Abstract
Fc receptor-like A (FCRLA) and FCRLB have homology to the transmembrane FCRL family members (FCRL1–6) and to the conventional receptors for the Fc portion of immunoglobulin, but uniquely are cytosolic proteins expressed in B cells. Here we describe the phenotype of Fcrlb gene targeted mice. B cell development and in vitro responses are normal; however, antibody responses to a T-dependent antigen are elevated. The gene encoding the inhibitory FcγRIIb is located nearby Fcrlb. Although Fcrlb gene targeting had no effect on the function or basal expression of FcγRIIb, its expression was reduced following activation. This abnormal regulation was due to co-inheritance of Fcgr2b and the mutant Fcrlb allele from the 129 ES cells. A promoter polymorphism in the 129/Sv Fcgr2b allele results in diminished upregulation of FcγRIIb following B cell activation. Thus, we speculate that the enhanced antibody response seen in the FCRLB-deficient mice may be due to the Fcgr2b promoter.
Keywords: Fc receptor, Gene targeting, FcγRIIb, Antibody response, Germinal center
INTRODUCTION
Receptors for the constant region of immunoglobulins (FcR), FcγRI, FcγRII, FcγRIII, FcεRI, are found on cells of both myeloid and lymphoid lineages [4;12;32]. Members of this “classical” FcR family are known to have important regulatory roles in both cell mediated and humoral immunity, including feedback suppression of B cell responses, regulation of hypersensitivity reactions, and the induction of cellular cytotoxicity. Other FcRs include the recently identified FcμR, which binds IgM [14;36], Fcα/μR [11;13;34], which binds IgA and IgM, the polymeric immunoglobulin receptor (polyIgR) [29] that mediates transcytosis of IgA across mucosal epithelial surfaces, the neonatal FcRn [15], a receptor related to MHC class I that mediates perinatal transfer of Ig and maintenance of basal immunoglobulin levels in adults, and the FcαR (CD89) [26], a receptor for IgA that is present on myeloid cells in humans and rats, but curiously not in mice [20]. The FcRn gene is located on chromosome 19q13.3 [10] and CD89 is found in the leukocyte receptor complex on chromosome 19q13.4 [17;38;39], whereas the genes encoding the other Fc receptors are located on human chromosome 1q32
There has been an unexpected recent harvest of FcR related genes from the human chromosome 1q region. Six human Fc receptor-like (FCRL 1–6) genes have been identified. They encode type I transmembrane proteins with similar extracellular Ig-like domains and cytoplasmic regions that contain consensus tyrosine based motifs, suggesting an inhibitory (ITIM) or activating (ITAM) signaling function for these receptors [5]. Until the very recent identification of MHC class II as a ligand for FCRL6 [33], the ligands for FCRL 1–6 have been unknown, but none of these receptors has been convincingly shown to bind immunoglobulins. During the characterization of these extended FcR family members, we identified two unusual relatives, FcRX and FcRY, in the human 1q region [6;21]. FcRX is also termed FREB (Fc receptor homolog expressed in B cells) and FcRL (FcR-like) because of its independent identification by other laboratories [7;25]. Similarly, FcRY is called FcRL2 and FREB2 [3;40]. The HUGO Gene Nomenclature Committee has recently adopted FCRLA and FCRLB as the approved human symbols for these genes; the mouse genes are designated Fcrla and Fcrlb [19].
Both FCRLA and FCRLB proteins have unusual features that distinguish them from other members of the FCRL family. Most notably, they are intracellular proteins rather than transmembrane receptors [3;7;25;40]. The only available information about the expression of these receptors at the protein level comes from studies in humans. Among hemopoietic cells FCRLA is expressed only in B cells, with the highest levels found in the germinal center B cells. Wilson and Colonna found that FCRLB expressing cells are also present in the germinal centers of tonsils [40]. However, the FCRLB+ cells were extremely rare, in tissue sections many germinal centers contained no FCRLB+ cells, and were non-proliferating. This is in striking contrast to FCRLA+ cells, which are abundant and enriched among proliferating germinal center centroblasts [7]. Moreover, FCRLA and FCRLB were not co-expressed in the same cells. Due to the lack of suitable mAb and the low levels of mRNA, FCRLB expression in mice has only been analyzed by RT-PCR. We found that Fcrlb transcripts could be detected in all B cell subsets in the spleen, although they were somewhat reduced in germinal center B cells, in keeping with our observation that Fcrlb expression is highest in non-proliferating cells [21]. By contrast, Wilson and Colonna found expression restricted to germinal center B cells and an undefined population of cells expressing B220, CD21, and CD23 [40]. The basis for this discrepancy in the Fcrlb expression profile is unclear, but may be due to the markers used for GC B cell isolation, peanut agglutinin versus the monoclonal antibody GL7.
Given the difficulty in analyzing FCRLB expression in vivo and in vitro, we reasoned that its function might be best elucidated by a genetic approach. Our analysis of Fcrlb gene targeted mice is described here.
METHODS
Generation of Fcrlb knockout mice
To isolate the genomic fragment containing the Fcrlb gene, we screened a BAC clone library of 129-derived R1 ES cells with a primer set (FcRY/s20086: 5’-TCAGGGAAGAGGTTATCAGG-3’; FcRY/as20404: 5’-CAACCCAACTCAAGAAATCC-3’). The isolated BAC clone was confirmed to contain the Fcrlb gene by sequencing the 5’ and 3’ end of the insert, as well as by digestion with multiple restriction enzymes. A 5.6-kb BglII fragment and a 3.2-kb SacI-NotI fragment were used as the 5’ and 3’ homology regions of the targeting vector, respectively, to replace exons 1, 2 and a portion of 3 along with a ~2-kb promoter region with the neomycin gene. 129-derived R1 ES cells were transfected with the linearized targeting vector and 2 days after transfection cultured in the presence of 600 µg/ml G418 and 2 µM of GANC (only for the first two days) as described previously [22]. Homologous recombination was verified by long range genomic PCR using primers flanking 5’ and 3’ homology regions, and neo primers. (See Fig S1B legend for details.) Chimeric mice were bred with C57BL/6 mice to obtain heterozygotes, which were then crossed to obtain homozygotes. Mouse genotypes were identified by genomic PCR using primers FcRY/s8055 (5’-TGGCTTCTCTTTAGTGATGC-3’), FcRY/as8642 (5’-ATGTGGTTGCTGGGACTTGA-3’) and neo/s at 95 °C for 2 min followed by 30 cycles of amplification at 95 °C for 10 s, 60 °C for 10 s and 72 °C for 90 s. The WT and KO allele give rise to a 590-bp and 880-bp band, respectively. Fcrlb mRNA expression was analyzed by PCR using primers s144 (5’-CAGGCAGAGTCATTATGTGG-3’), as561 (5’-GCCGTCGTGGTAGTAGTGAA-3’) and FW169 (5’-TTAGCACTCTCTGGTACCTGG-3’) at 95 °C for 2 min followed by 35 cycles of amplification at 95 °C for 15 s, 55 °C for 10 s and 72 °C for 2 min. Mice were housed in specific pathogen free conditions and all experiments were approved by the Animal Facility Committee of the RIKEN Yokohama Institute (Permission no. 20-025).
Flow cytometry analysis and proliferation assays
Flow cytometry analysis was performed as described previously [23]. For proliferation, purified spleen B cells were seeded in a 96 well plate at 5×105/ml, 100 µl/well, and stimulated for 2 days with F(ab’)2 or whole anti-IgM with or without soluble CD40 ligand. 3H-thymidine (1 µCi/well) was added during the last 8 h and thymidine uptake was measured as described previously. [23].
Immune response and ELISPOT assay
Immune responses were tested essentially as previously described [24]. Briefly, four WT and five Fcrlb-KO mice (8 wk-old) were injected i.p. with 100 µg of NP-CGG (Biosearch Technologies, Novato, CA) precipitated with alum. The mice were boosted with the same antigen at age 15 wks. Mice were bled weekly and serum titers of NP-specific IgG1 were analyzed by ELISA, using NP-specific monoclonal high (clone C6) and low (clone N1G9) affinity antibodies as standards.
