Abstract
Purpose
Repeated pregnancy loss (RPL) occurs in 1 out of 300 couples, and the cause of about 50% of them remains idiopathic. Mitochondria have an important role in human development through ATP production and their involvement in apoptosis.
Methods
96 RPL and 96 control females were used to investigate the frequency of deletions and point mutations in the displacement loop (D-loop) on mitochondria. Multiplex PCR and DNA sequencing methods were used to detect possible variations in the mitochondrial DNA (mtDNA).
Results
No deletions but a high frequency of point mutations were found in RPL females; among 129 variations observed in RPL, 22 mutations were significant (P < 0.05) and the insertion of C in nucleotide 114 was novel.
Conclusion
High rate of mutations in D-loop of mtDNA was observed in maternal blood, a fact that may have a direct or indirect role in inducing RPL. The results can be used in the assessment of RPL and designing possible treatments for improving assisted reproduction.
Keywords: Mitochondrial DNA, Deletion, Mutation, Repeated pregnancy loss
Introduction
Pregnancy loss can be defined as the unexpected spontaneous miscarriage before the fetus is capable of extra-uterine survival. Traditionally, recurrent abortion has been defined as the occurrence of three or more clinically recognized pregnancy losses before 20 weeks from the last menstrual period. About 1 in 300 couples and 0.5–2% of women worldwide have repeated pregnancy loss (RPL) [1]. Presumed etiological factors include endocrine disorders; such as hypothyroidism and luteal phase deficiency, chromosomal aberrations, uterine abnormalities, infectious disorders and thrombophilia. These factors cause in about 50% of RPLs and the reason for the others are still idiopathic [2, 3].
Recently, it has been hypothesized that mitochondria may be directly involved in human reproduction, this has taken attentions within the scientific and medical community [4]. Human mitochondrial DNA (mtDNA) contains a 16569-base pair double strand circular genome which encodes 13 proteins (from 37 genes) of the respiratory chain, 2 rRNAs and 22 tRNAs needed for mitochondrial protein synthesis [5, 6]. The double strand consists of heavy (H-strand that is Guanine-rich) and light strands (L-strand, that is cytosine-rich). This genome also includes a non-coding displacement region (D-loop) which consists of 1122 base pairs (bp) (16024- 577 nt of mtDNA) [7], acting as a promoter of mtDNA and containing essential transcription and replication elements. D-loop contains two hypervariable regions (HV1 at nucleotides 16024- 16383 and HV2 at nucleotides 57-372) [8]. Also, there is a repeat area of C nucleotide (poly-C) between 303 and 315 positions (D310) within this regulatory region.
Until now, over 200 mtDNA mutations have been reported in Mitomap database which are associated with a wide variety of human diseases. A recent study revealed a higher frequency of mtDNA variations in women with RPL [9]. Most of these mutations accumulate in the regulatory region or D-loop [10]. Since D-loop has a regulatory role on mtDNA replication and transcription, mutations in this region might have an important effect in copy number and gene expression of the mitochondrial genome, and could disturb mitochondrial function, oxidative phosphorylation (OXPHOS) and ATP production.
Previous studies, however, do not provide enough evidence for the association between apoptosis related genes, including mtDNA, and pregnancy loss. In this study we examined common mitochondrial deletions and D-loop nucleotide alterations in samples taken from RPL women and normal controls.
Materials and methods
Patients and samples
A total of 335 consecutive couples suffering from RPL were evaluated at a primary stage. They were referred to the Research and Clinical Center for Infertility, Yazd University of Medical Science between September 2006 and June 2008. Among them, 96 women were screened as idiopathic at reproductive age. Diagnosis of RPL was based on a documented history of at least three spontaneous, consecutive abortions before 20 weeks of gestation from the same partner. These women underwent a standard diagnostic procedure in order to rule out any known cause of RPL. The diagnostic procedure included paternal and maternal karyotype, uterine sonography, TORCH infection study (Toxoplasmosis, Rubella, Cytomegalovirus, Herpes Simplex virus type II and Listeria), assessment of hormonal status, IgM and IgG anticardiolipin, and antiphospholipids antibodies assay. According to other studies, thrombophilia could be also a determinant factor in RPL [11]. However, the scales of the current study did not allow investigation of this aspect and it was postponed to the future works. All of the cases, involved in primary RPL, had no history of child delivery. The control group consisted of 96 women with at least two live births and no history of pregnancy loss. All controls were at reproductive age and were waiting for their second or more delivery in delivery room. All of the participants were informed about research and signed the consent approved by the ethical committee.
