Abstract
Loss-of-function studies in human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) via nonviral approaches have been largely unsuccessful. Here we report a simple and cost-effective method for high-efficiency delivery of plasmids and siRNAs into hESCs and iPSCs. Using this method for siRNA delivery, we achieve >90% reduction in the expression of the stem cell factors Oct4 and Lin28, and observe cell morphological and staining pattern changes, characteristics of hESC differentiation, as a result of Oct4 knockdown.
Keywords: stem cells, siRNA, transfection
INTRODUCTION
Human embryonic stem cells (hESCs) are capable of self-renewal and differentiation into all derivatives of three primary germ layers, giving rise to every cell in the human body (Thomson et al. 1998). These features render hESCs a valuable model system for studying human embryogenesis, dissecting mechanism of human diseases, screening new drugs, and engineering tissues or even organs. Induced pluripotent stem cells (iPSCs) are believed to be essentially identical to hESCs and also bear great therapeutic potential since they can be generated from patients directly (Takahashi et al. 2007; Yu et al. 2007). To capitalize on the potential of hESCs and iPSCs, it is imperative to have an effective method to introduce genes into these cells or to knock down the expression of specific genes for genetic dissection of the molecular mechanisms underlying their proliferation and differentiation. RNA interference technology for loss-of-function studies is a powerful tool for these purposes.
Transient delivery of siRNA duplexes into cells facilitated by cationic lipid carriers such as Lipofectamine 2000 has been widely used for gene targeting in mammalian cells. However, this regime does not work well with hESCs, likely, in part, due to the low cloning (growing cell colonies from single cells) efficiency of hESCs. In most studies, hESCs are manipulated as small cell clumps, but not single cells, since single hESCs generated by enzymatic digestion suffer from low cell viability (Zaehres et al. 2005). This not only makes the process of clonal selection difficult, but also causes inaccessibility of the inner cells for transfection materials and reagents. In addition, the overgrowth of untransfected cells may overwhelm and dilute transfected cells, further reducing transfection efficiency. These limitations make hESC high transfection efficiency difficult to reach and high-efficiency siRNA-mediated gene knockdown hard to attain. To circumvent these problems, lentivirus delivery has been developed to target genes in hESCs (Zaehres et al. 2005). Although highly efficient transduction and gene knockdown have been achieved using this system, the process involves potential challenges in virus construction, packaging, and obtaining a sufficiently high titer, as well as the copy number and position effect of virus insertion into the genome. Here, we report a method that combines the usage of Accutase and ROCK inhibitors with single cell suspension transfection using Lipofectamine 2000 as a delivery reagent to achieve a >90% efficiency in transient silencing of endogenous Lin28 and Oct4 genes in both hESCs and iPSCs.
RESULTS
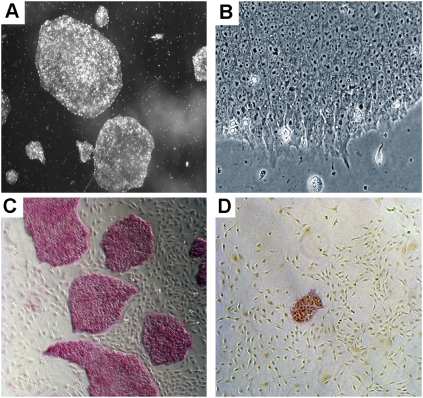
