before the protein name. Solid arrowhead, hollow arrowhead, and asterisk denote Ago2, Ago1, and Ago protein members, respectively.
By using the overexpressed Ago proteins, we identified independent antibodies for Western blot analysis as well (Supplemental Fig. 3A): anti-Ago2 antibody (clone C34C6) detects Ago2, while anti-Ago1 antibody (clone 4B8) detects Ago1. Another antibody (clone 2E12-1C9) recognizes all Ago isoforms.
We continued the characterization of the antibodies by evaluating their specificity for immunoprecipitation in animal liver lysates. We found that the anti-mAgo2 antibody (2D4) specifically immunoprecipitated mouse Ago2, the identity of which was confirmed by blotting with two other antibodies, C34C6 and 2E12-1C9 (Fig. 1B, lane 2). The anti-mAgo2 antibody 2D4 also immunoprecipitated rat Ago2 specifically (Supplemental Fig. 3B, lane 2). In addition, we evaluated whether anti-hAgo2 (4G8) could specifically immunoprecipitate monkey Ago2 from monkey liver lysates. This seemed possible because the protein fragment (human Ago2, 1–148) used to produce anti-hAgo2 antibody (4G8) (Azuma-Mukai et al. 2008) has 85% sequence identity to a corresponding region of rhesus monkey Ago2 (Supplemental Fig. 4). Indeed, the antibody cross-reacted with rhesus monkey Ago2 but not with rodent Ago2 (Fig. 1B; Supplemental Fig. 3B, lane 3). Furthermore, the anti-Ago1 antibody (clone 2A7) specifically immunoprecipitated both rodent and monkey Ago1 (Fig. 1B; Supplemental Fig. 3B, lane 5).
We also found that the anti-Ago1 antibody (2A7) did not immunoprecipitate rodent or monkey Ago2 and the anti-Ago2 antibodies did not pull down detectable levels of Ago1 (Fig. 1B; Supplemental Fig. 3B). These results suggest that there was insignificant association of Ago1 with Ago2 in the immunoprecipitates obtained with our method.
Addressing potential immunoprecipitation artifacts
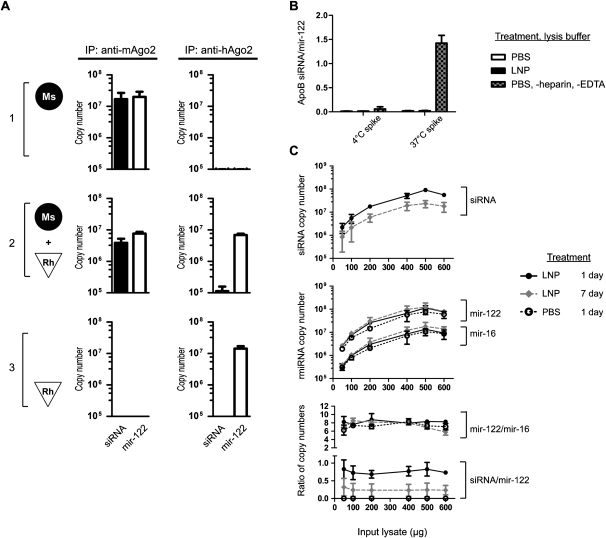
Faithful quantification of siRNA delivered into the RISC requires the control of potential artifacts. One reported artifact of immunoprecipitation is that RNA and protein may form complexes after cell lysis (Mili and Steitz 2004). Specifically for post-lysis Ago2–siRNA association, the siRNA duplex may be released from delivery vehicles circulating in the blood or from intracellular compartments such as endosomes upon tissue lysis, and then be loaded into Ago2. Additionally, siRNA strands may dissociate from other RNA-binding proteins such as Ago1 and re-associate with Ago2. To address these possibilities, we prepared three sets of tissue samples for analysis: liver tissue from Ssb siRNA-treated mouse, untreated monkey liver tissue, and a mix of the two tissues (1:1 by weight). Lysates were prepared from the three sets and subjected to immunoprecipitation using either anti-mouse Ago2 antibody (2D4) or anti-human/monkey Ago2 antibody (4G8). If there was significant association of siRNA with Ago2 after tissue lysis, siRNA from the mouse liver tissue would transfer to monkey Ago2 and be detected in anti-monkey Ago2 immunoprecipitates. Our experimental results argued against this possibility: The majority of siRNA remained associated with mouse Ago2 and <3% of siRNA transferred to monkey Ago2 (Fig. 2A). As an internal control, the conserved liver-specific mir-122 was measured in the Ago2 immunoprecipitates as well (Chang et al. 2004). The levels of mir-122 were comparable among the immunoprecipitation reactions except the two in which mismatched antibodies were used (Fig. 2A).