The ELISPOT assay was performed as described [24]. Briefly, Multiscreen HTS filter plate (Millipore) were coated with 50 µg/ml of NP3-BSA or NP30-BSA at 4°C overnight. The coated plate was then washed with PBS-T (PBS containing 0.1% Tween 20) 3 times and blocked with PBS containing 1% BSA for 1 h at RT. Splenocytes (5×105, 2.5×105and 1.25×105) were then seeded and incubated at 37°C for 100 min in a CO2 incubator. The plate was then washed twice with PBS-T containing 50 mM EDTA, 3 times with PBS-T and then blocked again with PBS containing 1% BSA for 1 h at RT. The plate was further incubated with alkaline phosphatase-conjugated goat anti-mouse IgG1 antibodies (1 µg/ml in PBS containing 1% BSA) at 37°C for 60 min in a CO2 incubator, washed 4 times with PBS-T and developed with BCIP/NBT reagent (MOSS INC) for 2–3 min. The plate was then washed 4 times with H2O, air dried and colonies were counted using an IMMUNOSPOT Analyzer (CTL Analyzers LLC, Cleveland, OH). The number of colonies obtained with different number of splenocytes was converted to number of colonies/105 cells and the average numbers are shown.
Expression of FcγRIIb
Spleen B cells were purified using IMAG negative sorting (BD) and then cultured for 2 days in a 24 well plate at 5×105 cells/ml, 1ml/well in the presence of 20 µg/ml of LPS or 10 µg F(ab’)2 anti-IgM plus 20 ng/ml of IL4. The cells were then stained with FITC-conjugated anti-FcγRIIb (2.4G2, Rat IgG2b, BD) plus APC-conjugated B220 and the FcγRIIb expression was analyzed on gated B220+ cells. As a control, the same cultured cells were stained with FITC-conjugated control rat IgG2b and APC-B220. 98% of the cultured cells were B200+ both in WT and KO mice.
Analysis of the polymorphism in the FcgRIIb promoter
129 mice are known to have a polymorphism in the Fcgr2b gene promoter, i.e., a 16-bp deletion when compared to C57BL/6 mice. We analyzed this deletion by genomic PCR using primers Fcgr2b/s (5’-GTTGATCTTCATTTTACAGAC-3’) and Fcgr2b/as (5’-TCTGTGCCCTAGTCCTGAATC-3’) at 95 °C for 3 min followed by 35 cycles of amplification at 95 °C for 5 s, 55 °C for 10 s and 72 °C for 30 s. The B6- and 129-derived genomes give rise to 164-bp and 148-bp PCR products, respectively, that were resolved on 2% agarose gels.
RESULTS
Generation of FCRLB-deficient mice
The Fcrlb targeting vector (Fig 1A and Supplemental Fig. 1A) was designed to replace ~1.5 kb of the 5’ flanking region of the Fcrlb gene as well as part of the coding region (exons 1, 2 and the 5’ end of exon 3) with a neo gene in the opposite transcriptional orientation. Homologous recombination in the R1 ES cells (129/Sv) containing the targeted allele was confirmed by Southern blots and genomic PCR (Fig S1). The ES clones were introduced into C57BL/6 blastocysts by an aggregation method and the resultant male chimeric mice were mated with C57BL/6 females to generate heterozygous offspring. These were intercrossed to obtain homozygous Fcrlb-gene targeted mice. Initial characterization of the mice was performed when they had been backcrossed to C57BL/6 mice for two generations. Detailed analysis of NP-antibody responses, B cell proliferative responses, and FcγRIIb expression was performed on mice backcrossed for eight generations.
Fig. 1. Fcrlb targeting strategy and expression of Fcrlb mRNA.
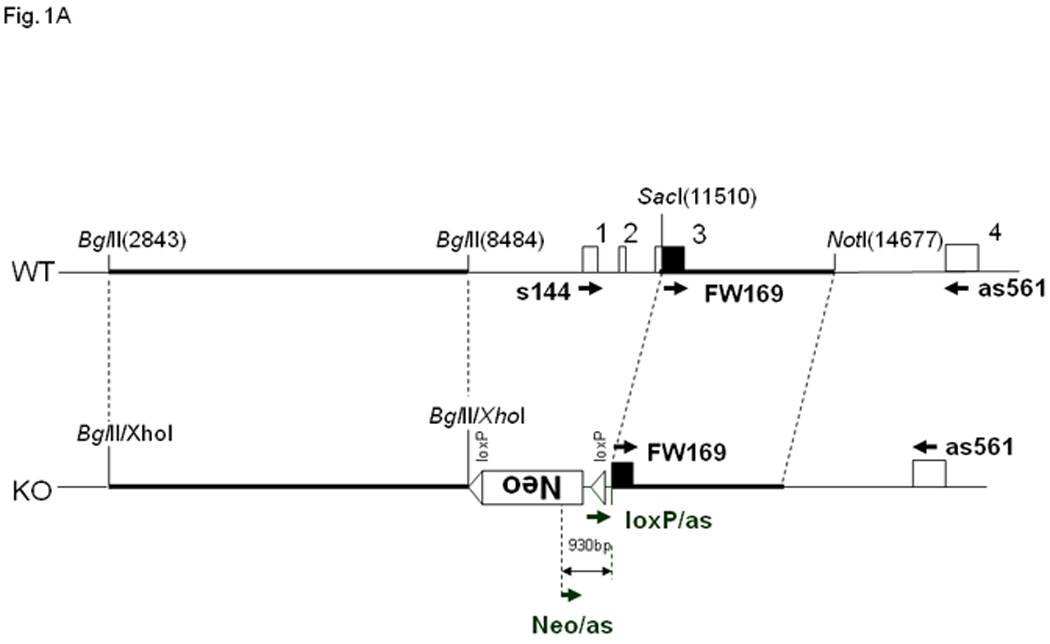
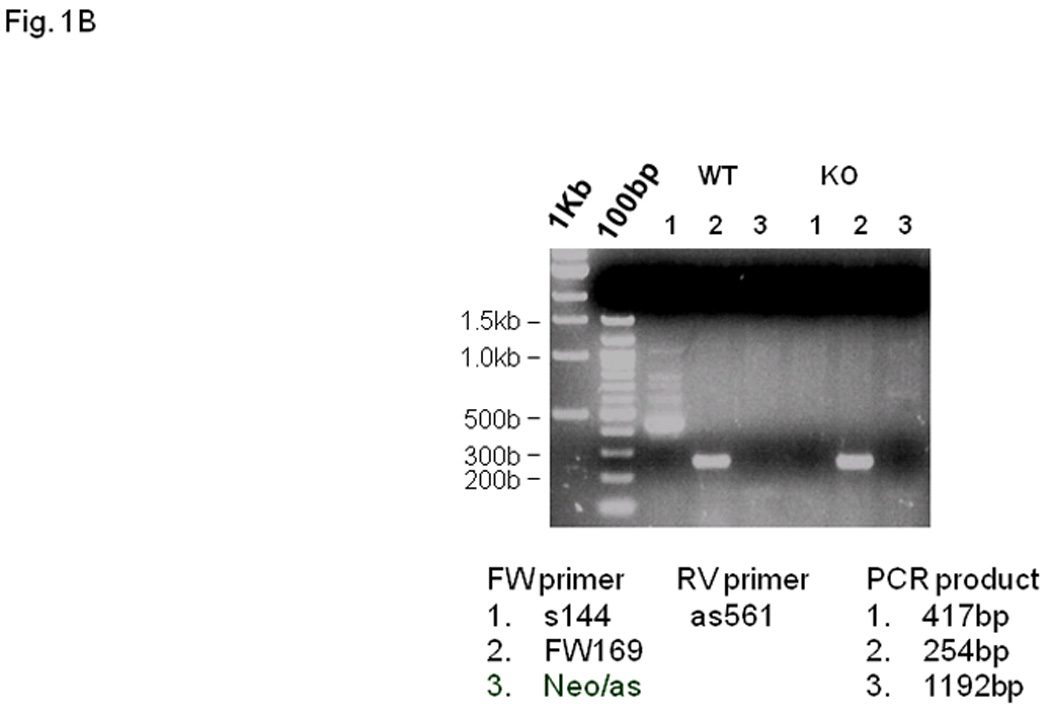
(A) Schematic representation of the wild type (WT) and targeted (KO) Fcrlb alleles. A detailed description of the targeting vector can be found in Fig. S1. Primers used for RT-PCR analysis of Fcrlb mRNA expression are illustrated by arrowheads. (B) RT-PCR analysis of Fcrlb gene expression in splenic B cells. Three different forward (FW) primers in conjunction with a common reverse (RV) primer were used in RT-PCR analysis of Fcrlb expression in purified splenic B cells from WT and KO mice. The expected sizes of the PCR products are indicated.
