Abstract
We describe methods for obtaining a quantitative description of RNA processing at high resolution in budding yeast. As a model gene expression system, we constructed tetON (for induction studies) and tetOFF (for repression, derepression, and RNA degradation studies) yeast strains with a series of reporter genes integrated in the genome under the control of a tetO7 promoter. Reverse transcription and quantitative real-time-PCR (RT-qPCR) methods were adapted to allow the determination of mRNA abundance as the average number of copies per cell in a population. Fluorescence in situ hybridization (FISH) measurements of transcript numbers in individual cells validated the RT-qPCR approach for the average copy-number determination despite the broad distribution of transcript levels within a population of cells. In addition, RT-qPCR was used to distinguish the products of the different steps in splicing of the reporter transcripts, and methods were developed to map and quantify 3′-end cleavage and polyadenylation. This system permits pre-mRNA production, splicing, 3′-end maturation and degradation to be quantitatively monitored with unprecedented kinetic detail, suitable for mathematical modeling. Using this approach, we demonstrate that reporter transcripts are spliced prior to their 3′-end cleavage and polyadenylation, that is, cotranscriptionally.
Keywords: single-molecule FISH, RNA quantification, splicing, transcription, yeast
INTRODUCTION
In eukaryotes, most nuclear pre-mRNAs undergo processing at their 5′ ends (capping), 3′ ends (cleavage and polyadenylation), and internally (splicing to remove noncoding introns), and there is good evidence to suggest that at least some of these processes are functionally coupled to transcription (for review, see Perales and Bentley 2009). To improve our understanding of these complex events and to facilitate mathematical modeling, methods are required for the quantitative measurement of RNA processing events with good time resolution during induction or repression of transcription.
The available techniques for quantifying transcripts have inherent advantages and disadvantages. Northern blotting can provide information on the size as well as the expression level of a transcript, but measures the relative abundance only of individual mRNAs and has a limited dynamic range. Microarray analyses can compare transcripts genome-wide, but have a poor dynamic range and are impractical for high-resolution kinetic studies. Measurements of mRNA transcripts as copies per cell have relied heavily on hybridization-based complexity assays (Iyer and Struhl 1996), serial analysis of gene expression (SAGE) (Velculescu et al. 1997), kRT-PCR (reverse transcriptase initiated PCR-kinetic PCR) (Holland 2002), and genomic run-on (Garcia-Martinez et al. 2004). However, using competitive PCR between genomic DNA and cDNA, Miura et al. (2008) concluded that the overall abundance of the yeast mRNA population is at least twice as high as previously estimated. Another approach is to visualize and count individual transcripts in single cells using fluorescence in situ hybridization (FISH) (Femino et al. 1998; Raj et al. 2006; Zenklusen et al. 2008). This approach also found that expression levels for particular yeast genes were higher than previously estimated and can vary substantially among cells in a population (Zenklusen et al. 2008).
Nutritionally modulated promoters such as MET3 or GAL1 (for review, see Maya et al. 2008) are not ideal for regulated gene expression studies as the changes in growth medium are likely to result in pleiotropic effects on cell metabolism. In particular, both a galactose to glucose shift and the presence or absence of methionine were reported to impact on RNA turnover in yeast (Dichtl et al. 1997; Bousquet-Antonelli et al. 2000). An alternative is to use promoters that respond to nonmetabolizable drugs. For example, tetracycline (tet) and its analog doxycyline (dox) have no significant effects on yeast cell growth, morphology or global transcription levels (Wishart et al. 2005).
As model gene expression systems we constructed tetON (for induction studies) and tetOFF (for repression, derepression, and RNA degradation studies) yeast strains, each of which expresses tetracycline-responsive repressor and trans-activator proteins (dual activator/repressor regulation [Belli et al. 1998b]). Using reporter genes in these strains we developed an RT-qPCR approach to determine mRNA abundance as average numbers of copies per cell in a population, and validated this approach by comparison with single-molecule FISH that measures transcript copy numbers in individual cells. We describe PCR-based methods to monitor RNA production, splicing, 3′-end maturation and degradation in a quantitative, tightly controlled manner and with rapid response times, as is required for mathematical modeling of these events. This combination of approaches allows a much more detailed kinetic analysis of the expression of an individual gene than is generally achieved by other methods. This is illustrated by high-resolution analysis of both steps of pre-mRNA splicing and 3′-end maturation of a reporter transcript, which permits cotranscriptional splicing to be demonstrated directly. Although developed and optimized for the tet-regulated expression system in yeast, these methods should have wide utility for gene expression studies with other regulated promoters and in other organisms.
RESULTS AND DISCUSSION
Construction of strains and tetracycline/doxycycline-regulated reporter genes
Tetracycline repressor (tetR) and trans-activator (tTA) fusion proteins bind at promoters containing tetO DNA sequences only in the absence of tetracycline or doxycycline, and are displaced by drug addition. Conversely, the modified tetR′ and tTA′ proteins bind tetO DNA only in the presence of the drug (Gossen et al. 1995; Gari et al. 1997; Belli et al. 1998a,b). For tight control and a better dynamic range of regulated transcription, dual activator/repressor regulation was used, in which each yeast strain coproduces both tetR and tTA′ for induction (tetON) or tetR′ and tTA for repression (tetOFF) (Belli et al. 1998b; for details, see Supplemental Material 1). In the original dual expression system of Belli et al. (1998b), the regulatable tetO promoter and the tTA or tTA′ transactivator gene were present together on a plasmid and the tetR/R′-repressor gene was integrated in the yeast genome. To place reporter genes or endogenous yeast genes under tet control, it would be preferable to integrate the tet0 promoter/tTA expression cassette in the genome, in order to maintain the chromatin environment and avoid possible plasmid stability/segregation problems.