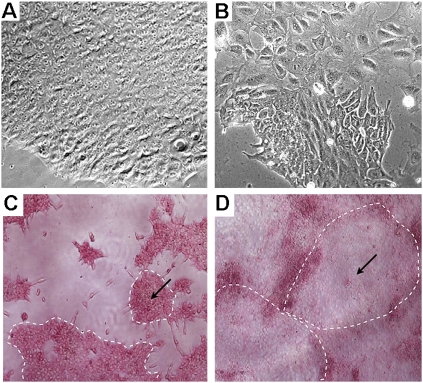
We have previously reported that preincubation of single cell suspension in transfection cocktails can dramatically increase transfection efficiency (Zhang et al. 2007). However, this strategy did not work well with hESCs, mainly due to their low viability in single cell suspension. We therefore initiated efforts to increase cell viability using cloning efficiency as a read-out. First, we used hESCs that were grown in Matrigel-coated plates under feeder-free conditions. Under these conditions, the cells were maintained as single layers that were easily dissociated into single cells (Fig. 1A,B). Second, we used Accutase (a mixture of proteolytic and collagenolytic enzymes) to dissociate the cells. It has been reported that using Accutase instead of trypsin can significantly improve cell viability (Bajpai et al. 2008). Third, we treated cells with a ROCK (Rho-associated kinase) inhibitor before and during transfection. ROCK inhibitors significantly increase the viability of hESCs during passaging, cryopreservation, and thawing, without adversely affecting their self-renewal or differentiation (Claassen et al. 2009; Watanabe et al. 2007). Combining the above three approaches, we were able to drastically increase the cloning efficiency. We obtained 1775 ± 305 colonies from a single cell suspension containing 1 × 105 cells, while only 7 ± 1 colonies were obtained from cells grown on feeders, dissociated by trypsin, and without the treatment with the ROCK inhibitor. Figure 1, C and D, shows representative images of cell colonies resulting from the different methods.
FIGURE 1.
Colony and cell morphology of hESCs grown under feeder-free, serum-free, and components-defined conditions. (A,B) Images of cell colonies taken at 20× and 200× magnifications, respectively. (C,D) Images of cell colonies resulted from cloning assays using our method and the traditional method, respectively. The pink color indicates undifferentiated alkaline phosphatase (AP) positive cells. Images were taken at a 40× magnification.
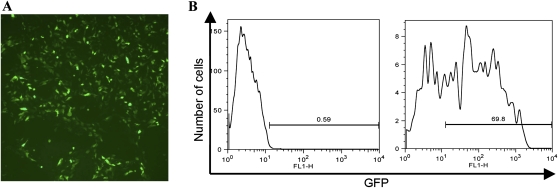
As a proof of principle that transfection in single cell suspension can, in general, augment the transfection efficiency in hESCs, we tested a GFP reporter plasmid (pSUPER-GFP) in H1 cells. Thirty-six hours after the transfection, cells were subjected to immunofluorescence and fluoresence-activated cell sorting (FACS). By immunofluorescence, the majority of growing cells were GFP positive (Fig. 2A). FACS analysis revealed that GFP-positive cells accounted for 69.8% of the population (Fig. 2B). In general, as we recover nearly 90% of viable cells 24 h after the transfection (data not shown), we conclude that this method is associated with low cytotoxicity. It is worthwhile to point out here that although a transfection efficiency of 50%–60% has been reported for hESCs (Hohenstein et al. 2008), this alternative method requires limited cell numbers, nucleoporators, and specialized reagents, which is in contrast to ours, where no special equipment or reagents are involved.
FIGURE 2.
(A) Immunofluorescence of H1 cells 36 h after transfection with a GFP reporter plasmid. (B) Results of FACS analysis of H1 cells mock transfected (left) or transfected with the GFP reporter plasmid (right).