DNA extraction
Blood was collected from the antecubital vein for the isolation of genomic DNA. The DNA from 96 females with idiopathic RPL was totally isolated from the blood samples using a Flexigene blood DNA kit (DNA fast, QIAGEN) according to the manufacturer’s protocol. The isolated DNA was kept at 4°C.
Multiplex PCR reactions
The deletion- prone region between nucleotide 5461 of light strand and nucleotide 15000 of heavy strand was examined in all the patients. Four fragments were studied in a multiplex PCR as described elsewhere [12]. The reaction mixture for multiplex PCR contained 10 pmol of each primer, 1unit Taq polymerase (Cinnagene, Iran), each dNTP at a final concentration of 200 μM, and 2.5 μl PCR buffer at a final volume of 25 μl. The PCR reactions were performed in a thermal cycler (TECHNE) for 35 cycles with denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and primer extension at 72°C for 35 sec. The amplified fragments were separated on 1.5% agarose gel. The distances between the primers were long enough to allow amplification only if a part of the DNA between respective primers was deleted.
PCR-Sequencing analysis
Two primer pairs of PCR were designed from the 5′ and 3′ flanking regions to amplify the mitochondrial D-loop region. The 20-nucleotide 5′ end primer of D-loop part one (5′-ATC ATT GGA CAA GTA GCA TC-3′) from nucleotide 15791 to 15810, and the 20-nucleotide 3′ end primer of it (5′-GCT CCG GCT CCA GCG TCT CG-3′) from nucleotide 91 to 110 were used to amplify the first part of D-loop genome. The 20-nucleotide 5′ end primer of D-loop part two (5′-GAT CAC AGG TCT ATC ACC CT-3′) from nucleotide 1 to 20, and 20-nucleotide 3′ end primer of it (5′-GAG CTG CAT TGC TGC GTG CT-3′) from nucleotide 761 to 780 were used to amplify the second part of the D-loop. Using these primers, we were able to evaluate nucleotides from 16791 to 780 covering the complete D-loop region. The PCR condition was an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 60 s, annealing at 58°C for 60 s, extension at 72°C for 35 s, and 5 min final extension. The PCR-amplified fragments were purified and sequenced by Macrogen Company (Seoul, South Korea). DNA sequences were determined using the same PCR primers in two directions for more precise results. The result of DNA sequence analysis was compared with the Cambridge reference sequence (http://www.mitomap.org/) using the Clustal X program. The sequence variants not found in the corresponding record of MITOMAP and other human databases were defined as novel alterations according to Bandelt’s method [13].
Statistical analysis
All of the statistical analyses were carried out with the SPSS software package 16.0 (SPSS Inc., Chicago, IL, USA). The prevalence of sequence variation between the case and control groups was compared using Chi-square, Fisher’s exact, and ANOVA tests. P < 0.05 was considered statistically significant.
Results
The characteristics of the females with RPL and the healthy females were evaluated as shown in Table 1. Our data shows that the mean age of the women with RPL at the time of blood sampling has been 28.73 and the mean gestational age at the time of miscarriages has been 10.35. The mean age of control group at the time of blood sampling has been 30.20 while they had 2–4 children.
Table 1.
Characteristics of women with RPL and control females
| Women with RPL | Controls | |
|---|---|---|
| Age at miscarriage study (years)a | 28.73 ± 5.86 | 30.20 ± 4.14 |
| No. of miscarriage b | 3 (3–11) | 0 |
| Gestational age at the time of miscarriagea | 10.35 ± 3.75 | 0 |
| Number of miscarriagea | 3.99 ± 1.65 | 0 |
| No. of live birthb | 0 | 2 (2–4) |
Mean ± standard deviationa; Median (range)b
The screening of 96 idiopathic cases of RPL by doing multiplex PCRs showed no common deletion in mitochondrial DNA.
D-loop region was evaluated by direct sequencing, which resulted in 153 different variations in our studied population. Among these, 89 variations were only seen in RPL cases (Table 2), 24 only in control samples, and also 40 were found in both (Table 3). A significant difference was seen between two groups in 7 SNPs (P < 0.05; T16126C, T16189C, C16223T, C16294T, T16311C, T16362C, T16519C). From the mutations which were only seen in RPL group, 15 SNPs were significant (T146C, C150T, C151T, T152C, T195C, T199C, C285T, C295T, C462T, T489C, C16069T, T16093C, C16148T, A16183C, C16261T) (Table 4) and one mutation was novel (C114insertion).
Table 2.