Lacking an antibody able to detect FCRLB expression, we performed RT-PCR to identify Fcrlb mRNA in purified splenic B cells from wild type and knockout mice to confirm the successful knockout of the Fcrlb gene. Using a sense primer (S144) located in exon 1 and an anti-sense primer (as561) located in exon 4 of the Fcrlb gene, a major band of the expected size (~400 bp) was amplified from RNA samples derived from wildtype B cells (Fig. 1B, WT lane 1). Several larger bands were also detected, most likely derived from incompletely spliced transcripts. No bands were observed when this same primer pair was used to amplify RNA from the knockout B cells (Fig. 1B, KO lane 1), as expected since exon 1 is within the region deleted by the targeting construct. When the upstream primer was replaced with a primer (FW169) in exon 3, which is retained in the mutant allele, a major band of ~250 bp was amplified from B cell RNA of both wildtype and knockout mice (Fig. 1B, lanes 2). The RNA species from the knockout B cells did not contain the neo sequence (Fig. 1B KO lane 3) or the loxP sequence (not shown) present in the targeting vector. Since the native promoter was deleted by the targeting vector, this RNA species is likely derived from cryptic transcription initiation within the remnant Fcrlb gene, or by splicing of an upstream sequence onto exon 3 of the gene. We attempted to define the origin of this transcript using 5’ RACE (rapid amplification of cDNA ends) but our repeated attempts were unsuccessful.
Phenotype of the FCRLB-deficient mice
We first performed a detailed analysis of the basal immune phenotype of the Fcrlb knockout mice. There were no significant differences in the levels of serum IgM, IgG subclasses, or IgA in 10–12 week old wildtype and knockout mice (Fig. S2). B cell development in the bone marrow, from the proB to the mature B cell stage was unaffected by the absence of FCRLB, and in the spleen, the percentage and absolute numbers of total B cells and B cell subsets (immature, transitional, mature, follicular and marginal zone) were normal (Representative flow cytometry data, Fig. S3A, B; Summary data, Fig. 2.) Although T cells do not express FCRLB, we examined this lineage in the event that FCRLB deficiency had any unanticipated indirect effect. T cell development in the thymus was normal, as was the frequency of CD4 and CD8 T cells in the spleen (Fig S3C).
Fig. 2. Analysis of lymphocyte subpopulations by flow cytometry.
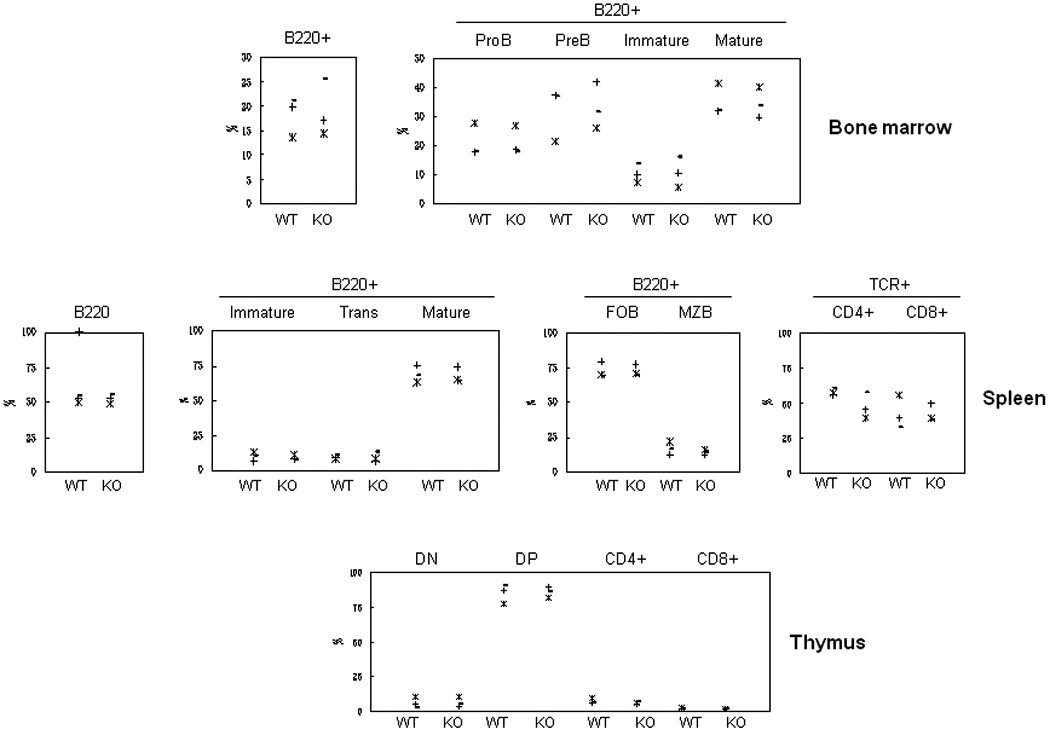
The frequency of the various lymphoid subpopulations was analyzed by flow cytometry using the markers described in the Materials and Methods. B cell populations analyzed were: Bone marrow, total B220+, proB, preB, immature and mature B; Spleen, total B220+, immature, transitional (Trans) and mature B, follicular B (FOB) and marginal zone B (MZB). T cell populations analyzed were: Spleen, CD4+ and CD8+; thymus, CD4 and CD8 double negative (DN) and double positive (DP), CD4+ and CD8+ single positive. Each symbol represents one mouse. Three pairs of mice were analyzed in these experiments. Representative flow cytometry profiles are shown in Fig. S3.
B cell function in the FCRLB-deficient mice was first tested in vitro. Purified splenic B cells were stimulated with the BCR agonist anti-IgM, the TLR4 agonist LPS, and a surrogate for T cell help, CD40L, alone or in combination (Fig. 3). No significant differences in the responses of the knockout and wildtype B cells to any of these stimuli were observed.
Fig. 3. In vitro B cell proliferation.