To introduce the tetracycline dual activator/repressor system into the yeast genome, the tTA or tTA′ hybrid trans-activator gene was integrated at the LYS2 locus and tetR′ or tetR hybrid repressor (Belli et al. 1998b) was integrated at either the LEU2 or URA3 locus in three different yeast strains (for details, see Materials and Methods). For initial tests of the system the tet07-CYC1 hybrid promoter (see Supplemental Material 2 for the sequence) was inserted in front of the nonessential STE3 gene (Supplemental Fig. S1). In the case of the tetOFF system, strains YMK118 (derived from CEN-PK2-1C) and YMK120 (W303) gave better levels of expression than YMK119 (BY4741), and YMK120 and its ura3 derivative, YIK120, were chosen for further study (for genotypes, see Supplemental Material 1).
In contrast, none of the corresponding tetON strains showed induction even after 15 h in the presence of dox (data not shown). After testing many constructs, finally, a functional tetON strain, YIK91, was produced with tTA′ on a centromeric plasmid expressed under the control of the strong S. cerevisiae ADH1 promoter (PADH1tTA′), and with the tetR-SSN6 repressor gene expressed from the Schizosaccharomyces pombe adh1 promoter (Padh1tetR-SSN6 from pCM247) (Belli et al. 1998b) integrated at the LEU2 locus of W303. The main differences between this and the Belli et al. tetON system are that the tetO7-regulated gene is integrated into the genome rather than being on a plasmid, and tTA′, although still plasmid based, is expressed from the stronger ADH1 promoter. In comparison, expressing tTA′ from the CMV promoter (Belli et al. 1998b) produced little or no expression in our strain, and the TEF2 promoter (Mumberg et al. 1995) supported only low level expression (Supplemental Fig. S2).
As model genes to analyze transcription, splicing, degradation, and 3′-end formation of pre-mRNA, a series of “RiboSys” reporter genes was constructed and each was integrated in the genomic his3 locus of the tetON and tetOFF strains, under control of the tetO7/CYC1-UAS promoter (Supplemental Material 2). The reporter genes are based on a previously described (Hilleren and Parker 2003) series of hybrid ACT1/PGK1 sequences in which the ACT1 intron was modified by inserting two copies of the λ boxB sequence (57 base pairs [bp] each), enabling it to be readily distinguished by RT-qPCR from the endogenous ACT1 intron without affecting splicing (data not shown). Variants of the “wild-type” reporter, Ribo1, contain a point mutation at the 5′ splice site (5′SS) or 3′ splice site (3′SS) or lack the intron (IL) (Fig. 1; Hilleren and Parker 2003).
FIGURE 1.
Diagram showing important features of the RiboSys reporter genes and the positions of hybridization probes and primers used in reverse transcription (RT) and RT-qPCR reactions. The reporter genes are modified from those developed by Hilleren and Parker (2003). Briefly, exon 1 contains the ACT1 5′ UTR and 30 codons from PGK1 and ACT1 sequences, and is followed by the ACT1 intron, then exon 2, which contains ∼100 bp of PGK1 fused in frame with three copies of the haemaglutinin epitope (3HA), followed by six MS2 coat protein binding sites (6MS2) at the start of the PGK1 3′ UTR. To further discriminate between the reporter transcripts and endogenous ACT1 and PGK1 transcripts in RT-qPCR assays we inserted two copies of the lambda N (box B; 57 bp) sequence 51 bp downstream from the ACT1 5′ splice site. Transcription of the RiboSys reporters is under control of the tetO7/CYC1-UAS promoter and the tetracycline (or doxycycline) regulatable dual activator/repressor system (Belli et al. 1998b). The intron is represented by a thin line with letters indicating splice signals (uppercase) and positions of splice site mutations (lowercase). Positions of flourophore probes used in FISH experiments are indicated below the reporter: 11 probes for mRNA and eight probes for intron (pre-mRNA) detection. Primers used for RT-qPCR assays are indicated by arrows and a northern probe by a black line. Primer pairs, p2_F and p2_R, amplify the 5′SS region of unspliced pre-mRNA. Exonic primer pairs, m2_F and m2_R, are used to quantify the spliced mRNA under conditions that do not produce a product from unspliced pre-mRNA. E2_F and E1_R measure all forms of this transcript regardless of their progress through the splicing reactions. The intron-containing products of the first and second steps of splicing are branched/lariat structures, containing a 2′-5′ phosphodiester bond that blocks the progress of reverse transcriptase from the 3′ end of the intron. The lariat intron-exon2 species can be measured in two ways. First, the product produced by primers 3′_F and 3′_R that flank the 3′SS, represents the sum of the unspliced pre-mRNA and the lariat intron-exon2 species. The difference between this and the RT-qPCR product of unspliced transcripts that crosses the 5′SS is due to the lariat intron-exon2 species. Second, oligonucleotide L1_R with the 5′ end complementary to the 5′ end of the intron and the 3′ end complementary to the intron sequence immediately 5′ of the branch site, but with a mismatch (A instead of T) opposite the branch site A (Vogel et al. 1997), anneals at the branch site of lariats and can be used to prime the RT reaction. and, with L1-F, primes RT-qPCR amplification of the circular part of the lariat. This measures the sum of the lariat-exon product of the first splicing reaction and the excised intron lariat product of the second splicing reaction. In normal conditions, the level of lariat species is very low, as the lariat intron-exon2 product of the first splicing reaction is rapidly processed in the second step and the excised intron is rapidly debranched and degraded. However, if the second splicing reaction is slow or blocked or if there is a defect in release and degradation of the excised intron, these species can be quantified. The sequences of all oligos are listed in Supplemental Table S3.