FIGURE 2.
Reliability and robustness of the immunoprecipitation step. (A) siRNA-treated mouse liver harvested at 24 h post-dosing (filled circle, set 1), untreated monkey liver tissue (hollow triangle, set 3), and a mix of the two tissues (set 2) were lysed, and Ago2 from the lysates was immunoprecipitated with either anti-mouse Ago2 or anti-human/monkey Ago2 antibody. After subtracting the background numbers of small RNAs that nonspecifically bind to a control IgG, the copy numbers of Ago2-associated Ssb siRNA and mir-122 from each immunoprecipitates are graphed as mean ± SD (two independent experiments, each performed in duplicate). (B) Synthetic ApoB siRNA duplex was spiked into the indicated liver lysates at a final concentration of 1 μM and incubated at either 4°C or 37°C for 4 h before immunoprecipitation. The ratio of ApoB siRNA guide strand to mir-122 from two independent experiments with duplicates are shown as the mean ± SD. (C) The indicated amounts of liver lysates were prepared from PBS-treated or siRNA-treated animals harvested at 1 d and 7 d after dosing and were then subjected to immunoprecipitation. Copy numbers (not normalized) of Ago2-associated siRNA guide strand, mir-122, and mir-16, as well as the ratios of mir-122/mir-16 and siRNA guide/mir-122 are presented as the mean ± SD (n = 2 per datum).
We took another approach to address potential artifacts by spiking a siRNA duplex against Apob directly into the liver lysates of either PBS (phosphate buffered saline)-treated or Ssb siRNA-treated mice prior to immunoprecipitation. Negligible amounts of the ApoB siRNA guide strand were detected in the Ago2 immunoprecipitates under our standard condition (4°C and the lysis buffer contains EDTA and heparin) (Fig. 2B). Upon raising the incubation temperature to 37°C and omitting heparin and EDTA in the lysis buffer, we observed significant loading of exogenous siRNA into Ago2, indicating that post-lysis loading of siRNA duplex could cause artifacts if not properly controlled for (Fig. 2B).
To improve the reliability of our method, we aimed to reduce the assay variability derived from difference in Ago2 protein expression level, lysate protein concentration determination, binding of antibodies, and the amount of magnetic beads. Because Ago proteins bind to endogenous miRNAs, we tested the possibility of normalizing siRNA level to those of miRNAs. When the amount of input mouse liver lysates varied from 100–600 μg, we observed a proportionality of immunoprecipitated siRNA guide strand and miRNAs (Fig. 2C). More importantly, the ratio of siRNA guide strand to mir-122 remained constant (Fig. 2C). The ratio between immunoprecipitated miRNAs (mir-122/mir-16) was not affected by the quantity of lysate input or the treatment (Fig. 2C), and the ratio correlated with the relative abundance of the miRNAs in the purified mouse total liver RNA (Supplemental Table 1).
In vivo quantification of Ago2-associated siRNA and target gene silencing
To further validate our method and gain quantitative insights into siRNA delivery in vivo, we performed a mouse study measuring both gene silencing and Ago2-associated siRNA strand levels from tissue samples. Mice were treated with LNP201b-Ssb siRNA at three escalating doses or with a control LNP, which incorporated a comparable amount of the same Ssb siRNA duplex (Supplemental Fig. 7A, mod2). Liver and spleen punch biopsies were collected at multiple time points post-dosing for analyses.
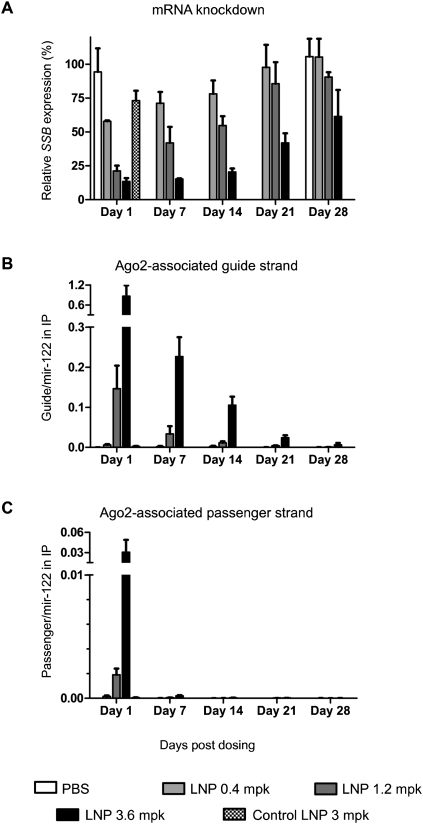
We found that in the liver samples, the amount of Ago2-associated siRNA guide strand coincided well with the efficacy of target knockdown in a dose- and time-dependent manner; the more siRNA detected in Ago2, the greater the reduction of target mRNA level (Fig. 3A,B). The control LNP delivered a negligible amount of guide strand into Ago2 and had no significant activity. In addition, the level of Ago2-associated siRNA guide strand showed a greater dynamic range than that of mRNA knockdown. Because of this, it was easier to discriminate the treatment condition effectiveness (for example, among days 1, 7, and 14 for 3.6 mg/kg dose). The greater dynamic range of this approach could enable the design of delivery studies requiring fewer animals.