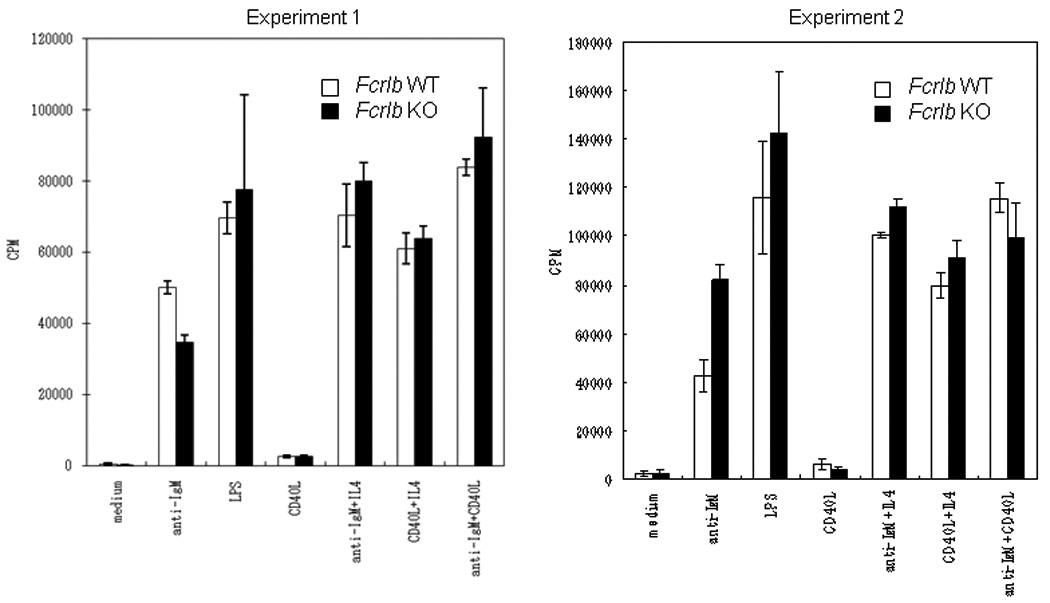
Purified splenic B cells were stimulated with anti-IgM, LPS, and CD40L, as well as with the indicated combinations of these stimuli. The proliferative response of KO and WT B cells was analyzed at 48 hours. Data from two independent experiments, each done with a pair of WT and KO mice are depicted. The bars indicate the standard deviation of four assay wells.
The in vivo function of the knockout B cells was tested after the mice had been backcrossed for eight generations. We examined the T-dependent response in detail, both in terms of primary and secondary responses and in terms of the affinity of the antibodies produced. Since FCRLB may be expressed in murine GC B cells, we considered that its absence might influence these parameters. The primary NP response in Fcrlb knockout mice was somewhat elevated compared to that in wildtype mice equivalent, and the secondary response was even more enhanced (Fig. 4). This increase was apparent in the total anti-NP IgG1 antibody levels and was even more striking in the high affinity anti-NP antibodies.
Fig. 4. Primary and secondary antibody responses after immunization with NP16CGG.
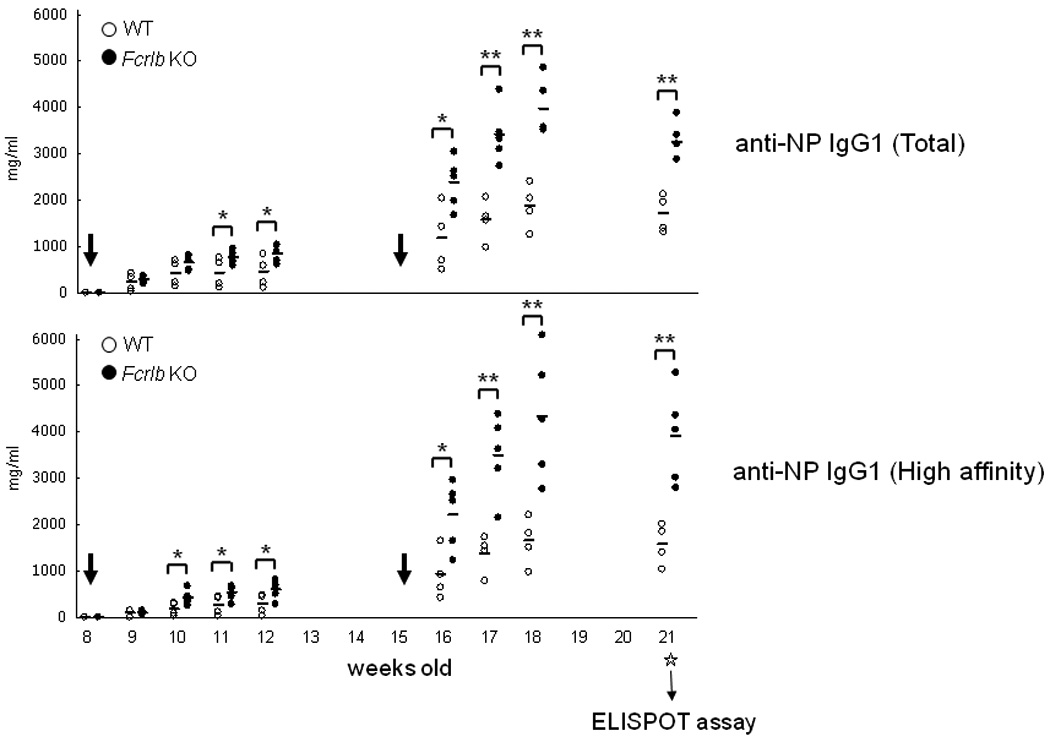
Mice were immunized when they were 8 weeks old and boosted at 15 weeks (indicated by the large arrows) with alum precipitated NP16CGG. Serum samples were collected at the indicated time points for analysis of total and high affinity NP-antibodies by an ELISA assay. Six weeks after the second immunization the mice were sacrificed and antibody secreting cells in spleen and bone marrow were enumerated by an ELISPOT assay (Fig. 5). Each symbol represents one mouse. *, p < 0.05; **, p <0.01 (unpaired t-test).
We next determined whether this increase in serum antibody titers was due to an increase in the numbers of antibody secreting cells or in the amount of antibody produced per cell. An ELISPOT assay was performed six weeks after the secondary immunization with NP16CGG, and the numbers of both total and high affinity AFC were increased in spleen and bone marrow of the FCRLB-deficient mice (Fig. 5).
Fig. 5. Enumeration of NP-specific antibody forming cells by ELISA in WT and KO mice.
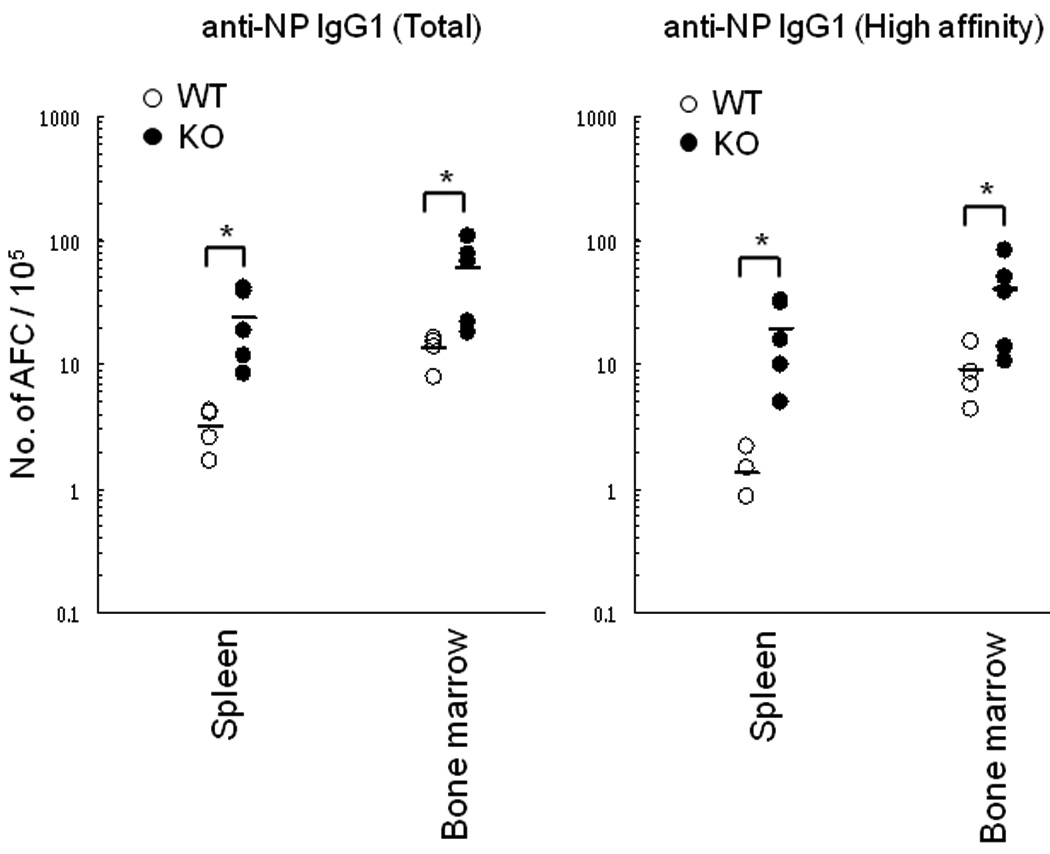
The frequency of antibody-forming cells (AFC) was determined six weeks after the secondary immunization with NP16CGG. Both total and high affinity IgG1 anti-NP AFC were increased in the Fcrlb KO mice. Each symbol represents one mouse. *, p < 0.05 (unpaired t-test).