Quantification of transcripts averaged over a population of cells
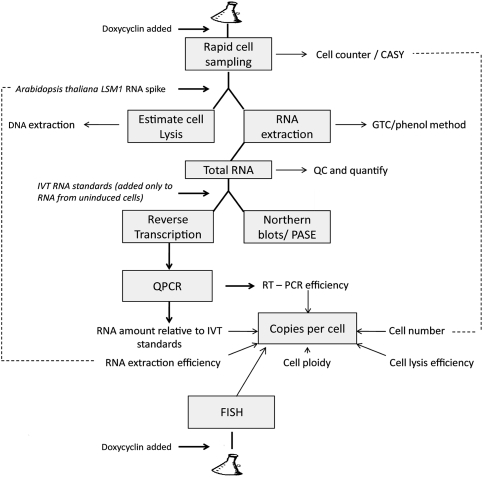
Previous applications of RT-qPCR to quantify transcripts have depended on the use of reference genes for normalization; however, this is unreliable, as few genes have constant levels of expression under different growth conditions (Teste et al. 2009). In order to obtain an estimate of the absolute number of copies of a transcript per cell, several stages in the isolation and assay of the RNA need to be quantified, including cell lysis, extraction of RNA from lysed cells, reverse transcription, and qPCR efficiency (summarized in Fig. 2; for full details, see Supplemental Material 3). Of these, the first is possibly the most difficult to control. An estimate of the efficiency of cell lysis can be obtained by determining the amount of genomic DNA recovered compared with the expected amount. The difference between the observed and the theoretical yield of DNA is largely due to lack of cell lysis and so an estimate of the efficiency of cell lysis can be derived.
FIGURE 2.
Scheme of work for estimating RNA copy number by determining the efficiency of cell lysis, RNA extraction, RT-qPCR, and FISH imaging. Briefly, the cell number is determined and the efficiency of cell lysis is estimated based on the amount of DNA recovered (total DNA is assayed spectroscopically and individual genes by RT-qPCR) compared with the expected amount of DNA, assuming that in an exponentially growing population of haploid cells the average cell contains 1.4 genomes worth of nuclear DNA. The S. cerevisiae genome is ∼0.012 pg (Fungal Genome Size Database www.zbi.ee/fungal-genomesize). Total RNA purified from the yeast lysates is measured optically. The efficiency of RNA recovery is estimated by measuring the recovery of a known amount of nonyeast RNA (in our case, Arabidopsis thaliana LSM1) added to the lysate. The reverse transcription and qPCR stages are standardized by comparison with known amounts of in vitro transcribed RNAs with the same sequence, mixed with total RNA purified from cells that do not express the reporter genes. The results from this procedure are compared with numbers obtained by visual inspection of individual transcripts detected by FISH in large numbers of single cells. The details of the various procedures are given in the Supplemental Material.
An estimate of the efficiency of RNA recovery from the cell lysate was obtained by adding a known amount of nonyeast in vitro transcribed RNA, and determining by RT-qPCR how much was recovered at the end of the RNA purification. This could only provide an estimate as it was not certain that the exogenous RNA (with no proteins bound to it) will behave in the same manner as endogenous mRNA from newly lysed yeast cells. The reverse transcription and qPCR reactions were standardized by comparison with known amounts of in vitro transcribed RNAs with the same sequences, after mixing with total RNA extracted from cells that lack the reporter gene.
The positions of hybridization primers used in the reverse transcription (RT) and qPCR reactions are shown in Figure 1, together with an explanation of how the products of the different stages of the splicing reaction were distinguished. This included an assay for branched, lariat RNA species that accumulate if the second splicing reaction is slow or blocked, or if there is a defect in release and degradation of the excised intron. It was shown previously by microarray analysis that doxycycline (at 10 times the concentration used here) has no significant effect on global transcription levels in yeast (Wishart et al. 2005). Therefore, assaying an endogenous transcript that is unaffected by dox provides a measure of the reproducibility of this technique during the time course of an experiment (Supplemental Fig. S3).
Image-based measurement of mRNAs in individual yeast cells
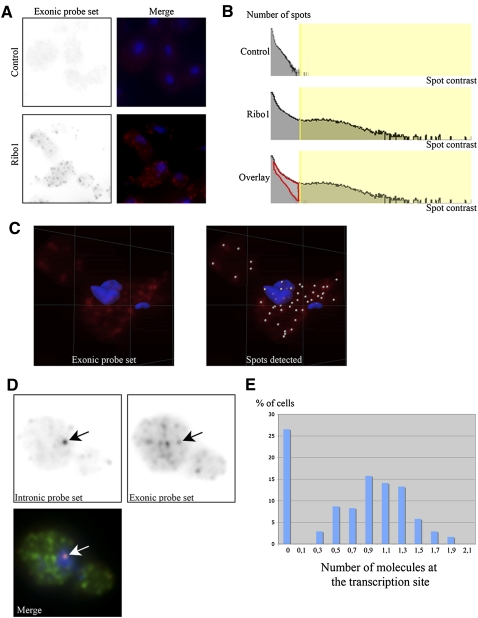
In order to determine transcript abundance in single cells a FISH approach was employed, using Cy3-labeled deoxyoligonucleotide probes complementary to the reporter transcripts. Previous studies have shown that single RNA molecules can be efficiently detected as isolated spots in microscopic images, using five to 10 oligo probes (Femino et al. 1998). Two sets of probes were used. The first set detects the intron, and consists of a pool of eight probes that cover the entire length of the Ribo1 intron. The second detects the exons, and contains a pool of six probes. One probe hybridizes six times against the MS2 tag, and a maximum of 11 oligonucleotides from this pool can thus hybridize at the same time on the mRNA or pre-mRNA. The exonic pool of probes labeled small and bright spots in the nucleus and the cytoplasm, which correspond to isolated, individual mRNA molecules (Femino et al. 1998). A semiautomated procedure was then developed to detect and automatically count single molecules of the reporter transcripts (seen as bright individual spots) in 3D stacks of cells expressing the various reporters and nonexpressing control cells (Fig. 