FIGURE 3.
Target (Ssb) gene silencing and quantification of Ago2-associated siRNA strands in mouse liver samples. (A) Dose-response and duration of knockdown in liver after a single intravenous administration of PBS or indicated LNP-siRNA. Expression levels of Ssb relative to housekeeping Ppib were determined by quantitative RT-PCR and expressed as percentage values in relation to PBS-treated animals. (B,C) Levels of Ago2-associated siRNA guide strand (B) or passenger strand (C) after normalizing their copy numbers to those of coimmunoprecipitated mir-122. The non-normalized copy numbers of the small RNAs are plotted in Supplemental Figure 5. Each bar, group mean ± SD (n = 4 per group).
With the assumption that the siRNA delivery into the majority of liver cells is not significantly different, we attempted to estimate the copy numbers of siRNA guide strand required for a given silencing efficacy in an average liver cell. For instance, in the above mouse study, ∼50% silencing was observed at two time points: day 14 for 1.2 mg/kg dose and day 21 for 3.6 mg/kg dose (Fig. 3A). By normalizing the copy number of the immunoprecipitated Ago2-bound siRNA guide strand to that of liver-specific mir-122 and by determining the total number of the mir-122 in a liver cell (for calculations, see Supplemental Table 2), we estimated about 370 copies of siRNA guide strand per mouse liver cell were required for 50% target Ssb mRNA knockdown. Note that among the estimated 370 siRNA copies, only a portion is associated with Ago2 and cleaves target mRNA, because all Ago subfamily members bind to siRNAs (Meister et al. 2004) and Ago2 can mediate cleavage-independent gene silencing as well (Wu et al. 2008).
By using absolute quantitative RT-PCR, we estimated that there were about 100 copies of Ssb target mRNA in a mouse liver cell. At the time points when 50% target mRNA inhibition was achieved, there were about 50 copies of Ssb mRNA remaining per cell. It is appealing to think that the number of target mRNA being cleaved, speculatively in the range of 50–100, reflects the number of cleavage-capable RISC.
At the 3.6 mg/kg dose, the most effective silencing occurred on day 1 post-dosing. There were about 18,000 copies of siRNA guide strand per cell and a 87% reduction in Ssb mRNA level. The knockdown of Ssb was sustained at >80% for ∼14 d, despite the steady reduction of the quantity of Ssb siRNA in Ago2 (Fig. 3A,B). After 14 d, the quantity of Ago2-bound siRNA continued to decrease, and the reduction of silencing efficacy was more appreciable. These findings imply that (1) there is a continuous degradation of Ago2-associated siRNA guide strand, and (2) delivering more copies of siRNA than the minimum required for achieving given efficacy may buffer the loss of siRNA and prolong the duration.
Another important question for siRNA therapeutics is whether administered siRNA competes with endogenous miRNAs for binding to Ago proteins. To address this, we quantified four miRNAs in the mouse liver Ago2 immunoprecipitates: highly abundant mir-122, intermediately abundant mir-16, and low abundance mir-26b and let-7a. For all the miRNAs examined, no significant difference was observed among the PBS-treated and siRNA-treated animals (Supplemental Fig. 5). This finding implies that the siRNA, at least at the doses we used, does not perturb the binding of miRNAs to Ago proteins. It also confirms that the coimmunoprecipitated miRNAs can be utilized for quantification normalization.
In addition to the guide strand and miRNAs, we also quantified the passenger strand of the Ssb siRNA in the Ago2 immunoprecipitates. The quantity of the passenger strand was more than 30-fold less than that of the guide strand on day 1 post-dosing and declined more rapidly afterward (Fig. 3C). We thus conclude that for the siRNA we used, the loading of the passenger strand into Ago2 is unfavorable (see below). The selectivity against the passenger strand may help to improve siRNA specificity by reducing the off-target effects induced by the Ago-associated passenger strand (Chen et al. 2008).