Does the inhibitory FcγRIIb have a role in the phenotype of the Fcrlb knockout mice?
The Fcgr2b gene, which is ~37.5 kb telomeric from the Fcrlb gene, encodes an ITIM-bearing receptor that can potently inhibit B cell responses. This inhibition occurs when IgG antigen-antibody complexes crosslink the BCR with the inhibitory FcγRIIb, which recruits SHIP1 to its ITIM motif and ultimately dampens BCR signaling by preventing intracellular calcium mobilization [1]. From the outset we recognized the possibility that our targeting of the Fcrlb gene might influence expression of the Fcgr2b gene in cis, and that this could have profound effects on B cell responses. This concern was obviated, however, by our initial analysis. A classical assay to demonstrate the inhibitory effect of FcγRIIb is to compare proliferation of B cells stimulated with intact anti-IgM antibodies, which can colligate the BCR and FcγRIIb, to cells stimulated with F(ab’)2 antibodies, which cannot. The proliferation of both wild type and knockout B cells was reduced approximately sevenfold when intact anti-IgM antibodies were used as the stimulus (Fig. 6). Co-stimulation with CD40L can partially rescue this FcγRIIb-mediated inhibition, and again, the response of wildtype and knockout B cells was identical. These results suggested that the in vitro function of FcγRIIb on B cells was not influenced by targeting the Fcrlb gene. Further evidence in support of this idea was obtained by flow cytometry. The levels of FcγRIIb detected by the 2.4G2 mAb were the same, or if anything slightly higher, on the knockout resting B cells compared to the wildtype B cells (Fig. 7). Thus the basal expression of FcγRIIb was unaffected by our gene targeting strategy. Interestingly, however, its expression was significantly dysregulated following B cell activation. Compared to wildtype, the level of FcγRIIb on the knockout B cells was more than 1.5 fold less following LPS stimulation and more than 2 fold less after stimulation with anti-IgM plus IL4.
Fig. 6. FcγRIIb-mediated inhibition of BCR induced B cell proliferation is intact in Fcrlb KO mice.
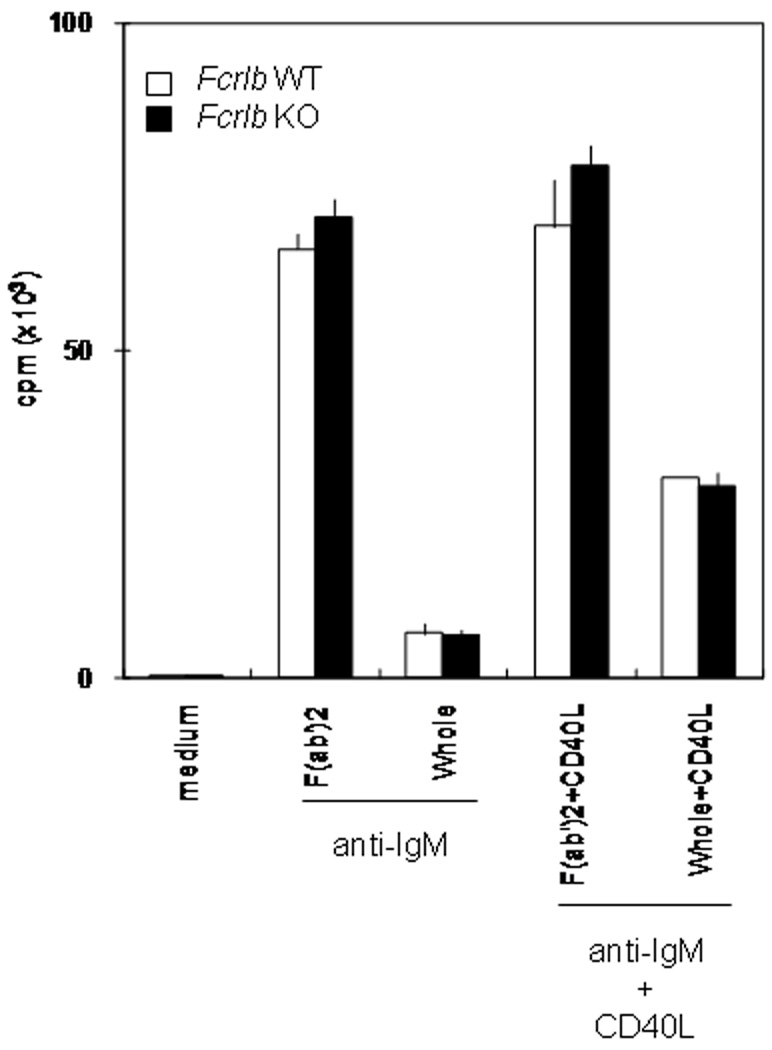
Purified splenic B cells from WT or Fcrlb KO mice were stimulated with F(ab’2) anti-IgM antibodies or with intact anti-IgM antibodies (to co-engage the BCR and FcγRIIb) in the presence or absence of CD40 ligand (CD40L). Proliferation was measured at 48 hours.
Fig. 7. FcγRIIb expression by splenic B cells before and after stimulation.
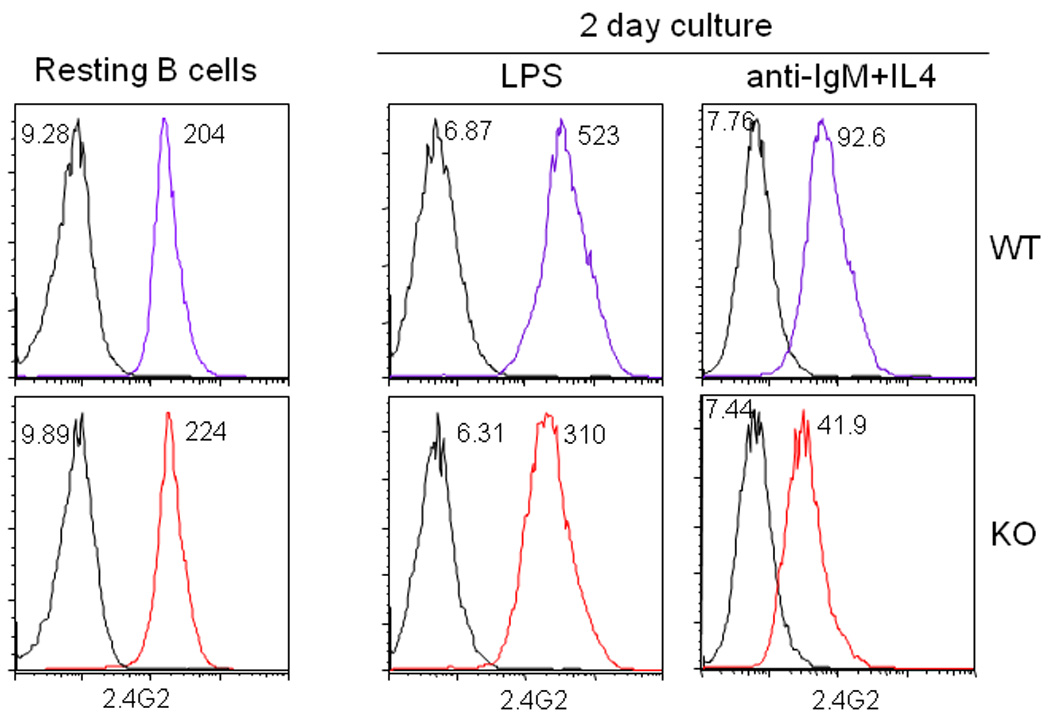
Purified splenic B cells were analyzed by flow cytometry for FcγRIIb expression using the 2.4G2 mAb. FcγRIIb expression on resting B cells was identical in WT and KO mice, however it was lower in the KO B cells after stimulation with LPS or anti-IgM plus IL4.