Distribution of point mutations in RPL and control females
| Sample/ Description | Number | Min. | Max. | Mean | Std. Deviation |
|---|---|---|---|---|---|
| RPL | 96 | 3 | 17 | 8.79 | 3.356 |
| Normal | 96 | 2 | 10 | 4.90 | 1.756 |
ANOVA test # 0.0001
Table 3.
Individual single nucleotide polymorphism loci in RPL and control groups
| Row | SNP | RPL | Control | Odds Ratio | P-value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| 1 | A73G | 69 | 71.87 | 73 | 76.04 | 0.81 | 0.5106 |
| 2 | G143A | 2 | 2.08 | 1 | 1 | 2.02 | 0.5606 |
| 3 | A189G | 4 | 4.17 | 6 | 6.25 | 0.65 | 0.5159 |
| 4 | A197G | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 5 | A200G | 5 | 5.20 | 1 | 1 | 5.22 | 0.0970 |
| 6 | G207A | 6 | 6.25 | 4 | 4.17 | 1.53 | 0.5159 |
| 7 | A235G | 2 | 2.08 | 1 | 1 | 2.02 | 0.5606 |
| 8 | C324G | 4 | 4.17 | 7 | 7.29 | 0.55 | 0.3515 |
| 9 | G499A | 1 | 1 | 2 | 2.08 | 0.51 | 0.5606 |
| 10 | AC524add | 4 | 4.17 | 3 | 3.12 | 1.35 | 0.7002 |
| 11 | C16071T | 3 | 3.12 | 1 | 1 | 3.06 | 0.3122 |
| 12 | T16126Ca | 28 | 29.17 | 3 | 3.12 | 12.76 | 0.0001 |
| 13 | G16129A | 8 | 8.33 | 3 | 3.12 | 2.82 | 0.1204 |
| 14 | C16134T | 1 | 1 | 3 | 3.12 | 0.33 | 0.3122 |
| 15 | G16145A | 12 | 12.50 | 7 | 7.29 | 1.82 | 0.2268 |
| 16 | A16163G | 3 | 3.12 | 4 | 4.17 | 0.74 | 0.7002 |
| 17 | T16172C | 4 | 4.17 | 1 | 1 | 4.13 | 0.1740 |
| 18 | C16186T | 4 | 4.17 | 1 | 1 | 4.13 | 0.1740 |
| 19 | T16189Ca | 23 | 23.96 | 4 | 4.17 | 7.25 | 0.0001 |
| 20 | T16217C | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 21 | C16223Ta | 14 | 14.58 | 4 | 4.17 | 3.93 | 0.0132 |
| 22 | T16249C | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 23 | A16265C | 2 | 2.08 | 2 | 2.08 | 1 | 1.0000 |
| 24 | G16274A | 3 | 3.12 | 3 | 3.12 | 1 | 1.0000 |
| 25 | C16278T | 3 | 3.12 | 1 | 1 | 3.06 | 0.3122 |
| 26 | C16286T | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 27 | C16292T | 2 | 2.08 | 1 | 1 | 2.02 | 0.5606 |
| 28 | C16294Ta | 7 | 7.29 | 1 | 1 | 7.47 | 0.0302 |
| 29 | C16296T | 4 | 4.17 | 1 | 1 | 4.13 | 0.1740 |
| 30 | T16298C | 2 | 2.08 | 1 | 1 | 2.02 | 0.5606 |
| 31 | A16300G | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 32 | A16309G | 4 | 4.17 | 3 | 3.12 | 1.35 | 0.7002 |
| 33 | T16311Ca | 10 | 10.41 | 2 | 2.08 | 5.47 | 0.0170 |
| 34 | G16319A | 2 | 2.08 | 2 | 2.08 | 1 | 1.0000 |
| 35 | A16343G | 2 | 2.08 | 1 | 1 | 2.02 | 0.5606 |
| 36 | T16362Ca | 11 | 11.46 | 1 | 1 | 12.29 | 0.0028 |
| 37 | G16390A | 3 | 3.12 | 7 | 7.29 | 0.41 | 0.1938 |
| 38 | A16399G | 1 | 1 | 1 | 1 | 1 | 1.0000 |
| 39 | T16519Ca | 44 | 45.83 | 8 | 8.33 | 9.31 | 0.0001 |
| 40 | C16527T | 1 | 1 | 1 | 1 | 1 | 1.0000 |
aStatistically significant
Table 4.