It has been reported that the LNP can also deliver siRNA to mouse spleen, but compared with that observed in the liver, the silencing effect was weak (Morrissey et al. 2005; Abrams et al. 2010). We extended the analysis of Ago2-bound siRNA strands to the mouse spleen samples harvested 1 d post-dosing. We found that (1) the knockdown of the Ssb target in spleen was indeed marginal; (2) the active LNP did deliver siRNA into Ago2 in a dose-dependent manner, whereas the less-active control LNP failed to do so; and (3) the prevailing bias for guide strand loading was maintained in the spleen (Supplemental Fig. 6). We surmise that the insignificant knockdown of Ssb mRNA in the spleen is due to the failure to deliver the threshold level of siRNA needed for silencing in the spleen cells expressing Ssb.
Chemical modifications of siRNA confer strand selection
The siRNA used in the above mouse study is chemically modified (Supplemental Fig. 7A, mod2), and the guide strand was almost exclusively incorporated into Ago2. To understand the basis for this exclusivity, we probed the effect of chemical modifications on siRNA loading in cultured HEK 293T cells using the same Ago2 immunoprecipitation and small RNA quantification approach.
Whereas the unmodified siRNA showed an approximately threefold preference for the guide strand, likely due to the thermodynamic asymmetry of the sequence (Schwarz et al. 2003), chemical modifications on the siRNA conferred enhanced guide strand selection: ∼17-fold for mod1 and ∼33-fold for mod2 (Supplemental Fig. 7A,C). All three siRNA duplexes were effective at reducing Ssb mRNA level (Supplemental Fig. 7B). The difference between mod1 and mod2 is the extent of backbone modification, which does not appear to be the primary determinant for selectivity. In both modifications the ends of the passenger strand were blocked with inverted abasic caps (B). We surmise that the 5′-B modification controls strand selection by blocking 5′ phosphorylation, which is important for RISC loading (Nykanen et al. 2001; Czauderna et al. 2003; Chen et al. 2008).
A quantitative view of mouse liver siRNA delivery at early time points
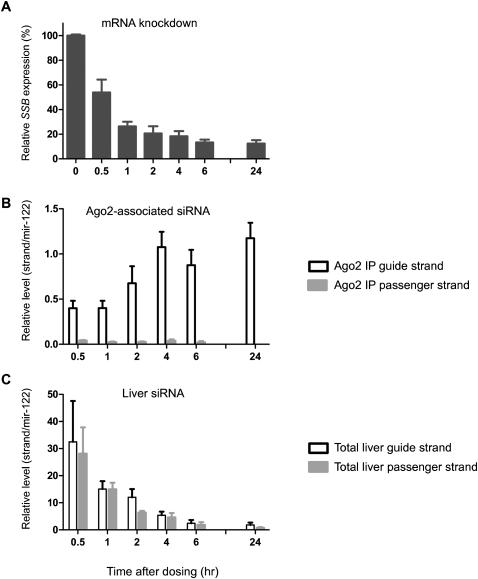
In our mouse study discussed above, the silencing effect and the level of siRNA guide strand in Ago2 peaked as early as 1 d after dosing. To evaluate the siRNA delivery at earlier time points, we performed another mouse experiment and examined the kinetics of mRNA knockdown, siRNA delivery in the liver, and siRNA association with Ago2 (Fig. 4).
FIGURE 4.
Kinetics of silencing, siRNA tissue uptake, and Ago2 loading in mouse liver. Mice were treated with a single intravenous dose (3.6 mg/kg) of LNP201b-Ssb siRNA and harvested at the indicated time points. (A) Silencing of the Ssb target mRNA. Expression levels of Ssb relative to housekeeping Ppib were determined by quantitative RT-PCR and expressed as percentage values in relation to PBS-treated animals. (B) Normalized quantities of Ago2-assocaited siRNA guide and passenger strands. (C) Normalized amounts of siRNA guide and passenger strands in liver homogenates.
We observed that the silencing of Ssb mRNA was already evident at 30 min and reached a steady state ∼6 h post-dosing (Fig. 4A). The loading of siRNA into Ago2 was also apparent at 30 min and increased until 4 h post-dosing (Fig. 4B). The majority of the siRNA associated with Ago2 was the guide strand (Fig. 4B). In contrast, siRNA detected in total liver homogenates at early time points was in much higher abundance. The majority of the total liver siRNA was likely in duplex form, and the levels of siRNA strands decreased sharply over time (Fig. 4C). At 24 h post-dosing, the levels of the guide strand in Ago2 and in the liver appeared to converge. We estimated that the amount of the siRNA guide strand in Ago2 at 24 h was only ∼4% of that in liver at 0.5 h and conceivably even lower if compared to the amount of siRNA dosed. This implies that there is ample room for improving siRNA delivery.