3). Briefly, a Gaussian filter of 0.300 μm was applied to the images to remove variation in intensities smaller than the size of single molecules. All local maxima were then determined and a spot was defined as a sphere of the desired diameter (0.3 μm in this case), centered on each maximum. To remove the background, the spots were then thresholded on the basis of their local contrast (the standard deviation of the signal in the volume of the spot). For this, we used a control strain that did not contain the reporter gene, and selected a minimal local contrast that left less than one spot per cell in the control strain. Eye examination of the results indicated that this procedure automatically detected single RNA molecules with a reasonable efficiency. To quantify this better, we plotted histograms of the number of spots detected as a function of local spot contrast (see Fig. 3B). With the entire exonic probe set, the spots detected in Ribo1 expressing cells were of much higher local contrast than in nonexpressing cells. Using a contrast threshold that excluded most spots in the nonexpressing cells (Fig. 3B, yellow vertical bar) (the threshold is chosen such that one molecule per cell is detected in the control), the specific spots could be sorted from the background. In addition, by overlaying normalized histograms for the control and Ribo1 expressing strains, it could be seen that the threshold excluded a small fraction of spots from the Ribo1 sample (Fig. 3B, outlined in red). By measuring the total number of specific spots (the ones above the threshold and those outlined in red), we could estimate that <20% of the specific spots were lost by the thresholding procedure, yielding a detection efficiency higher than 80%. It should be noted that this approach is limited to RNAs with a copy number above 1, which is similar to the threshold chosen here, and that above 60 per cell many fluorescent spots merge, making identification of individual RNA molecules less reliable.
FIGURE 3.
Image based measurement of Ribo1 RNAs in individual yeast cells. (A) Detection of single molecules of the Ribo1 reporter RNA in TetOFF strains. Control strain lacking the reporter gene (top), or expressing Ribo1 (bottom) were hybridized in situ with the exonic probe set. Right panels display maximal image projections of the RNA signal (red) overlayed with the nuclei (blue). Each field is a projection of a 3D stack (6 × 6 × 6 μm). (B) Efficiency of RNA detection. Histograms of the number of spots versus spot contrast across an entire 3D stack (63 × 63 × 6 μm) are shown for control and Ribo1 expressing cells. The area shaded in yellow corresponds to the spots included in the analysis after thresholding. Bottom panel: overlay of the two histograms revealing the amount of Ribo1 mRNA molecules lost by the thresholding procedure (red area; <20% of the total number of spots identified) (C) Maximal image projection (3.5 × 3.5 × 6 μm) of a cell with the spots identified indicated with small spheres on the right panel. (D) Single plane of a Ribo1 expressing cell with both the exonic (Cy5, green) and the intronic signals (Cy3, red). Blue: Dapi. Each field is 3.6 × 3.6 μm. Arrows indicate the position of the intronic focus that identifies the putative transcription site. (E) Histogram plotting the number of molecules of Ribo1 RNA, identified with the exonic probe set, at the intronic foci (putative transcription site). Cells with zero molecules have no signal for the intronic pool of probe.
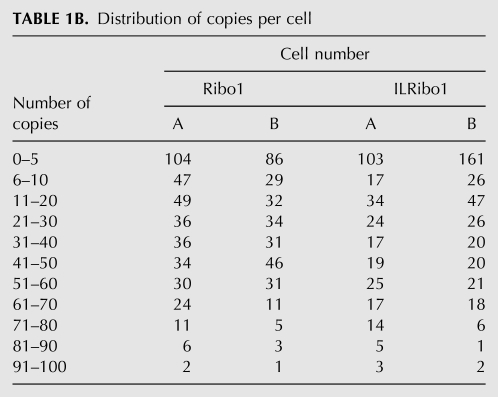
In order to compare the two methods of estimating copy numbers for Ribo1 and ILRibo1, duplicate cultures of the tetOFF strains were propagated in one lab, where the RT-qPCR method was used, and samples of these cultures were fixed with paraformaldehyde and sent to be assayed by FISH in another lab. The numbers obtained by RT-qPCR and by FISH are in good agreement (Table 1A), with the difference being similar to the variation seen when comparing duplicate cultures using a single method. Of course the RT-qPCR approach measures the copy number averaged over the entire population of cells, and the FISH approach shows that the copy number varies greatly from cell to cell, with the distribution being from 0 to 100 copies per cell for Ribo1 and ILRibo1 (Table 1B). This is likely due to the promoter cycling between On and Off states, as previously proposed and modeled (Blake et al. 2006; Zenklusen et al. 2008; Aitken et al. 2010). It therefore seems likely that much of the variability in copy-number measurements between duplicate cultures is due to biological variation. Furthermore, cultures grown in rich (YPD) medium showed a much greater variation between biological replicates than cultures grown in synthetic medium (data not shown). This suggests that the frequency of promoter cycling between On and Off states may be sensitive to the growth rate or metabolic state of the cells, which may be induced by subtle differences between batches of rich media.
TABLE 1A.
Copy-number measurement by RT-qPCR and FISH using the same cultures
TABLE 1B.