Individual single nucleotide polymorphism loci in RPL group
| row | SNPs | Frequency | Percentage | P-valuea |
|---|---|---|---|---|
| 1 | C64T | 2 | 2.08 | 0.4973 |
| 2 | A93G | 3 | 3.12 | 0.2460 |
| 3 | C114ins.b | 4 | 4.17 | 0.1210 |
| 4 | T146C | 14 | 14.58 | 0.0001 |
| 5 | C150T | 5 | 5.20 | 0.0234 |
| 6 | C151T | 7 | 7.29 | 0.0070 |
| 7 | T152C | 31 | 32.29 | 0.0001 |
| 8 | T195C | 13 | 13.54 | 0.0001 |
| 9 | C198T | 2 | 2.08 | 0.4973 |
| 10 | T199C | 5 | 5.20 | 0.0234 |
| 11 | T204C | 4 | 4.17 | 0.1210 |
| 12 | T217C | 2 | 2.08 | 0.4973 |
| 13 | C271T | 3 | 3.12 | 0.2460 |
| 14 | C285T | 7 | 7.29 | 0.0070 |
| 15 | C295T | 13 | 13.54 | 0.0001 |
| 16 | A335G | 1 | 1.04 | 1.0000 |
| 17 | C340T | 3 | 3.12 | 0.2460 |
| 18 | A385G | 2 | 2.08 | 0.4973 |
| 19 | C456T | 2 | 2.08 | 0.4973 |
| 20 | C462T | 12 | 12.50 | 0.0003 |
| 21 | T482C | 2 | 2.08 | 0.4973 |
| 22 | T489C | 19 | 19.79 | 0.0001 |
| 23 | C497T | 3 | 3.12 | 0.2460 |
| 24 | A503G | 2 | 2.08 | 0.4973 |
| 25 | G228A | 2 | 2.08 | 0.4973 |
| 26 | A16051G | 4 | 4.17 | 0.1210 |
| 27 | C16069T | 13 | 13.54 | 0.0001 |
| 28 | T16093C | 5 | 5.20 | 0.0234 |
| 29 | C16111T | 3 | 3.12 | 0.2460 |
| 30 | C16148T | 5 | 5.20 | 0.0234 |
| 31 | G16153A | 2 | 2.08 | 0.4973 |
| 32 | A16182C | 2 | 2.08 | 0.4973 |
| 33 | A16183C | 7 | 7.29 | 0.0070 |
| 34 | C16193T | 2 | 2.08 | 0.4973 |
| 35 | C16193ins | 2 | 2.08 | 0.4973 |
| 36 | C16188T | 2 | 2.08 | 0.4973 |
| 37 | A16194C | 3 | 3.12 | 0.2460 |
| 38 | C16222T | 4 | 4.17 | 0.1210 |
| 39 | T16224C | 3 | 3.12 | 0.2460 |
| 40 | C16248T | 2 | 2.08 | 0.4973 |
| 41 | C16256T | 3 | 3.12 | 0.2460 |
| 42 | C16261T | 10 | 10.42 | 0.0011 |
| 43 | T16288C | 2 | 2.08 | 0.4973 |
| 44 | C16290T | 3 | 3.12 | 0.2460 |
| 45 | C16295T | 3 | 3.12 | 0.2460 |
| 46 | T16304C | 2 | 2.08 | 0.4973 |
| 47 | A16318T | 3 | 3.12 | 0.2460 |
| 48 | C16320T | 2 | 2.08 | 0.4973 |
| 49 | C16327T | 3 | 3.12 | 0.2460 |
| 50 | C16344T | 2 | 2.08 | 0.4973 |
| 51 | C16355T | 3 | 3.12 | 0.2460 |
| 52 | T16381C | 2 | 2.08 | 0.4973 |
| 53 | G16384A | 2 | 2.08 | 0.4973 |
| 54 | Other mutationsc | 36 | 1.04 | 1.0000 |
aFisher exact test (2-tailed) is done for the values less than 5; bNovel mutations; cThese mutations were seen in one case
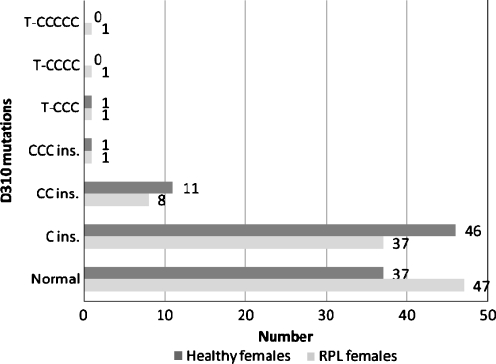
Six types of D310 mutations were seen in 49 (51.04%) of the females with RPL. The most common type of these mutations was C insertion at nucleotide 310 that was found in 37 cases (38.54%). The others were CC and CCC insertion, and substitution of T with CCC, CCCC and CCCCC (Fig. 1).