Finally, we did a Western blot analysis of Ago2 in liver lysates and found that LNP-siRNA treatment had no obvious effect on Ago2 protein abundance (Supplemental Fig. 8).
Extending the Ago2–siRNA complex quantification to rat and nonhuman primate
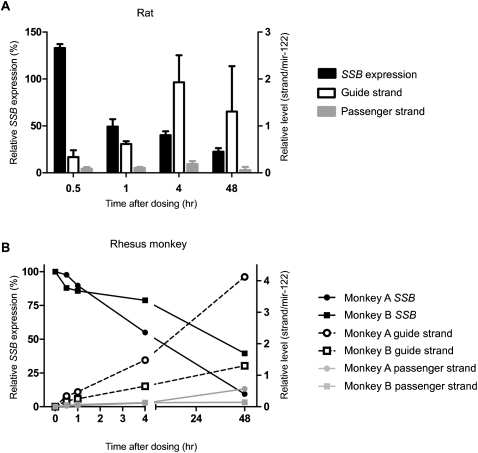
The rat and rhesus monkey are other important preclinical species, and we extended the quantification of Ago2–siRNA complex to their liver tissues. Similar to what we observed for the mouse, the quantity of Ago2-associated Ssb siRNA guide strand in the livers of rat and monkey correlated well with the extent of target silencing (Fig. 5). Moreover, a similar correlation was observed when we tested a siRNA against a different target, Apob (data not shown). These studies indicate that the in vivo quantification of RISC delivery can be applied to animals other than the mouse.
FIGURE 5.
Quantification of RISC in rat and rhesus monkey. Rats (A) or two rhesus monkeys (B) were treated with a single intravenous dosing of 3.6 mg/kg LNP201b-Ssb siRNA and analyzed at indicated time points. Ssb mRNA expression (left y-axis) relative to housekeeping Ppib was normalized to that of PBS-treated animals (defined as 100%). Levels of siRNA strands (right y-axis) were normalized to those of respective coimmunoprecipitated mir-122. For rats each bar represents the group mean SD (n = 3 per group). For monkeys the data are shown as for the individual animal.
DISCUSSION
We conducted a quantitative siRNA in vivo delivery analysis that bridges dosing and efficacy. The quantitative information obtained from this type of analysis will facilitate the development of siRNA therapeutics and establish pharmacokinetics/pharmacodynamics relationships. Moreover, the combined approaches of lipid-mediated delivery and quantitative analysis present an appealing platform for investigation of silencing mechanisms in vivo.
Quantification of the RISC assembled in vitro has been demonstrated by the Zamore laboratory using bead-immobilized 2′-O-methyl-modified oligonucleotides that were antisense to the siRNA strand within the RISC (Schwarz et al. 2003; Hutvágner et al. 2004). Because the siRNA strand was 5′-32P-radiolabeled, the amount of radioactivity pulled down by the beads was a direct measurement of the amount of siRNA incorporated into the RISC in vitro. Utilizing this method on an elevated scale in animals, however, would be difficult due to the requirement for radiolabeled siRNA.
Our quantitative analysis of the hepatic delivery of siRNA in mouse reinforced several concepts in the field. The first is the requirement for a threshold quantity of siRNA for efficacy. Our findings support this notion by estimating that about 370 copies of siRNA per mouse liver cell are required for a 50% Ssb mRNA inhibition. Interestingly, this estimated value is similar to the one reported previously and derived from experiments in which siRNA against a different target was delivered into cultured cells via either lipofectamine or microinjection and quantified using a non–PCR-based method (Veldhoen et al. 2006; Mescalchin et al. 2007). It remains to be investigated whether this threshold value is influenced by siRNA modification, target mRNA, and abundance of Ago protein members. In theory, this value is readily achievable by liposomal delivery vehicles because, by approximation, each LNP may contain up to 4000 siRNAs (Peer et al. 2008).
The second concept is the stability of the RISC. Our results imply that siRNA in Ago2 undergoes continuous degradation. Our data also suggest that the duration of silencing is a balanced outcome of initial loading of the RISC and siRNA degradation for the nondividing hepatocyte. Recently, it has been demonstrated the miRNA homeostasis in Caenorhabditis elegans is controlled by a 5′→3′ exoribonulease XRN-2 and target binding modulates miRNA stability (Chatterjee and Grosshans 2009). It would be important to investigate whether this scenario pertains in animals for siRNA stability.