Distribution of copies per cell
Consistently, FISH with the intronic probes showed that 73% of these haploid cells had a single focus of signal, which was always in the nucleus, and which colocalized with a spot labeled by the exonic probes and was thus most likely the transcription site (Fig. 3D). To quantify the number of RNA molecules present at the intronic foci, we first calculated the average intensity of the cytoplasmic spots labeled by the exonic probe set, which corresponded to single, full-length mRNA molecules. We then divided the intensity of the spot that colocalized with the intronic foci by this value. This showed that the signal corresponded to one molecule of fully transcribed pre-mRNA (Fig. 3E), consistent with RT-qPCR quantification of 1.28 ± 0.1 copies of pre-mRNA per cell in the tetOFF strain when derepressed, and indicating that only a few polymerases were loaded on the gene at any time. Moreover, the lack of intronic signal elsewhere in the cells was compatible with splicing taking place at the transcription site, cotranscriptionally.
To determine whether unspliceable pre-mRNAs are retained at a particular area in the cell, we analyzed the localization of the 5′SSRibo1 mutant by in situ hybridization. Using the intronic pool of probes, we found that the mutant unspliced pre-mRNAs accumulated at the nuclear periphery (Supplemental Fig. S4), as previously described (Galy et al. 2004).
Repression and RNA degradation kinetics
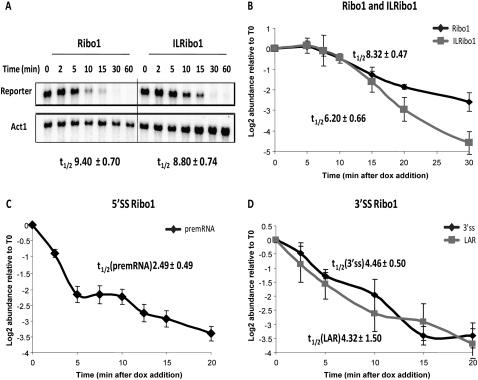
A particular advantage of tetOFF strains is the ability to measure RNA degradation rates following the turn off of expression, by simply adding dox without the need to change the growth medium. By northern analysis, the Ribo1 and ILRibo1 mRNAs had similar apparent half-lives of 9.4 and 8.8 min, respectively (Fig. 4A), compared to 8.3 and 6.2 min when assayed by RT-qPCR (Fig. 4B) (measured as the slope from the maximum at 5 min). The RT-qPCR assay seems likely to be more reliable, as more replicate assays were performed and the assay was standardized against known amounts of synthetic RNA standards. The 5′SS and 3′SS mutant reporter transcripts are defective in the first and second step of splicing, respectively, and have much shorter apparent half-lives (Fig. 4C,D). Interestingly, degradation begins immediately after dox addition, whereas there is a delay of several minutes before degradation of Ribo1 and ILRibo1 mRNAs is apparent. This most likely reflects very rapid targeting of the aberrant, unspliced transcripts for degradation by the RNA surveillance systems (Bousquet-Antonelli et al. 2000; Hilleren and Parker 2003; Houseley and Tollervey 2009).
FIGURE 4.
Kinetics of repression and turnover of Ribo1 reporter transcripts using the RiboSys tetOFF strains. The tetOFF strains with intron-containing Ribo1 or intonless ILRibo1 (A,B), mutant 5′SSRibo1 (C), or mutant 3′SSRibo1 (D) reporters were grown in SD minimal medium with doxycycline (4 μg/ mL) added at time 0 to initiate repression of transcription. (A) Northern blot analysis of Ribo1 and ILRibo1 reporters showing relative mRNA levels following repression. Apparent half-lives of the reporter transcripts are shown below the blot. (B–D) Repression of RiboSys reporters showing relative abundance (log2) of the Ribo1 and ILRibo1 mRNAs (B), 5′SSRibo1 pre-mRNA (C), and 3′SSRibo1 lariat-exon2 intermediate (D) assayed by RT-qPCR of the 3′SS region (3′SS) or using lariat-specific RT-qPCR (LAR). The apparent half-lives, calculated from the linear parts of the curves, are displayed.
Knowledge of the number of molecules per cell and RNA half-life permit calculation of the transcript synthesis rate, at least for unspliced RNAs (Perez-Ortin et al. 2007). For ILRibo1 this gives values of 1.9–2.5 and 4.9–6.3 (molecules/cell/min), respectively, at steady state in the tetOFF and tetON strains. Assuming that initiation of transcription is the limiting step in the rate of synthesis, this indicates an initiation event every 27 and 11 sec, respectively (Iyer and Struhl 1996), reflecting the generally higher copy numbers in the tetON strains.
Derepression and induction kinetics
The validated RT-qPCR method for copy-number measurement was then used to follow the kinetics of derepression and induction in the tetOFF and tetON strains, respectively. Note that the RT-qPCR assay of Ribo1 exon2 in the tetOFF strain (Fig. 5A) (exon) reaches slightly higher levels than that of Ribo1 mRNA, and approximately equates to the amount of mRNA plus pre-mRNA and lariat-exon2 (i.e., all the RNA species that contain exon2), supporting the specificity and reliability of the RT-qPCR assays of the different RNA species. In the tetOFF strain under repressed conditions (Fig. 5A) (0 min) less than one copy per cell of Ribo1 mRNA could be detected. Higher levels of reporter transcripts were reproducibly detected in the tetON strains (Fig. 5D–F), in both uninduced (0 min) and induced conditions, and with more rapid accumulation than in the tetOFF strains (Fig. 5A–C; note the different scales). The unspliced 5′SSRibo1 transcripts (Fig. 5B,E) reached lower maximum copy numbers than for Ribo1 mRNA, which may be explained by the faster turnover of this species (Fig. 4C). With 3′SSRibo1 expression (Fig. 5C,F), the lariat-exon 2 product of step 1 splicing was readily measured, but a pronounced transient accumulation of pre-mRNA suggests that there is a delay before the first step of splicing occurs, apparently more so than with Ribo1.
FIGURE 5.