Fig. 1.
Frequency of D310 mutations between RPL and healthy females. (P = 0.591)
Discussion
The mechanisms of pregnancy loss which could be induced by various agents are not completely understood. The biological processes for maintaining a stable pregnancy are mediated by a series of differential gene expressions. The aberrant expression of apoptotic related genes is seen in some cases of RPL [14]. Several of these genes are involved in the internal apoptotic pathway and have interaction with mitochondria which having a critical role in this pathway. MtDNA is inherited from the mother’s ovum [15], and it is unusual for sperm cells to contribute to mitochondria when fertilizing the oocyte. Mature oocytes have at least 100,000 copy of mtDNA [16]. MtDNA sequence variants rapidly segregate between progenies during oogenesis or early embryogenesis [17]. Mitochondria are the bioenergetics and metabolic centers of eukaryotic cells and responsible for producing most of the cellular ATP through oxidative phosphorylation. The role of mitochondria is more important during the processes with high-energy consumption such as cell proliferation and development. Mitochondrial function serves as an important marker for oocyte quality, explaining some cases of fertilization failure [18]. Maintaining a functional complement of maternally derived mitochondria is vital for the early embryo. Decreased mitochondrial transcription may result in a poor fertilization rate and compromised embryonic development [19]. The disability of mitochondria to produce ATP or to activate the apoptosis cascade has been suggested as a possible cause for early human embryo loss [20].
In mtDNA, D-loop part acts as a promoter of mtDNA and contains essential elements for transcription and replication of mitochondrial genes. D-loop has been demonstrated as a mutation ‘hot spot’ in some disorders such as cancers. The rate of mutations in D-loop is much higher than in other parts of mtDNA, and such alterations can affect the transcription or translation of mtDNA [21].
In recent study, no common mitochondrial deletions but a significant high rate of point mutations in D-loop region of mtDNA were seen. There were 22 significant mutations in RPL group and finally the prevalence of D310 mutations was not significantly different between two groups.
The RPL women did not have any deletions in mitochondrial genome. Deletions have been found in mtDNA in various cancers [21, 22] and mitochondrial myopathies [23]. Such deletions can interfere with cell proliferation and could be possibly observed in somatic cell disorders. At level of germinal cells however, they can lead to the early apoptosis, elimination of the cells and fetal loss. Although, we did not find any deletions in RPL women, investigating this issue in the aborted fetus as well could provide further information.
The minimum and maximum numbers of nucleotide changes in RPL females were 3 and 17, respectively. This was significantly decreased to 2 and 10 in the normal group (Table 2). Previous investigations have shown a high rate of mutations in D-loop region in some cancers such as colorectal or gastric cancers [21, 22]; some of these mutations are associated with special community haplogroups. Therefore, it could be concluded that D-loop mutations disrupt apoptosis in two direction; they can either cause an uncontrolled cell proliferation (cancers) and arrest or a decreased cell proliferation (degenerative disorders or growth arrest in fetus). Although a high rate of point mutations is seen in maternal blood, the women have no clinical manifestations. Further biochemical studies on these women’s blood as well as studying aborted materials for such mutations can reveal other effects of D-loop alterations.
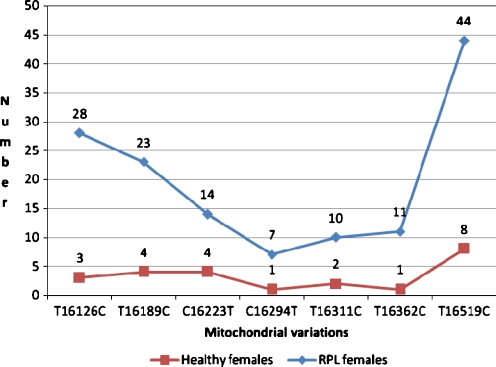
Analysis of the mtDNA regulatory region revealed 129 variations in RPL women which 40 of them were seen in both groups. SNPs A263G and C315insertion were seen in all the samples which shows a genomic diversity as a community haplogroup. Eighty nine and 24 of the variations were only found in RPL and control groups respectively; whereas 40 variations were found in both groups. Our results, also demonstrate that most of D-loop variations in RPL females were homoplasmic. We found 7 point mutations consisting of T16126C, T16189C, C16223T, C16294T, T16311C, T16362C, and T16519C more frequent in RPL females compared to the controls (Fig. 2).