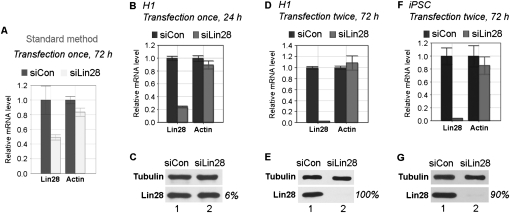
Inspired by the above observations, we next set out to test siRNAs in H1 cells. First, we used a siRNA specific for Lin28 (siLin28) (Qiu et al. 2010). Lin28 is an RNA-binding protein that is highly expressed in undifferentiated hESCs and is among four factors (including Oct4, Sox2, and Nanog) used for making iPSCs (Yu et al. 2007). Using the standard transfection method, we obtained ∼50% knockdown efficiency on Lin28 at the RNA level 72 h following siLin28 transfection, compared with control siRNA (siCon) transfection (Fig. 3A, cf. light bars with dark bars). In contrast, we were able to reduce Lin28 expression by 77% at the RNA level 24 h after transfecting the same siRNA, while as expected, the untargeted β-actin mRNA was not affected (Fig. 3B, cf. gray bars with black bars). At the protein level, there was a 6% decrease at the 24-h time point (Fig. 3C, bottom blot, cf. lane 2 with lane 1). When samples were collected and analyzed 72 h after siRNA transfection, even greater knockdown efficiencies were obtained: 98% at the RNA level (Fig. 3D) and essentially 100% at the protein level (Fig. 3E). In this particular experiment (Fig. 3D,E), a second siRNA transfection (double transfection) was conducted 48 h following the first siRNA transfection, and samples were collected and analyzed 24 h after the second transfection. Although in the case of siLin28 a single transfection is often sufficient to achieve a >90% knockdown (data not shown), double transfection ensures a >95% efficiency.
FIGURE 3.
siRNA knockdown of Lin28 in H1 (A–E) and iPSC (F,G) cells, using standard (A) or new (B–G) method. (A,B,D,F) Results of qRT-PCR. Relative mRNA levels are shown after normalization against β-tubulin mRNA. mRNA levels in cells transfected with siCon were arbitrarily set as 1. Numbers are mean ±SD (n = 3). (C,E,G) Representative Western blot results. Antibodies used are shown on the left. Protein bands on Western blots were determined using Bio-Rad Quantity One software, and calculated after normalization against β-tubulin loading control. Lin28 protein levels in cells transfected with siCon were arbitrarily set as 100%. Percentages at right indicate knockdown efficiencies.
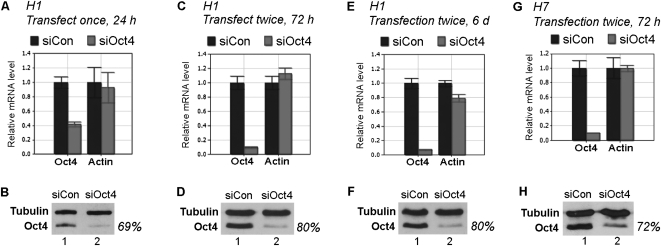
To determine whether this method is also effective for silencing other genes, we tested a siRNA specific for Oct4, a transcription factor that is crucial for ESC self-renewal and pluripotency (Pei 2009). We observed a 59% and 69% decrease in Oct4 expression at the RNA and protein level, respectively, 24 h after a single transfection (Fig. 4A,B). Double transfection increased the efficiency to 90% and 80% at the RNA and protein levels, respectively (Fig. 4C,D), and the knockdown level remained low for at least 6 d following the initial transfection (Fig. 4E,F).
FIGURE 4.
siRNA knockdown of Oct4 in H1 and H7 cells. (A,C,E,G) Results of qRT-PCR. Relative mRNA levels are shown after normalization against β-tubulin mRNA. mRNA levels in cells transfected with siCon were arbitrarily set as 1. Numbers are mean ±SD (n = 3). (B,D,F,H) Representative Western blot results. Antibodies used are shown on the left. Oct4 protein levels in cells transfected with siCon were arbitrarily set as 100%. (A–F) Results from H1 transfection. (G,H) results from H7 transfection.
To ask whether this method also works in other hESC lines, we used the H7 line, a female embryo-derived line (H1 cells were derived from a male embryo). We observed a 90% and 72% knockdown of Oct4 at the RNA and protein level, respectively (Fig. 4G,H).