To further evaluate a possible role for FcγRIIb in the Fcrlb knockout phenotype, we tested the mice for autoantibody production. ELISA titers of anti-nuclear antibodies were marginally increased as the mice aged (Fig. 8). A similar marginal increase in ANA titers has been observed in C57BL/6 mice congenic for the 129 strain-derived chromosome 1 interval from 87.9 to 100 cM, a region that contains the Fcgr2b gene [2].
Fig. 8. Anti-nuclear antibody titers.
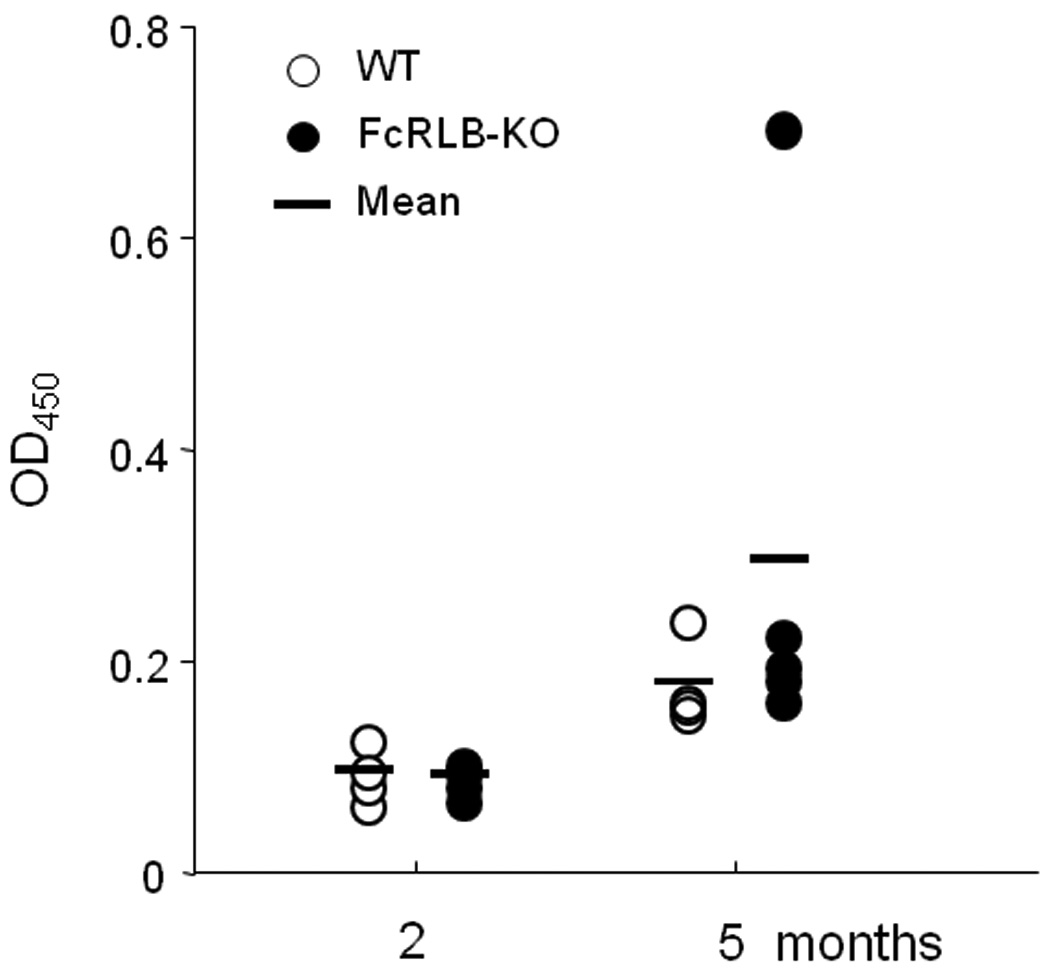
Serum samples from wildtype (open circles) and Fcrlb−/− (filled circles) mice at two and five months of age were analyzed for ANA titers by ELISA. Data from individual mice are depicted.
In search of a possible explanation for the abnormal regulation of FcγRIIb expression in activated B cells, we focused on the Fcgr2b promoter. A polymorphism in this promoter has been identified in certain autoimmune mouse strains and in some cases is thought to promote autoantibody formation due to B cell hyperactivation [8;9;16;30;41]. This same polymorphism is present in the 129 mouse strain, from which the ES cells used for Fcrlb targeting were derived. A genomic PCR assay was designed to distinguish 129 and C57BL/6 promoters by size of the amplified fragment and, indeed, the Fcrlb gene targeted mice possessed the 129-derived Fcgr2b promoter polymorphism (Fig. S4).
DISCUSSION
Our studies here report for the first time on the phenotype of the knockout of one of the intracellular FcRL family members, Fcrlb. The mutant mice developed normally and the absence of FCRLB had no obvious effect on the differentiation of B and T lineage cells. The in vitro responses of the knockout B cells stimulated with the BCR agonist anti-IgM, the TLR4 agonist LPS, and a surrogate for T cell help, CD40L, as well as various combinations of these stimuli, were also normal. However, the in vivo antibody response to a T dependent antigen was increased in the knockout mice.
Fcrlb is related to Fcrla, another member of the FcR like gene family. In humans both FCRLA and FCRLB encode intracellular proteins that are preferentially, although not exclusively expressed in germinal center B cells [6;7;25;40]. The human FCRLA is found in the endoplasmic reticulum where it binds intracellular immunoglobulin and may be involved in retention of the secretory form of antibody in B cells (T. Santiago, L. Hendershot, A. Taranin, and PDB manuscript submitted). Human FCRLB is also expressed in the germinal center but only by very few of the B cells [40]. In considering a function for FCLRB, we reasoned that, like FCRLA, it might retain secreted immunoglobulin in B cells. Therefore, in the absence of FCRLB there might be more Ig secreting cells, or more immunoglobulin secreted per cell. Our data showing increased serum antibody responses and an increase in AFC following immunization in the FCRLB-deficient mice were consistent with this hypothesis.
As a computational approach to determining FCRLB function, we sought evidence for SNPs in the human FCRLB gene. Several FCRLB SNPs have been described in the coding region, but none in the 5’ flanking region, and some of these result in non-synonymous amino acid substitutions. http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=127943. However, there have been no published studies describing immunological characterization of individuals with different FCRLB SNP haplotypes. Surprisingly, though, one of these FCRLB alleles has been nominally associated as a risk factor for cardiovascular disease [18;35], although among 74 such risk factor genes, FCRLB was not in the top ten based on probability analyses. By contrast there is significant association with SLE and an Fcgr2b promoter polymorphism [27;28;37]
Targeting the Fcrlb gene had no effect on the basal expression levels of FcγRIIb, the inhibitory Fc receptor expressed on B cells and encoded by a neighboring gene. However, expression of FcγRIIb on the knockout B cells was reduced compared to wildtype B cells following activation, consistent with previous studies [30;41]. Further analysis revealed the co-inheritance of the targeted Fcrlb and FcgR2b129 alleles. The latter contains a promoter polymorphism that results in decreased expression of the receptor on activated B cells [30;31;41]. Given the very limited expression of Fcrlb in B cells compared with the powerful effects of FcγRIIb on B cell responses, the elevated antibody response in the FCRLB-deficient mice may be attributable to the presence of this promoter polymorphism. The modest increase in anti-nuclear antibodies in the Fcrlb knockout mice harboring the FcgR2b129 allele is consistent with this interpretation, since a similar observation was made in C57BL/6 mice congenic for 129-derived chromosome 1 loci in this region [2]. Ultimately, however, these speculations will have to be tested by defining the individual contribution of the Fcrlb and FcgR2b genes by performing gene targeting of Fcrlb in ES cells of C57BL/6 origin. Thus our findings, while not entirely conclusive in terms of FCRLB function, nonetheless confirm the functional consequences of the FcgR2b promoter polymorphism on antibody responses. Moreover, investigators interested in using gene targeting to define functions of other members of the FCRL family, notably FCRL1 and FCRL5 which are expressed on B cells, should bear these new findings in mind.