Fig. 2.
Distribution of significant variations in RPL and healthy females
The T16126C variant has been previously reported in breast and endometrial cancers and glioblastoma multiform [24, 25]. The T16189C variant has been associated with type 2 diabetes a well as some metabolic syndrome [26, 27]. The C16223T variant have been described in infantile sudden death syndrome, schizophrenic patients, age-related macular degeneration, and idiopathic cardiomyopathy [28–31]. The C16294T variant is associated with Parkinson disease, infantile sudden death syndrome and age-related macular degeneration [29, 30]. The T16311C variant is seen in primary prostatic cancer [32]. The T16362C variant is used in phylogenic studies and mitochondrial haplogroups determination [33]. The T16519C variant that worsens pancreatic cancer prognosis seems to be a predisposing genetic factor for diabetes mellitus and gasterointestinal disorders [34, 35]. This variant has been the most common variant among both RPL and control groups in our study (45.83%).
Among 89 point mutations that were only detected in RPL group, C114 insertion was novel. Also, 15 variations consisting of T146C, C150T, C151T, T152C, T195C, T199C, C285T, C295T, C462T, T489C, C16069T, T16093C, C16148T, A16183C, and C16261T were significant in this group (Table 4).
The T152C variant that has an important role in respiratory morbidity among children [36] had the second significant frequency (32.29%) in our study [36]. Such polymorphism is also described in H, U and K haplogroups. The role of haplogroups is emphasized for various disorders such as age-related macular degeneration [30]. This mutation can increase the risk of Parkinson disease as a variation associated with haplogroup H [37]. The T489C variant is found more frequent in some populations and disorders [38, 39]. The T146C variant is important in phylogenic analysis and has been reported in ovarian cancer and mitochondrial myopathies [40, 41]. In turn, the other variations can have important roles in RPL, independently or as a part of haplogroups. Because of the location that some critical haplogroups polymorphisms have in other parts of mitochondrial genome, haplogroup determination was not feasible.
In our study, D310 mutations were seen in 49 (51.04%) cases of RPL females and 59 (61.45%) of the controls (Fig. 1).The D310 area has been recently identified as a frequent hot spot of deletion/insertion mutation in some disorders such as tumors [42, 43]. This polymorphic c-stretch (CCCCCCCTCCCCC) is involved in the formation of the persistent RNA-DNA hybrid which is essential for the mtDNA heavy strand replication [44]. The difference of D310 mutation between two studied groups was not significant; therefore, we conclude that this variation is not suitable to be used as a marker in RPL evaluation strategies.
The role of mtDNA mutations in embryogenesis and reproduction is still under investigation. Such mitochondrial dysfunction affecting ATP production during development may result in empty sacs and other features of fetal loss. Thus , it is possible that some mtDNA mutations cause a developmental arrest before pregnancy is clinically recognized [45].
Two mechanisms for inducing mtDNA mutations could be considered. It is demonstrated that the disorder of mtDNA can be induced by the defects of nuclear DNA [46]. Another putative mechanism is associated with reactive oxygen species (ROS). The mitochondrial genome is extremely susceptible to damages from continuous exposure to ROS. These molecules endogenously are produced in mitochondrial respiratory chain and are evidenced to increase the ratio of point mutations in mtDNA [47]. This effect is probably through inhibition of the repair system in mtDNA, detoxification of ROS, or increase in ROS production. Such effects could be considered as possible causes for D-loop point mutations of mtDNA.
Due to difficulties for obtaining aborted materials, our work was only focused on maternal blood. More studies are necessary to show the primary or secondary role of D-loop mutations in embryonic development.
Conclusion
The observed correlation between some mitochondrial genome variations and RPL shows important role of this organelle in fetal development. Further complementary studies on mtDNA mutations can lead to a molecular diagnostic panel for RPL patients. Variations in the noncoding region have been previously reported as polymorphisms and unlikely to cause miscarriage. However, more analysis may also discover a role for these regions in RPL. Furthermore, preclinical abortions which are usually reported as infertile couples, and also failure of in vitro fertilization are our next targets for expanding this research to infertility.
Acknowledgements
This study is supported by the National Institute of Genetic Engineering and Biotechnology, Tehran, Iran; and Research and Clinical Center for Infertility of Shahid Sadughi University of Medical Science, Yazd, Iran
Footnotes
Capsule
No deletions but a high rate of D-loop point mutations were observed in mtDNA of RPL women. This condition may have a role in inducing RPL.