To determine whether Oct4 knockdown leads to cell differentiation, we performed morphologic analysis. We initially noticed morphological changes within 72 h following transfection (data not shown), and the changes became more prominent at day 6 after the first transfection. As shown in Figure 5B, cells located at the center of the colony exhibited enlarged cell size, increased cytoplasmic area, and a decreased nuclear-to-cytoplasmic ratio, characteristic of differentiated cells. In contrast, no morphological changes were detected in the control siRNA-transfected cells (Fig. 5A). To provide further evidence for cell differentiation, we examined the expression of alkaline phosphatase (AP), which is expressed in undifferentiated hESCs. Six days after the first transfection of siOct4, most cells at the center of the colonies lost staining for AP (Fig. 5D), while those around the edges of the colonies remained AP positive (Fig. 5D). In contrast, cells transfected with control siRNA remained AP positive throughout the colonies (Fig. 5C). Thus, siOct4 transfection leads to both cell morphological change and loss of AP staining, consistent with hESC differentiation.
FIGURE 5.
Effects of Oct4 knockdown on cell differentiation. H1 cells were transfected with siCon (A,C) or siOct4 (B,D). Cells were analyzed by phase contrast microscopy (A,B, 200× magnification) or stained for alkaline phosphatase expression (C,D, 40× magnification). (C,D) Cell colonies were outlined with arrows pointing to the center of the colonies. As differentiated cells had enlarged cell size and increased cytoplasmic area, the colonies thus appeared larger and fused together with each other.
Finally, we tested this method in iPSCs. Human iPSCs first generated in 2007 (Takahashi et al. 2007; Yu et al. 2007) are of great therapeutic potential owning to their patient specificity. The iPSC line we used was derived from reprogramming of human fibroblasts using the Yamanaka factors (Takahashi et al. 2007). We tested silencing of endogenous Lin28 using siLin28. As shown in Figure 3, F and G, 72 h following the first transfection, Lin28 mRNA was reduced by 97%, and Lin28 protein reduced by 90%.
DISCUSSION
We have described a new transfection method that is highly effective at inducing gene silencing of two endogenous pluripotency factors in two hESC lines and one iPSC line. While comparably efficient to the other delivery methods (Zaehres et al. 2005; Hohenstein et al. 2008), our method is simpler and far less expensive. The method should allow for fast and convenient evaluation of silencing of genes involved in self-renewal and survival of hESCs. In general, the maximal knock-down of the target genes is achieved within the first 24–72 h, with morphological changes also evident within this time frame. The knock-down effect is lessened afterward, but the initial effect can be effective enough, in the case of certain genes, to drive hESC differentiation. This is apparently the case for the Oct4 knock-down. The persisting low-level expression of Oct4 for at least 6 d (Fig. 4E,F) likely reflects the fact that the initial knock-down of Oct4 expression was sufficient to drive hESC differentiation. The differentiated hESCs then, in turn, down-regulates Oct4 expression. In addition, we show that two consecutive siRNA transfections at a 48-h interval were well tolerated by the hESCs (Figs. 3D–G, 4C–H). Thus, additional rounds of siRNA transfection may extend the knockdown period of time even further, in case it is necessary for observing effects of other genes involved in hESC self-renewal/differentiation. Our method will be equally useful in understanding mechanisms of cell de-differentiation, as well as the epigenetics behind iPSCs, thereby facilitating the clinical potential of iPSCs.
As evident from the above analysis, transient gene knockdown is very useful in addressing gene function: The transient gene knockdown as the method presented here is quick and less consuming, allows tracking the direct gene partner and effect, and is useful especially for those genes crucial for cell survival; the constitutive gene knockdown needs to put in more effort and is an important tool for addressing long-term effect, signaling networks, and long-term physiological aspects.
So far, we have found a ratio of 9–36 pmol of siRNA (or 0.4 μg of plasmid DNA) per 5 μL of Lipofactamine and a ratio of 1 μL of transfection solution per 15 μL of fresh growth medium to work well in obtaining >90% gene-silencing efficiencies using both siLin28 and siOct4 (or 70% plasmid DNA transfection efficiency). However, these ratios may be adjusted based on siRNA/plasmid DNA, cell type, and culture condition to achieve maximum transfection efficiency, while avoiding off-target effects. We have also found that increasing the ratio between the transfection solution and fresh growth medium (for example, 1 μL of transfection solution per 4 μL of fresh growth medium) tends to increase transfection efficiency, which is sometimes compromised by increased cytotoxicity (data not shown). We have noticed that siRNA knockdown efficiencies vary between different siRNAs, as well as target genes using our method. Although a single round of transfection of siLin28 is often sufficient to inhibit Lin28 expression by >90%, additional rounds of siRNA transfection are necessary for some other siRNAs, including siOct4 (data not shown).