The last concept is the saturation of silencing. Within the dose regimen we tested, there is no indication that maximal RISC loading was achieved, but a plateau for efficacy was observed. It is likely that RISC machinery can be saturated at higher doses, as inferred from in vitro transfection experiments (McManus et al. 2002). One interesting question is the source of loadable Ago2 protein. It has been reported that cell–cell contact globally activates miRNA biogenesis via enhanced processing and more efficient formation of the RISC, but not through increasing abundance of Ago proteins (Hwang et al. 2009). Because there was no evident induction of Ago2 expression upon siRNA treatment in our study either, there could be a cellular reservoir of unoccupied Ago2. This pool of Ago2 could be active but not utilized by the endogenous miRNA pathway, possibly because the concentration of the miRNA precursors is below or near the Kd of the loading machinery. With the influx of administered siRNA (Fig. 4C), however, this fraction of Ago2 can be loaded with siRNA due to elevated siRNA concentration. Alternatively, the unoccupied Ago2 may be in an inactive form and/or in a different compartment and enter into the RISC after exogenous siRNA reaches the cytoplasm.
A potential consequence for RISC machinery saturation is the competition between siRNA and endogenous miRNAs for Ago2. It has been reported that transfected siRNAs could compete with endogenous miRNA for silencing complex and disturb gene regulation by miRNAs (Castanotto et al. 2007; Khan et al. 2009). However, we did not witness any significant changes in the levels of coimmunoprecipitated miRNAs after siRNA treatment. Our finding is consistent with another in vivo report that siRNA treatment caused no changes in total hepatic levels of miRNAs nor any increase in expression of mir-122 targets (John et al. 2007). The discrepancies may be attributed to differences in siRNA concentration between in vivo and in vitro transfections. Our method for quantification of RISC may enable further investigation on this issue.
In our mouse study, nonproportional amounts of the RISC were required for increased levels of silencing: about 370 copies for 50% inhibition and about 2200 copies for 80% inhibition on day 14. One possibility to account for this observation is target accessibility. If the RISC needs to transiently interact with mRNA and “scan” for the complementary site (Ameres et al. 2007), more copies of the RISC may be required to find the few remaining target mRNAs.
Target accessibility may also account for the nominal silencing activity observed in the spleen. The spleen is more heterogeneous with respect to cell types than the liver, and the cells that are more amenable to siRNA delivery may be different from those expressing target mRNA. To examine this possibility, the quantification of RISC loading could be extended to specific cell types isolated from disaggregated tissues.
It should be acknowledged that stem–loop RT-PCR may not efficiently detect certain truncated yet active siRNA species, insofar as it relies on the intact 3′ end of the small RNA (Wu et al. 2007; Stratford et al. 2008). Consequently, alternative detection methods should be exploited to confirm the results obtained through stem–loop RT-PCR. In addition, it would be intriguing to examine the small RNA loading in all Ago paralogs and probe whether it is feasible to design siRNAs that preferentially enter into Ago2. The studies described here serve as a starting point for further quantitative analyses in support of fulfilling the promises of novel siRNA therapeutics.
MATERIALS AND METHODS
Animals
Female Crl:CD-1/ICR mice were obtained from Charles River Laboratories and were between 6 and 10 wk old at time of study (25–30 g). Female Crl:CD (SD) rats were also obtained from Charles River Laboratories, and they were 5–6 wk old at time of study (125–150 g). Rhesus monkeys were obtained from New Iberia Research Center and were between 3 and 4 yr old at time of study (4–5 kg). All studies were performed in Merck Research Laboratories' AAALAC-accredited West Point (PA) animal facility using protocols approved by the Institutional Animal Care and Use Committee.
Monoclonal antibodies
Antibodies for Ago2 (clone 2D4 and 4G8) and for Ago1 (clone 2A7) were purchased from Wako Chemicals USA, Inc. Anti-Ago antibody (clone 2E12-1C9) and anti-Flag antibody were from Sigma. Anti-Ago2 rabbit antibody (clone C34C6) was from Cell Signaling Technology. Monoclonal antibody against Ago1 (4B8) was purchased from Ascenion GmbH (Beitzinger et al. 2007). Anti-HA antibody was from Covance.