References
- 1.Wilcox AJ, et al. Incidence of early loss of pregnancy. N Engl J Med. 1988;319(4):189–94. doi: 10.1056/NEJM198807283190401. [DOI] [PubMed] [Google Scholar]
- 2.Clifford K, et al. An informative protocol for the investigation of recurrent miscarriage: preliminary experience of 500 consecutive cases. Hum Reprod. 1994;9(7):1328–32. doi: 10.1093/oxfordjournals.humrep.a138703. [DOI] [PubMed] [Google Scholar]
- 3.Hatasaka HH. Recurrent miscarriage: epidemiologic factors, definitions, and incidence. Clin Obstet Gynecol. 1994;37(3):625–34. doi: 10.1097/00003081-199409000-00016. [DOI] [PubMed] [Google Scholar]
- 4.May-Panloup P, et al. Mitochondria and reproduction. Med Sci (Paris) 2004;20(8–9):779–83. doi: 10.1051/medsci/2004208-9779. [DOI] [PubMed] [Google Scholar]
- 5.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106(1):18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 6.Thorburn DR, Dahl HH. Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet. 2001;106(1):102–14. doi: 10.1002/ajmg.1380. [DOI] [PubMed] [Google Scholar]
- 7.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki M, et al. Alterations in the mitochondrial displacement loop in lung cancers. Clin Cancer Res. 2003;9(15):5636–41. [PubMed] [Google Scholar]
- 9.Kaare M, et al. Do mitochondrial mutations cause recurrent miscarriage? Mol Hum Reprod. 2009;15(5):295–300. doi: 10.1093/molehr/gap021. [DOI] [PubMed] [Google Scholar]
- 10.Parsons TJ, et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15(4):363–8. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 11.Nelen WL, et al. Hyperhomocysteinemia and recurrent early pregnancy loss: a meta-analysis. Fertil Steril. 2000;74(6):1196–9. doi: 10.1016/s0015-0282(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 12.Massoud Houshmand, M.H.S., Baharak Hooshiar Kashani, Mehdi Shafa Shariat Panahi, Mohammad Mehdi Banoei, Anna Isaian, Mostafa Moin, Abolhasan Farhoudi Role of mitochondria in Ataxia-Telangiectasia:Investigation of mitochondrial deletions and Haplogroups. Iranian Journal of Biotechnology (IJB). 2006;4(1):64–8.
- 13.Bandelt HJ, Salas A, Bravi CM. What is a ‘novel’ mtDNA mutation–and does ‘novelty’ really matter? J Hum Genet. 2006;51(12):1073–82. doi: 10.1007/s10038-006-0066-5. [DOI] [PubMed] [Google Scholar]
- 14.Baek KH. Aberrant gene expression associated with recurrent pregnancy loss. Mol Hum Reprod. 2004;10(5):291–7. doi: 10.1093/molehr/gah049. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda H, et al. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci U S A. 1995;92(10):4542–6. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoubridge EA, Wai T. Mitochondrial DNA and the mammalian oocyte. Curr Top Dev Biol. 2007;77:87–111. doi: 10.1016/S0070-2153(06)77004-1. [DOI] [PubMed] [Google Scholar]
- 17.Shoubridge EA. Mitochondrial DNA segregation in the developing embryo. Hum Reprod. 2000;15(Suppl 2):229–34. doi: 10.1093/humrep/15.suppl_2.229. [DOI] [PubMed] [Google Scholar]
- 18.Santos TA, Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85(3):584–91. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Au HK, YT, Kao SH, Tzeng CR, Hsieh RH. Abnormal mitochondrial structure in human unfertilized oocytes and arrested embryos. Ann N Y Acad Sci. 2005;(1042):177–85. [DOI] [PubMed]
- 20.Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- 21.Akouchekian M, et al. High rate of mutation in mitochondrial DNA displacement loop region in human colorectal cancer. Dis Colon Rectum. 2009;52(3):526–30. doi: 10.1007/DCR.0b013e31819acb99. [DOI] [PubMed] [Google Scholar]
- 22.Kamalidehghan B, et al. Tumoral cell mtDNA approximately 8.9 kb deletion is more common than other deletions in gastric cancer. Arch Med Res. 2006;37(7):848–53. doi: 10.1016/j.arcmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Yuzaki M, et al. Multiple deletions in mitochondrial DNA at direct repeats of non-D-loop regions in cases of familial mitochondrial myopathy. Biochem Biophys Res Commun. 1989;164(3):1352–7. doi: 10.1016/0006-291x(89)91818-4. [DOI] [PubMed] [Google Scholar]
- 24.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–62. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 25.Kirches E, et al. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93(4):534–8. doi: 10.1002/ijc.1375. [DOI] [PubMed] [Google Scholar]