The method described here is equally useful for both plasmid DNA and siRNA transfection into hESCs and iPSCs. In addition, this method can be applied to other hard-to-transfect cells, including primary cells, except that ROCK inhibitors will not be necessary and Accutase will be replaced by trypsin (Zhang et al. 2007). Finally, the protocol described here that enables the dramatic increase in the efficiency of single cell cloning should be of great value for the studies involved in clonal selection in hESCs and iPSCs.
MATERIALS AND METHODS
Cell culture
The hESC lines H1 and H7 (listed in the NIH hESC registry under the names of WA01 and WA07, respectively) were obtained from WiCell. The iPSC line was generated at the Yale Stem Cell Center using Yamanaka factors (Takahashi et al. 2007). The cells were cultured in an undifferentiated state in Matrigel (BD Pharmingen CA)-coated plates under feeder-free and defined component conditions (Yao et al. 2006) at 37°C in hESC culture incubators (5% CO2, 5% O2, and 90%–95% humidity). Cell growth media were composed of Dulbecco's modified Eagle medium (DMEM)/F12 (Invitrogen), supplemented with 1% MEM-nonessential amino acids (Invitrogen), 1 mM L-glutamine, 1% penicillin-streptomycin (P/S), 50 ng/mL basic fibroblast growth factor (Millipore), N2 supplements (1×), and B27 supplements (1×) (Invitrogen). Cells were maintained with daily medium change and passaged weekly by dissociation into small clumps using 1 mg/mL dispase (StemCell Technology). The hESCs and iPSCs used were between passages 30 and 60 and 10 and 15, respectively. Free of mycoplasma contamination, the cells exhibited normal karyotype, expressed common ES markers, and were able to differentiate into the three germ layers (data not shown).
siRNAs, antibodies, and plasmids
siLin28 (ON-TARGETplus SMARTpool), siOct4 (ON-TARGETplus SMARTpool), and siCon were purchased from Dharmacon. Antibodies used in the study were anti-Lin28 (Abcam, ab46020), anti-β-tubulin (Abcam, ab6046), and anti-Oct4 (Chemicon, AB3209). Plasmid pSUPER-GFP (5.4 kb, driven by a CMV promoter, Oligoangine) was a gift from Dr. Katherine Uyhazi.
Cloning assay
On the day of cloning, healthy and exponentially growing cells (60%–70% confluence on day 5 after passaging) were fed 6–8 h prior to transfection, followed by incubation in the presence of ROCK Inhibitor Y-27632 (Calbiochem) (working concentration at 10 μM in ES growth medium) for 1 h. These steps were found to be necessary to preserve cell viability after transfection. Cells were then dissociated into single-cell suspension using Accutase (STEMCELL) and pelleted by centrifugation. Cell pellets containing 1 × 105, 1 × 104, or 1 × 103 cells were each resuspended in 2 mL of ES growth medium and plated onto one well of a Matrigel-coated six-well plate. Colonies were counted 14 d after plating, and cloning efficiencies calculated.
siRNA and plasmid DNA transfection
A cell pellet containing 1 × 106 cells produced by the above-described method was resuspended in 100 μL of siRNA/lipid or plasmid DNA/lipid solution (see below), and the suspension incubated at room temperature for 10–15 min (note, incubation >15 min will decrease cell viability). At the end of incubation, 1.5 mL of prewarmed fresh growth medium (containing ROCK inhibitor at a final concentration of 10 μM) was added and the suspension transferred into a Matrigel-coated well of a six-well plate, followed by incubation in a hESC incubator overnight. The medium was replaced with fresh growth medium the second day of the transfection. In the case of double transfection, the cells were transfected the second time 48 h following the first transfection using the same protocol as described above.