Plasmids
Full-length cDNAs encoding mouse Ago1-4 (accession nos. NP_700452, NP_694818, NP_700451, and NP_694817), rat Ago2 (accession no. NP_067608), and human Ago1-4 (accession nos. NP_036331, NP_036286, NP_079128, and NP_060099) were transferred into a mammalian expression vector derived from pIRESpuro3 (Clontech) from respective Entry vectors by Gateway cloning.
siRNA and liposomes
Chemically modified siRNA against Ssb was synthesized by Merck/Sirna Therapeutics. The sequence and chemical modifications are illustrated in Supplemental Figure 7A. siRNA-LNPs were assembled as previously described (Abrams et al. 2010), except that the molar ratio of cLinDMA:cholesterol:polyethylene glycerol was 57:38:5 for LNP201b. For the control LNP, the cationic lipid cLinDMA was replaced by (3b)-3-[4-({(2S)-1-(morpholin-4-yl)-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-2-yl}oxy)butoxy]cholest-5-ene, and the molar ratio of cationic lipid:cholesterol:polyethylene glycerol was 50.3:44.3:5.4.
Quantification of tissue siRNA and mRNA
The Ssb mRNA levels were measured by relative quantitative RT-PCR as previously described (Abrams et al. 2010). To estimate the copy number of Ssb mRNA per cell, the protocol was modified to be absolutely quantitative by using an in vitro transcribed sense RNA fragment as the standard. Briefly, a 250-bp synthetic minigene containing the Ssb qPCR amplicon surrounded by 5′ and 3′ flanking sequences of Ssb mRNA was cloned into the plasmid pIDTBlue (Integrated DNA Technologies). The insert and the upstream T7 RNA Polymerase promoter were amplified by conventional PCR and subjected to in vitro transcription (IVT) using the MAXIscript kit (Ambion). A standard curve was generated by subjecting the IVT product to serial dilutions in a matched concentration of irrelevant rat total RNA. A linear equation derived from parallel qPCR analysis of the standard curve was used to transform sample Ct values into absolute copy numbers per nanogram of RNA. The copy numbers of Ssb mRNA per cell was then calculated by assuming that each cell contains 20 pg of RNA. The siRNA concentration in tissues was determined using a modified stem–loop RT-PCR procedure also as described (Abrams et al. 2010), except that mir-122 replaced RNU6B as a normalization control.
Tissue lysate preparation
Six-millimeter tissue punch biopsies were excised from the liver or spleen of individual animal and flash-frozen at time of harvest. The frozen tissues were pulverized in Cryobags (Covaris) using a Cryoprep system (Covaris) at impact level of 4. Pulverized tissues were transferred to 17-mm glass tubes on dry ice, mixed with 1 mL ice-cold lysis buffer (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 0.5% Triton X-100, 2 mM EDTA, 1 mg/mL heparin, and protease inhibitor cocktail; Roche), and homogenized with a Covaris E-210 instrument at the setting of 10% duty cycle, intensity 8, and 200 cycles/burst for 30 sec. After processing, lysates were centrifuged at 21,000g for 45 min at 4°C. The supernatants were aliquoted, and total protein concentrations were determined by the Biorad Bradford dye reagent with bovine serum albumin as the standard. In the spike-in experiment, a siRNA against ApoB was added to the lysate at a final concentration of 1 μM, and the mixtures were incubated at either 4°C or 37°C for 4 h before immunoprecipitation.
Immunoprecipitation
For immunoprecipitation in 96-well microplate format, 200 ng of antibody was absorbed to 50 μL of magnetic Protein G Dynabeads (Invitrogen) during a 2-h incubation at 4°C in 100 μL lysis buffer on a micromixer. The beads were washed twice with 100 μL lysis buffer to remove unbound antibody. Six microliters of lysates (360 μg total protein) was then incubated with the antibody-bound beads overnight at 4°C with mixing. The beads were washed five times with lysis buffer, followed by one washing with a buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 0.2% Triton X-100. The small RNAs coimmunoprecipitated with the beads were extracted by incubation with Cells-to-Ct reagents (Ambion) following the manufacturer's protocol. In Figure 1B immunoprecipitation was performed in microcentrifuge tube format with the following changes: 5 μg of antibody was absorbed to 50 μL of Protein A/G beads (Santa Cruz), 400 μL lysates at 2 mg/mL were used, and the beads were heated at 70°C for 10 min in 2xNuPAGE sample buffer before an aliquot of the supernatant was taken for gel electrophoresis.
Quantification of small RNAs in immunoprecipitates
Custom Taqman small RNA assay reagents for Ssb siRNA guide strand and passenger strand were obtained through the early access program from Applied Biosystems (Cheng et al. 2009). Taqman miRNA assays were also purchased from Applied Biosystems. Five microliters of the Cells-to-Ct eluents was assayed following the vendor's protocol with respective small RNA as the standard. Small RNA copy numbers were calculated using a linear equation derived from the respective standard curve. The copy number of mir-122 per liver cell was estimated by quantifying mir-122 from 10 ng of purified total liver RNA with the assumption that each liver cell contains 20 pg of RNA.