Supplementary Material
(A) Schematic representation of the wild type (WT) and targeted (KO) Fcrlb alleles and of the primers used in PCR analysis of the targeted ES cell clones are depicted. TK, thymidine kinase gene from herpes simplex virus. (B) Results of long range genomic PCR assays to confirm homologous recombination of the targeting vector into the Fcrlb locus. Left blot, 5’ homology arm using primers s1864 and Neo/s. Right blot, 3’ homology arm using primers as15239 and Neo/as. The primer sequences are: 5’ primer, s1864 (5’-GGATGCGGAGGATACTGAAC-3’); 3’ primer, as15239 (5’-TCCCCTTCGTAGTGCGTAGT-3’); neo/s (5’-TCGCCTTCTATCGCCTTCTT-3’); neo/as (5’-ATAGCCGAATAGCCTCTCCA-3’). PCR was performed at 95 °C for 2 min followed by 30 cycles of amplification at 95 °C for 10 s, 58 °C for 20 s and 68 °C for 11 min using LA-Taq polymerase (TAKARA Bio., Japan).
Levels of the indicated serum Ig isotypes were determined by ELISA from 10–12 week old mice. Data from individual mice and the mean are depicted.
B cell subpopulations in bone marrow (A) and spleen (B) and T cell subpopulations in spleen and thymus (C) were analyzed by flow cytometry using the indicated markers.
The location of the Fcgr2b promoter and transcription start site, the PCR primers used in this assay, and the location with the 129 deletion polymorphism are indicated in the top part of the figure. Genomic PCR results from B6, 129, and Fcrlb +/+, +/−, and −/− mice on the B6 background are shown in the bottom of the figure.
ACKNOWLEDGEMENTS
We would like thank our UAB and RCAI colleagues for helpful discussions during the preparation of this manuscript. These studies were supported in part by an RCAI International Collaboration Award (J-YW and PDB) an NIH NIAID R56 AI069365 (PDB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bolland S, Pearse RN, Kurosaki T, Ravetch JV. SHIP modulates immune receptor responses by regulating membrane association of Btk. Immunity. 1998;8:509–516. doi: 10.1016/s1074-7613(00)80555-5. [DOI] [PubMed] [Google Scholar]
- 2.Carlucci F, Cortes-Hernandez J, Fossati-Jimack L, Bygrave AE, Walport MJ, Vyse TJ, Cook HT, Botto M. Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J.Immunol. 2007;178:2352–2360. doi: 10.4049/jimmunol.178.4.2352. [DOI] [PubMed] [Google Scholar]
- 3.Chikaev NA, Bykova EA, Najakshin AM, Mechetina LV, Volkova OY, Peklo MM, Shevelev AY, Vlasik TN, Roesch A, Vogt T, Taranin AV. Cloning and characterization of the human FCRL2 gene. Genomics. 2005;85:264–272. doi: 10.1016/j.ygeno.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Daeron M. Fc receptor biology. Annu.Rev.Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 5.Davis RS. Fc receptor-like molecules. Annu.Rev.Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 6.Davis RS, Li H, Chen CC, Wang Y-H, Cooper MD, Burrows PD. Definition of an Fc receptor related gene (FcRX) expressed in human and mouse B cells. International Immunology. 2002;14:1075–1083. doi: 10.1093/intimm/dxf074. [DOI] [PubMed] [Google Scholar]
- 7.Facchetti F, Cella M, Festa S, Fremont DH, Colonna M. An unusual Fc receptor-related protein expressed in human centroblasts. Proc.Natl.Acad.Sci.U.S.A. 2002;99:3776–3781. doi: 10.1073/pnas.022042699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Hirose S, Abe M, Sanokawa-Akakura R, Ohtsuji M, Mi X, Li N, Xiu Y, Zhang D, Shirai J, Hamano Y, Fujii H, Shirai T. Polymorphisms in IgG Fc receptor IIB regulatory regions associated with autoimmune susceptibility. Immunogenetics. 2000;51:429–435. doi: 10.1007/s002510050641. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Hirose S, Sanokawa-Akakura R, Abe M, Mi X, Li N, Miura Y, Shirai J, Zhang D, Hamano Y, Shirai T. Genetically determined aberrant down-regulation of FcgammaRIIB1 in germinal center B cells associated with hyper-IgG and IgG autoantibodies in murine systemic lupus erythematosus. Int.Immunol. 1999;11:1685–1691. doi: 10.1093/intimm/11.10.1685. [DOI] [PubMed] [Google Scholar]
- 10.Kandil E, Egashira M, Miyoshi O, Niikawa N, Ishibashi T, Kasahara M. The human gene encoding the heavy chain of the major histocompatibility complex class I-like Fc receptor (FCGRT) maps to 19q13.3. Cytogenet.Cell Genet. 1996;73:97–98. doi: 10.1159/000134316. [DOI] [PubMed] [Google Scholar]
- 11.Kikuno K, Kang DW, Tahara K, Torii I, Kubagawa HM, Ho KJ, Baudino L, Nishizaki N, Shibuya A, Kubagawa H. Unusual biochemical features and follicular dendritic cell expression of human Fcalpha/mu receptor. Eur.J.Immunol. 2007;37:3540–3550. doi: 10.1002/eji.200737655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinet JP. The high-affinity IgE receptor (FcεRI): from physiology to pathology. Annu.Rev.Immunol. 1999;17:931–972. doi: 10.1146/annurev.immunol.17.1.931. [DOI] [PubMed] [Google Scholar]
- 13.Kinet JP, Launay P. Fc α/μR: single member or first born in the family? Nat.Immunol. 2000;1:371–372. doi: 10.1038/80805. [DOI] [PubMed] [Google Scholar]
- 14.Kubagawa H, Oka S, Kubagawa Y, Torii I, Takayama E, Kang DW, Gartland GL, Bertoli LF, Mori H, Takatsu H, Kitamura T, Ohno H, Wang JY. Identity of the elusive IgM Fc receptor (FcmuR) in humans. J.Exp.Med. 2009;206:2779–2793. doi: 10.1084/jem.20091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lencer WI, Blumberg RS. A passionate kiss, then run: exocytosis and recycling of IgG by FcRn. Trends Cell Biol. 2005;15:5–9. doi: 10.1016/j.tcb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Lin Q, Xiu Y, Jiang Y, Tsurui H, Nakamura K, Kodera S, Ohtsuji M, Ohtsuji N, Shiroiwa W, Tsukamoto K, Amano H, Amano E, Kinoshita K, Sudo K, Nishimura H, Izui S, Shirai T, Hirose S. Genetic dissection of the effects of stimulatory and inhibitory IgG Fc receptors on murine lupus. J.Immunol. 2006;177:1646–1654. doi: 10.4049/jimmunol.177.3.1646. [DOI] [PubMed] [Google Scholar]
- 17.Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP. Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics. 2000;51:659–669. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- 18.Luke MM, O'Meara ES, Rowland CM, Shiffman D, Bare LA, Arellano AR, Longstreth WT, Jr, Lumley T, Rice K, Tracy RP, Devlin JJ, Psaty BM. Gene variants associated with ischemic stroke: the cardiovascular health study. Stroke. 2009;40:363–368. doi: 10.1161/STROKEAHA.108.521328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maltais LJ, Lovering RC, Taranin AV, Colonna M, Ravetch JV, Dalla-Favera R, Burrows PD, Cooper MD, Davis RS. New nomenclature for Fc receptor-like molecules. Nat.Immunol. 2006;7:431–432. doi: 10.1038/ni0506-431. [DOI] [PubMed] [Google Scholar]