To prepare siRNA/lipid solutions for each well of a six-well plate, 9 or 36 pmol of siRNAs were diluted in 50 μL of OPTI-MEMI (Invitrogen) and incubated at room temperature for 5 min. In a separate tube, 5 μL of Lipofectamine 2000 (Invitrogen) was diluted in 50 μL of OPTI-MEMI and incubation carried out for 5 min at room temperature. The contents of the two tubes were combined by gentle pipetting and incubated at room temperature for 30–50 min. The resulting 100 μL of transfection solution was used to resuspend the cell pellet (1 × 106) produced after Accutase dissociation (see above).
For siRNA transfection using the standard method, cells were seeded in a 6-well plate 5 d prior to transfection. On the day of transfection, 100 μL of siRNA/lipid solution diluted in 1.5 mL of growth medium was added to each well of cells grown as single-layer colonies attached to the plate. ROCK inhibitors were not involved before, during, or after the transfection. RNAs were extracted for analysis 72 h after the transfection.
The protocols were the same for the transfection of plasmid DNA, except that for each well of a six-well plate transfection, 1.6 μg of a reporter plasmid instead of siRNA was used to make the transfection solution.
RNA extraction, qRT-PCR, protein extraction, and Western blot analyses
These were done as previously described (Xu and Huang 2009). The PCR primers specific for the human genes are: β-actin forward: 5′-ATCAAGATCATTGCTCCTCCTGAG; β-actin reverse: 5′-CTGCTTGCTGATCCACATCTG; tubulin forward: 5′-CGTGTTCGGCCAGAGTGGTGC, tubulin reverse: 5′-GGGTGAGGGCATGACGCTGAA; Lin28 forward: 5′-CGGGCATCTGTAAGTGGTTC, Lin28 reverse: 5′-CAGACCCTTGGCTGACTTCT; Oct4 forward: 5′-GTGGAGGAAGCTGACAACAA, Oct4 reverse: 5′- GCCGGTTACAGAACCACACT.
Alkaline phosphatase staining
Cells were stained using Leukocyte Alkaline phosphatase kit (Fisher) according to the manufacturer's protocols.
ACKNOWLEDGMENTS
We thank Diane Krause for critical reading of the manuscript, Zheng Wang for helping with AP staining, and Katerine Uyhazi and Octavian Henegariu for plasmids. This material is based on work supported by the State of Connecticut under the Connecticut Stem Cell Research Grants Program (09SCAYALE14 to Y.H., 06SCD01 and 08SCD04 to H.L.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut, or Connecticut Innovations, Inc.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2350710.
REFERENCES
- Bajpai R, Lesperance J, Kim M, Terskikh AV 2008. Efficient propagation of single cells Accutase-dissociated human embryonic stem cells. Mol Reprod Dev 75: 818–827 [DOI] [PubMed] [Google Scholar]
- Claassen DA, Desler MM, Rizzino A 2009. ROCK inhibition enhances the recovery and growth of cryopreserved human embryonic stem cells and human induced pluripotent stem cells. Mol Reprod Dev 76: 722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenstein KA, Pyle AD, Chern JY, Lock LF, Donovan PJ 2008. Nucleofection mediates high-efficiency stable gene knockdown and transgene expression in human embryonic stem cells. Stem Cells 26: 1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei D 2009. Regulation of pluripotency and reprogramming by transcription factors. J Biol Chem 284: 3365–3369 [DOI] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y 2010. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res 38: 1240–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y 2007. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25: 681–686 [DOI] [PubMed] [Google Scholar]
- Xu B, Huang Y 2009. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res 37: 4256–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Chen S, Clark J, Hao E, Beattie GM, Hayek A, Ding S 2006. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci 103: 6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ 2005. High-efficiency RNA interference in human embryonic stem cells. Stem Cells 23: 299–305 [DOI] [PubMed] [Google Scholar]
- Zhang M, Guller S, Huang Y 2007. Method to enhance transfection efficiency of cell lines and placental fibroblasts. Placenta 28: 779–782 [DOI] [PubMed] [Google Scholar]