Kinetics of transcription and splicing of RiboSys reporter transcripts in tetOFF and tetON strains. A time course is shown of the levels of pre-mRNA (pre), lariat intron-exon2 splicing intermediate (lar-exon2; measured by 3′SS assay and subtracting the amount of unspliced pre-mRNA) (see Fig.1), exon2 (exon) and spliced mRNA in copies per cell. (A–C) tetOFF cultures were grown in SD medium to midlog phase (OD600 0.5) with transcription repressed by the presence of dox (4 μg/ mL). After 90 min of transcriptional repression, cells were harvested by centrifugation and resuspended into medium without dox (time 0). (D–F) tetON cultures (YIK91 with various Ribo1 reporters integrated at the his3 locus) were grown in SD minimal medium (-trp) with doxycline (4 μg/ mL) present to induce transcription. (A,D) Ribo1; (B,E) 5′SSRibo1; and (C,F) 3′SSRibo1. In each panel the data represent three experiments, each assayed in triplicate, and error bars indicate standard deviation.
With both tetON and tetOFF strains the lag period between dox addition or removal, respectively, and first detection of transcripts varied slightly between cultures grown at different times. The reason for this is not known but, combined with the variation in the copy number noted above, may indicate sensitivity of the transcription machinery to subtle differences in growth medium, other environmental conditions or the growth phase of the cells.
3′-End formation
In yeast, the 3′ ends of all mRNAs are produced by cleavage and polyadenylation. The efficient formation of the mRNA 3′ ends is important for mRNA stability as well as for capping, splicing, transcription, and translation (Minvielle-Sebastian and Keller 1999). Polyadenylation of the Ribo1 transcripts is driven by 3′ UTR sequences derived from the PGK1 gene. Initially, using total RNA obtained from tetOFF strains grown under nonrepressive conditions, we determined the sites of poly(A) addition, using a ligation mediated cDNA amplification strategy to determine sequences encompassing the poly(A) sites (Fig. 6A). Analysis of 51 independent clones revealed a major poly(A) site located at positions 1407–1409 (relative to the PGK1 start codon) that was used in 85% of cases and a minor site at positions 1396–1400 was used in 14% of the sequenced clones. Next, we used the same assay to follow the poly(A) status of reporter transcripts during derepression in tetOFF strains (Fig. 6B). The poly(A) length ranged from approximately 20 to 70 adenosines (A's) at steady state (30 min and later). The abundance of transcripts carrying longer or shorter tails, respectively, dropped sharply beyond those values. Linear poly(A) shortening during the early phase of reporter derepression was used to deduce the apparent rate of deadenylation, which was found to be 0.8 adenosines min−1 for the Ribo1 reporter and 1.5 A's min−1 for ILRibo1 (Fig. 6C). These values are consistent with the deadenylation rate of 2–3 A's min−1 that was reported for the PGK1 3′ UTR in the context of the related PGK1pG construct (Tucker et al. 2001). However, the mRNA half-lives associated with the Ribo1 reporters are short (<10 min) (see Fig. 4) compared to PGK1 mRNA that is very stable (t1/2 45 min) (Parker and Jacobson 1990). The lack of correlation between deadenylation rates and mRNA half-lives indicates that deadenylation-independent mechanisms may determine the turnover of Ribo1 reporter transcripts.
FIGURE 6.
3′-End formation and deadenylation (A) Flow diagram of the PCR-based method that was applied to analyze transcript 3′-end formation. In a first step a 5′ phosphorylated DNA linker oligonucleotide is ligated by T4 RNA ligase to available 3′-OH groups. This is followed by first-strand cDNA synthesis using the linker-specific RevB oligonucleotide and subsequent PCR analysis. The obtained PCR product is further amplified by nested PCR using RevA and PGK1-B oligonucleotides. The obtained PCR product can be transferred into a plasmid and analyzed by DNA sequencing. Alternatively, the nested PCR can be performed with radioactively labeled oligonucleotides and PCR products can be separated on denaturing polyacrylamide gels. (B) Time course of adenylation/deadenylation following derepression of ILRibo1 and Ribo1 in the tetOFF strain, analyzed as described in A. Radioactive PCR products were resolved on 6% (w/v) polyacrylamide/ 8.3 M urea gels. The length of the poly(A) tract associated with the PCR products is indicated by numbers. To control for specificity of PCR amplification a minus reverse transcriptase (−RT) control was included. (C) Graph depicting the length of the shortest poly(A) tails observed at individual time points during reporter derepression. Data are the mean of three (− Intron) and four (+ Intron) experiments and error bars represent S.D. Deadenylation rates were derived from initial linear phases of poly(A) shortening.
High-resolution kinetic analyses reveal splicing prior to 3′-end cleavage
There has been much debate about the extent to which splicing takes place co- or post-transcriptionally in yeast (Gornemann et al. 2005; Lacadie and Rosbash 2005; Tardiff et al. 2006). However, the assays used to date, including chromatin immunoprecipitation to measure cotranscriptional recruitment of splicing factors or of RNA-binding proteins that distinguish spliced from unspliced transcripts, are indirect and therefore may be inaccurate. The modified RT-qPCR approach described here permits more quantitative measurements of transcript accumulation and both steps of splicing. As RNA species are measured in copies per cell, different RNA species can be compared with each other and between different samples and cell cultures. Furthermore, identification of the positions of 3′-end cleavage/polyadenylation permits RT-qPCR analysis of the kinetics of 3′-end formation and how this relates to splicing. As shown in Figure 7, by taking samples at short time intervals (30 sec), it is possible to detect a peak of full-length nascent (pre-mRNA) transcripts at 9 min, prior to their 3′-end cleavage, which takes place within 30 sec. The lariat-exon2 product of the first step of splicing can also be detected at 9 min, prior to 3′-end processing. Furthermore, at 9.5 min spliced mRNA that is not yet 3′-end processed can be detected, prior to the appearance of polyadenylated mRNA at 10 min. These data clearly demonstrate that splicing takes place on a large proportion of transcripts prior to 3′-end cleavage, as was suggested by the FISH analysis; and we conclude that Ribo1 transcripts are, to a large extent, spliced cotranscriptionally.