- 20.Maruoka T, Nagata T, Kasahara M. Identification of the rat IgA Fc receptor encoded in the leukocyte receptor complex. Immunogenetics. 2004;55:712–716. doi: 10.1007/s00251-003-0626-1. [DOI] [PubMed] [Google Scholar]
- 21.Masuda K, Davis RS, Maruyama T, Zhang J, He T, Cooper MD, Wang J, Burrows PD. FcRY, an Fc receptor related gene differentially expressed during B lymphocyte development and activation. Gene. 2005;363:32–40. doi: 10.1016/j.gene.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, Wang J. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J.Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 24.Masuda K, Ouchida R, Yokoi M, Hanaoka F, Azuma T, Wang JY. DNA polymerase eta is a limiting factor for A:T mutations in Ig genes and contributes to antibody affinity maturation. Eur.J.Immunol. 2008;38:2796–2805. doi: 10.1002/eji.200838502. [DOI] [PubMed] [Google Scholar]
- 25.Mechetina LV, Najakshin AM, Volkova OY, Guselnikov SV, Faizulin RZ, Alabyev BY, Chikaev NA, Vinogradova MS, Taranin AV. FCRL, a novel member of the leukocyte Fc receptor family possesses unique structural features. Eur.J.Immunol. 2002;32:87–96. doi: 10.1002/1521-4141(200201)32:1<87::AID-IMMU87>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Monteiro RC, Van De Winkel JG. IgA Fc receptors. Annu.Rev.Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 27.Niederer HA, Clatworthy MR, Willcocks LC, Smith KG. FcgammaRIIB, FcgammaRIIIB, and systemic lupus erythematosus. Ann.N.Y.Acad.Sci. 2010;1183:69–88. doi: 10.1111/j.1749-6632.2009.05132.x. [DOI] [PubMed] [Google Scholar]
- 28.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat.Rev.Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 29.Norderhaug IN, Johansen FE, Schjerven H, Brandtzaeg P. Regulation of the formation and external transport of secretory immunoglobulins. Crit Rev.Immunol. 1999;19:481–508. [PubMed] [Google Scholar]
- 30.Pritchard NR, Cutler AJ, Uribe S, Chadban SJ, Morley BJ, Smith KG. Autoimmune-prone mice share a promoter haplotype associated with reduced expression and function of the Fc receptor FcgammaRII. Curr.Biol. 2000;10:227–230. doi: 10.1016/s0960-9822(00)00344-4. [DOI] [PubMed] [Google Scholar]
- 31.Rahman ZS, Manser T. Failed up-regulation of the inhibitory IgG Fc receptor Fc gamma RIIB on germinal center B cells in autoimmune-prone mice is not associated with deletion polymorphisms in the promoter region of the Fc gamma RIIB gene. J.Immunol. 2005;175:1440–1449. doi: 10.4049/jimmunol.175.3.1440. [DOI] [PubMed] [Google Scholar]
- 32.Ravetch JV, Bolland S. IgG Fc receptors. Annu.Rev.Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 33.Schreeder DM, Cannon JP, Wu J, Li R, Shakhmatov MA, Davis RS. Cutting Edge: FcR-Like 6 Is an MHC Class II Receptor. J.Immunol. 2010 doi: 10.4049/jimmunol.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibuya A, Sakamoto N, Shimizu Y, Shibuya K, Osawa M, Hiroyama T, Eyre HJ, Sutherland GR, Endo Y, Fujita T, Miyabayashi T, Sakano S, Tsuji T, Nakayama E, Phillips JH, Lanier LL, Nakauchi H. Fcα/μ receptor mediates endocytosis of IgM-coated microbes. Nat.Immunol. 2000;1:441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 35.Shiffman D, O'Meara ES, Bare LA, Rowland CM, Louie JZ, Arellano AR, Lumley T, Rice K, Iakoubova O, Luke MM, Young BA, Malloy MJ, Kane JP, Ellis SG, Tracy RP, Devlin JJ, Psaty BM. Association of gene variants with incident myocardial infarction in the Cardiovascular Health Study. Arterioscler.Thromb.Vasc.Biol. 2008;28:173–179. doi: 10.1161/ATVBAHA.107.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shima H, Takatsu H, Fukuda S, Ohmae M, Hase K, Kubagawa H, Wang JY, Ohno H. Identification of TOSO/FAIM3 as an Fc receptor for IgM. Int.Immunol. 2010;22:149–156. doi: 10.1093/intimm/dxp121. [DOI] [PubMed] [Google Scholar]
- 37.Su K, Wu J, Edberg JC, Li X, Ferguson P, Cooper GS, Langefeld CD, Kimberly RP. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcgammaRIIb alters receptor expression and associates with autoimmunity. I. Regulatory FCGR2B polymorphisms and their association with systemic lupus erythematosus. J.Immunol. 2004;172:7186–7191. doi: 10.4049/jimmunol.172.11.7186. [DOI] [PubMed] [Google Scholar]
- 38.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol.Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 39.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm.Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 40.Wilson TJ, Colonna M. A new Fc receptor homolog, FREB2, found in germinal center B cells. Genes Immun. 2005;6:341–346. doi: 10.1038/sj.gene.6364185. [DOI] [PubMed] [Google Scholar]
- 41.Xiu Y, Nakamura K, Abe M, Li N, Wen XS, Jiang Y, Zhang D, Tsurui H, Matsuoka S, Hamano Y, Fujii H, Ono M, Takai T, Shimokawa T, Ra C, Shirai T, Hirose S. Transcriptional regulation of Fcgr2b gene by polymorphic promoter region and its contribution to humoral immune responses. J.Immunol. 2002;169:4340–4346. doi: 10.4049/jimmunol.169.8.4340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of the wild type (WT) and targeted (KO) Fcrlb alleles and of the primers used in PCR analysis of the targeted ES cell clones are depicted. TK, thymidine kinase gene from herpes simplex virus. (B) Results of long range genomic PCR assays to confirm homologous recombination of the targeting vector into the Fcrlb locus. Left blot, 5’ homology arm using primers s1864 and Neo/s. Right blot, 3’ homology arm using primers as15239 and Neo/as. The primer sequences are: 5’ primer, s1864 (5’-GGATGCGGAGGATACTGAAC-3’); 3’ primer, as15239 (5’-TCCCCTTCGTAGTGCGTAGT-3’); neo/s (5’-TCGCCTTCTATCGCCTTCTT-3’); neo/as (5’-ATAGCCGAATAGCCTCTCCA-3’). PCR was performed at 95 °C for 2 min followed by 30 cycles of amplification at 95 °C for 10 s, 58 °C for 20 s and 68 °C for 11 min using LA-Taq polymerase (TAKARA Bio., Japan).
Levels of the indicated serum Ig isotypes were determined by ELISA from 10–12 week old mice. Data from individual mice and the mean are depicted.
B cell subpopulations in bone marrow (A) and spleen (B) and T cell subpopulations in spleen and thymus (C) were analyzed by flow cytometry using the indicated markers.
The location of the Fcgr2b promoter and transcription start site, the PCR primers used in this assay, and the location with the 129 deletion polymorphism are indicated in the top part of the figure. Genomic PCR results from B6, 129, and Fcrlb +/+, +/−, and −/− mice on the B6 background are shown in the bottom of the figure.