Cell culture and transfections
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a 5% CO2-containing atmosphere. For transfection, cells were plated on 10-cm Corning CellBIND dishes. Plasmids overexpressing Ago proteins were transfected at 10 μg each using FuGENE 6 (Roche) according to the manufacturer's protocol. Ssb siRNA duplexes were reversely transfected at 0.2 nM using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instruction.
Western blot analysis
Aliquots of the tissue lysates (30 μg each) or immunoprecipitates were heated for 5 min at 100°C and resolved by electrophoresis on 4%–12% NuPAGE gels (Invitrogen). The polypeptides were transferred to a nitrocellulose membrane (Invitrogen) with a wet transfer apparatus (Bio-rad). The membrane was probed with primary antibodies, and the immune complexes were visualized with appropriate IRDye 800CW-conjugated secondary antibodies (LI-COR Biosciences) using an Odyssey Infrared Imaging System (LI-COR Biosciences).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Joseph Davide, Lisa Hettrick, Michael Lyman, Kim Michel, Kenneth Lodge, and members of the Central Pharmacology group for helping with the animal experiments or providing tissue samples. We also thank Jason Murphy and James Cunningham for LNPs and other colleagues in the department for reagents and helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2255810.
REFERENCES
- Abrams MT, Koser ML, Seitzer J, Williams SC, DiPietro MA, Wang W, Shaw AW, Mao X, Jadhav V, Davide JP, et al. 2010. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: Effect of dexamethasone co-treatment. Mol Ther 18: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G 2007. Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol 4: 76–84 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ 2009. The promises and pitfalls of RNA-interference-based therapeutics. Nature 457: 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ 2007. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res 35: 5154–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1: 106–113 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179 doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G 2008. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 14: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Li M, Liang Y, Wang Y, Wong L, Chen C, Vlassov AV, Magdaleno S 2009. Stem-loop RT-PCR quantification of siRNAs in vitro and in vivo. Oligonucleotides 19: 203–208 [DOI] [PubMed] [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J 2003. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res 31: 2705–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD 2004. Sequence-specific inhibition of small RNA function. PLoS Biol 2: E98 doi: 10.1371/journal.pbio.0020098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2009. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci 106: 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, Soutschek J, Vornlocher HP, Manoharan M, et al. 2007. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature 449: 745–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, et al. 2008. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 452: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- McManus MT, Haines BB, Dillon CP, Whitehurst CE, van Parijs L, Chen J, Sharp PA 2002. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol 169: 5754–5760 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Mescalchin A, Detzer A, Wecke M, Overhoff M, Wunsche W, Sczakiel G 2007. Cellular uptake and intracellular release are major obstacles to the therapeutic application of siRNA: Novel options by phosphorothioate-stimulated delivery. Expert Opin Biol Ther 7: 1531–1538 [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10: 1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Uejima H, Nagami-Okada T, Siomi H, Siomi MC 2008. In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol Biol 442: 29–43 [DOI] [PubMed] [Google Scholar]
- Morin A, Gallou-Kabani C, Mathieu JR, Cabon F 2009. Systemic delivery and quantification of unformulated interfering RNAs in vivo. Curr Top Med Chem 9: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. 2005. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol 23: 1002–1007 [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M 2008. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science 319: 627–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sepp-Lorenzino L, Ruddy M 2008. Challenges and opportunities for local and systemic delivery of siRNA and antisense oligonucleotides. Clin Pharmacol Ther 84: 628–632 [DOI] [PubMed] [Google Scholar]
- Stratford S, Stec S, Jadhav V, Seitzer J, Abrams M, Beverly M 2008. Examination of real-time polymerase chain reaction methods for the detection and quantification of modified siRNA. Anal Biochem 379: 96–104 [DOI] [PubMed] [Google Scholar]
- Svensson RU, Shey MR, Ballas ZK, Dorkin JR, Goldberg M, Akinc A, Langer R, Anderson DG, Bumcrot D, Henry MD 2008. Assessing siRNA pharmacodynamics in a luciferase-expressing mouse. Mol Ther.16: 1995–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Tseng YC, Mozumdar S, Huang L 2009. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev 61: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen S, Laufer SD, Trampe A, Restle T 2006. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: Quantitative analysis of uptake and biological effect. Nucleic Acids Res 34: 6561–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG 2009. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov 8: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T 2002. The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N 2007. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE 2: e1020 doi: 10.1371/journal.pone.0001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2008. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol 18: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y 2010. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]