FIGURE 7.
Kinetic analysis of splicing and 3′-end formation of Ribo1 transcripts demonstrates cotranscriptional splicing. RNA, sampled at 30-sec intervals during a time course of induction of Ribo1, was reverse transcribed with either oligo (dT) to copy cleaved and polyadenylated transcripts, or with a primer downstream of the mapped cleavage/poly(A) sites to copy uncleaved transcripts (there is a short delay between transcription through the poly(A) site and 3′-end cleavage). qPCR was performed to measure unspliced and spliced transcripts. (A) Schematic of the assay, showing the approximate positions of primers used for qPCR of unspliced pre-mRNA (across 5′SS), lariat intron-exon (across 3′SS then subtract the value for pre-mRNA) and spliced mRNA (across exon junction) or used to prime cDNA synthesis. pA indicates the 3′-end cleavage/polyadnylation site. Results are shown for: pre-mRNA (B), lariat intron-exon2 (C), and mRNA (D). As a control, it was demonstrated that no cDNA was produced when oligo (dT)-selected polyadenylated RNA was reverse transcribed using the downstream primer (data not shown).
Furthermore, the half-life of each step of splicing can be approximately estimated as 30 sec, equating to 60 sec for the conversion of pre-mRNA to mRNA. This is similar to a measurement based on indirect ChIP analyses in yeast (Tardiff et al. 2006) and significantly faster than the 5–10 min measured for splicing an intron in human cells, irrespective of intron length (Singh and Padgett 2009). Tardiff et al. (2006) determined that splicing begins when transcription is >600 bp beyond the 3′SS and that maximal cotranscriptional splicing requires ∼1 kilobase (kb) past the 3′SS. The Ribo1 major and most distal 3′ cleavage site was mapped at 735–737 nucleotides (nt) past the 3′SS. It can also be seen that cleavage and polyadenylation take place within 30 sec of transcription through this region. This compares with an estimated 55 sec for 3′-end cleavage and polyadenylation of HIV-1 (Human Immunodeficiency Virus type 1) mRNA, measured by FRAP (fluorescence recovery after photobleaching) (Boireau et al. 2007). This level of quantification and kinetic resolution will permit mathematical modeling to determine more accurate rates for the two steps of splicing and estimates of how much splicing occurs pre- and post-transcriptionally.
CONCLUSIONS
The regulated genes studied here are chromosomally located, rather than being on plasmids as in previous tet-regulated constructs (Belli et al. 1998b). Yen et al. (2003) developed a similar tetOFF strain with tet02 or tet07 promoters integrated in the genome but, to our knowledge, this is the first report of such a tetON strain. The tet-regulated expression of other genes could be studied by integrating other ORFs with the tet07 promoter at the HIS3 locus, using the pMK121 or pMK123 plasmids that we describe. The tet07 promoter could also be integrated upstream of any gene in the genome, although the efficiency of tet-repression is variable with different yeast genes (Mnaimneh et al. 2004; Wishart et al. 2006), and in our experience this is also true for tet-regulated induction.
The RT-qPCR approach for estimating RNA copy number could also be used to measure the regulated expression of endogenous genes from their native promoters and, in principle, it should be adaptable to other organisms, including the analysis of tissue-specific gene expression. Using this approach to estimate the copy number of constitutively produced transcripts requires careful consideration of the synthetic RNA used to standardize the RT-qPCR assays. As the efficiency of RT-qPCR differs for purified transcripts compared to transcripts assayed in cell extracts, ideally the in vitro transcribed RNAs should be distinguishable from the endogenous transcripts without affecting the properties of the qPCR reaction, although it may be possible to use a synthetic RNA that is identical to the endogenous transcript and measure the increase in abundance.
The absolute measurement of RNA levels facilitates modeling studies of RNA metabolism, for example, allowing comparisons between different growth conditions, strains, or cell types. Previous efforts to model RNA processing pathways in yeast have depended on northern hybridization, other gel-based analyses (Cao and Parker 2001, 2003) or imaging (Zenklusen et al. 2008). The use of RT-qPCR is more sensitive and offers a greater dynamic range than these approaches, and the ease of scale permits more replicates and therefore a better measure of accuracy and reproducibility. As illustrated here, the more quantitative RNA measurements combined with rapid sampling can produce very-high-resolution data for in vivo kinetic studies, revealing kinetic details of RNA processing events not previously detected in yeast. By comparing pre-mRNAs with different introns, and the effects of trans-acting mutations that affect different stages of RNA processing, this approach should permit the identification of rate-limiting steps in the production and turnover of intron-containing transcripts and distinguish whether the kinetics of transcription and splicing are functionally coupled.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions
Full details of yeast strains, plasmids, and primer sequences are described in Supplemental Material 1. Sequences of the tet-CYC1 promoter and reporter constructs are in Supplemental Material 2. Briefly, to construct the tetOFF strains, the gene encoding the tTA hybrid trans-activator was integrated at the LYS2 locus, deleting LYS2. For tetOFF, the gene encoding tetR′ hybrid repressor was integrated along with URA3 as a marker at the ura3 or leu2 locus. For the successful tetON strain YW156, the gene encoding tetR hybrid repressor, expressed from the strong Schizosaccharomyces pombe adh1 promoter, was integrated at the leu2 locus, and the gene encoding the tTA′ hybrid trans-activator was cloned with the strong S. cerevisiae ADH1 promoter on a centromeric plasmid. Plasmids pMK121 and pMK123 were constructed that contain the tetO7/CYC1-UAS promoter followed by a multiple cloning site, plus Nat (pMK121) or KanR (pMK123) as a yeast transformation marker, all bordered by HIS3 flanking sequences, allowing integration at the his3 locus.