- 26.Park KS, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008;51(4):602–8. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 27.Weng SW, et al. Association of mitochondrial deoxyribonucleic acid 16189 variant (T->C transition) with metabolic syndrome in Chinese adults. J Clin Endocrinol Metab. 2005;90(9):5037–40. doi: 10.1210/jc.2005-0227. [DOI] [PubMed] [Google Scholar]
- 28.Bandelt HJ, et al. ‘Distorted’ mitochondrial DNA sequences in schizophrenic patients. Eur J Hum Genet. 2007;15(4):400–2. doi: 10.1038/sj.ejhg.5201781. [DOI] [PubMed] [Google Scholar]
- 29.Arnestad M, et al. Are substitutions in the first hypervariable region of the mitochondrial DNA displacement-loop in sudden infant death syndrome due to maternal inheritance? Acta Paediatr. 2002;91(10):1060–4. doi: 10.1080/080352502760311557. [DOI] [PubMed] [Google Scholar]
- 30.Udar N, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50(6):2966–74. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 31.Ozawa T, et al. Patients with idiopathic cardiomyopathy belong to the same mitochondrial DNA gene family of Parkinson’s disease and mitochondrial encephalomyopathy. Biochem Biophys Res Commun. 1991;177(1):518–25. doi: 10.1016/0006-291x(91)92014-b. [DOI] [PubMed] [Google Scholar]
- 32.Baek KH, et al. Comparison of gene expression at the feto-maternal interface between normal and recurrent pregnancy loss patients. Reprod Fertil Dev. 2002;14(3–4):235–40. doi: 10.1071/rd02008. [DOI] [PubMed] [Google Scholar]
- 33.Non AL, Kitchen A, Mulligan CJ. Identification of the most informative regions of the mitochondrial genome for phylogenetic and coalescent analyses. Mol Phylogenet Evol. 2007;44(3):1164–71. doi: 10.1016/j.ympev.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Navaglia F, et al. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126(4):593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 35.Camilleri M, et al. Mitochondrial DNA and gastrointestinal motor and sensory functions in health and functional gastrointestinal disorders. Am J Physiol Gastrointest Liver Physiol. 2009;296(3):G510–6. doi: 10.1152/ajpgi.90650.2008. [DOI] [PubMed] [Google Scholar]
- 36.Schmuczerova J, et al. Genetic variability of HVRII mtDNA in cord blood and respiratory morbidity in children. Mutat Res. 2009;666(1–2):1–7. doi: 10.1016/j.mrfmmm.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Khusnutdinova E, et al. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson’s disease. Ann N Y Acad Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 38.Dato S, et al. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur J Hum Genet. 2004;12(12):1080–2. doi: 10.1038/sj.ejhg.5201278. [DOI] [PubMed] [Google Scholar]
- 39.Kurtz A, et al. Somatic mitochondrial DNA mutations in neurofibromatosis type 1-associated tumors. Mol Cancer Res. 2004;2(8):433–41. [PubMed] [Google Scholar]
- 40.Trappen PO, et al. Somatic mitochondrial DNA mutations in primary and metastatic ovarian cancer. Gynecol Oncol. 2007;104(1):129–33. doi: 10.1016/j.ygyno.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Filosto M, et al. Lack of paternal inheritance of muscle mitochondrial DNA in sporadic mitochondrial myopathies. Ann Neurol. 2003;54(4):524–6. doi: 10.1002/ana.10709. [DOI] [PubMed] [Google Scholar]
- 42.Tang M, et al. Mitochondrial DNA mutation at the D310 (displacement loop) mononucleotide sequence in the pathogenesis of gallbladder carcinoma. Clin Cancer Res. 2004;10(3):1041–6. doi: 10.1158/1078-0432.ccr-0701-3. [DOI] [PubMed] [Google Scholar]
- 43.Mao L. A new marker determining clonal outgrowth. Clin Cancer Res. 2002;8(7):2021–3. [PubMed] [Google Scholar]
- 44.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273(46):30614–21. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 45.Blerkom J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction. 2004;128(3):269–80. doi: 10.1530/rep.1.00240. [DOI] [PubMed] [Google Scholar]
- 46.Zeviani M, et al. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339(6222):309–11. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 47.Ozawa T. Genetic and functional changes in mitochondria associated with aging. Physiol Rev. 1997;77(2):425–64. doi: 10.1152/physrev.1997.77.2.425. [DOI] [PubMed] [Google Scholar]