Cultures were grown in synthetic dropout (SD) medium to midlog phase, transcription was repressed or induced by the addition of doxycycline to 4 μg/mL and aliquots were snap-frozen by pipetting into an equal volume of methanol sitting on dry ice. The frozen cells in the methanol slurry were pelleted by centrifugation and stored at −80°C.
RT-qPCR measurements
An overview of the scheme of work for measuring the efficiency of cell lysis, RNA extraction, and RT-qPCR normalization for RNA copy-number estimation is shown in Figure 2. Full details of the various procedures are given in Supplemental Material 3. Primer sequences are listed in Supplemental Material 1. Transcription profiles obtained using calculation of RNA copies per cell were confirmed by measuring the changes in reporter level compared to the quantity of TDH1 mRNA (that is unaffected by doxycycline) and also by comparison with Northern blot analysis (Supplemental Fig. S5). This verifies that including efficiencies of cell lysis and RNA recovery in the former approach does not misreport the end result.
Image-based measurement of mRNA in individual yeast cells
Yeast strains were grown in SD medium and fixed with paraformaldehyde in midlog phase. Probes were 40–55 nt long and each contained four amino-allyl dT that were conjugated to Cy3 (Femino et al. 1998). The sequence of the probes and the protocols for in situ hybridization and to detect and automatically count single mRNA molecules are described in Supplemental Material 4.
Polyadenylation status analysis
The assay (for overview, see Fig. 6A) is described in detail in Supplemental Material 5.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was funded by EC grant no. LSHG-CT-2005-518280 for the RiboSys project and the Wellcome Trust, and benefitted from facilities in the BBSRC-funded Edinburgh Centre for Systems Biology. J.D. Beggs holds the Royal Society Darwin Trust Research Professorship.
Authors' contributions: R.D. Alexander—construction of the RiboSys reporter genes, copy number, and kinetic data by RT-qPCR; Figures 1, 2, 4B–D, 5, 7 and Supplemental Figure S4. J.D. Barrass—development of the copy-number estimation and lariat RT-qPCR methods. M. Kos and D. Tollervey—construction of the tetOFF strains and extensive efforts to produce tetON strains; Supplemental Figures S1, S2. M.-C. Robert and E. Bertrand—FISH analyses; Figure 3 and Supplemental Figure S5. Beatriz Dichtl, L. Mariconti, and Bernhard Dichtl—3′-end cleavage sites and PASE assays; Figure 6. T. Obtulowicz, M. Koper, I. Karkusiewicz, and J. Kufel—construction of the successful tetON strain, analyses by northern and qPCR comparing with TDH1; Figure 4A and Supplemental Figure S6. J.D. Beggs—coordination of the project and preparation of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2162610.
REFERENCES
- Aitken S, Robert MC, Alexander RD, Goryanin I, Bertrand E, Beggs JD 2010. Processivity and coupling in messenger RNA transcription. PLoS ONE 5: e8845 doi: 10.1371/journal.pone.0008845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli G, Gari E, Aldea M, Herrero E 1998a. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Belli G, Gari E, Piedrafita L, Aldea M, Herrero E 1998b. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res 26: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Stevens A, Tollervey D 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J 16: 7184–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH 1998. Visualization of single RNA transcripts in situ. Science 280: 585–590 [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Aranda A, Perez-Ortin JE 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 15: 303–313 [DOI] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Alea M, Herrero E 1997. A set of vectors with tetracycline-regulatable promoter system for modulated gene expression in S. cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769 [DOI] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Holland MJ 2002. Transcript abundance in yeast varies over six orders of magnitude. J Biol Chem 277: 14363–14366 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci 93: 5208–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Maya D, Quintero MJ, de la Cruz Munoz-Centeno M, Chavez S 2008. Systems for applied gene control in Saccharomyces cerevisiae. Biotechnol Lett 30: 979–987 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastian L, Keller W 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol 11: 352–357 [DOI] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Yoshida M, Uematsu C, Kito K, Sakaki Y, Ito T 2008. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics 9: 574 doi: 10.1186/1471-2164-9-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Parker R, Jacobson A 1990. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT α1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci 87: 2780–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin JE, Alepuz PM, Moreno J 2007. Genomics and gene transcription kinetics in yeast. Trends Genet 23: 250–257 [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309 doi: 10.1371/journal.pbio.0040309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA 2009Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste MA, Duquenne M, Francois J, Parrou JL 2009. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Molecular Biology 10: 99 doi: 10.1186/1471-2199-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE Jr, Hieter P, Vogelstein B, Kinzler KW 1997. Characterization of the yeast transcriptome. Cell 88: 243–251 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hess WR, Borner T 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res 25: 2030–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart JA, Hayes A, Wardleworth L, Zhang N, Oliver SG 2005. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast 22: 565–569 [DOI] [PubMed] [Google Scholar]
- Wishart JA, Osborn M, Gent ME, Yen K, Vujovic Z, Gitsham P, Zhang N, Ross MJ, Oliver SG 2006. The relative merits of the tetO2 and tetO7 promoter systems for the functional analysis of heterologous genes in yeast and a compilation of essential yeast genes with tetO2 promoter substitutions. Yeast 23: 325–331 [DOI] [PubMed] [Google Scholar]
- Yen K, Gitsham P, Wishart J, Oliver SG, Zhang N 2003. An improved tetO promoter replacement system for regulating the expression of yeast genes. Yeast 20: 1255–1262 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]