Abstract
The nucleolytic activity of animal Argonaute proteins is deeply conserved, despite its having no obvious role in microRNA-directed gene regulation. In mice, Ago2 (also known as Eif2c2) is uniquely required for viability, and only this family member retains catalytic competence. To investigate the evolutionary pressure to conserve Argonaute enzymatic activity, we engineered a mouse with catalytically inactive Ago2 alleles. Homozygous mutants died shortly after birth with an obvious anaemia. Examination of microRNAs and their potential targets revealed a loss of miR-451, a small RNA important for erythropoiesis. Though this microRNA is processed by Drosha (also known as Rnasen), its maturation does not require Dicer. Instead, the pre-miRNA becomes loaded into Ago and is cleaved by the Ago catalytic centre to generate an intermediate 3′ end, which is then further trimmed. Our findings link the conservation of Argonaute catalysis to a conserved mechanism of microRNA biogenesis that is important for vertebrate development.
Argonaute proteins are the key effectors of small RNA-mediated regulatory pathways that modulate gene expression, regulate chromosome structure and function, and provide an innate immune defence against viruses and transposons1. The structure of Ago proteins is well conserved, consisting of an amino-terminal domain, the Mid domain, and their signature PAZ and Piwi domains. Structure–function relationships in this family are becoming increasingly well understood2. The PAZ and Mid domains help to anchor the small RNA guide, with PAZ binding the 3′ end using a series of conserved aromatic residues and the Mid domain providing a binding pocket for the 5′ end. The Piwi domain contains an RNase H motif that was cryptic in the primary sequence but easily recognizable in the tertiary structure. Loading of a highly complementary target into an Ago brings the scissile phosphate, opposite nucleotides 10 and 11 of the small RNA guide, into the enzyme active site, allowing cleavage of the RNA to leave 5′ P and 3′ OH termini3–7.
Ago proteins can be divided into three clades. The Piwi clade is animal-specific, and forms part of an elegant innate immune system that controls the activity of mobile genetic elements8. The Wago clade is specific to worms and acts in a variety of different biological processes9. The Ago clade is defined by similarity to Arabidopsis Ago1 (ref. 10). Ago-clade proteins are found in both plants and animals where one unifying thread is their role in gene regulation. In plants, some Ago family members bind to microRNAs and are directed thereby to recognize and cleave complementary target messenger RNAs11,12.
Animal microRNAs function differently from their plant counterparts, with nearly all microRNA–target interactions providing insufficient complementarity to orient the scissile phosphate properly for cleavage. Here, target recognition relies mainly on a ‘seed’ sequence corresponding to miRNA nucleotides 2–8. Although complementarity of the target to other segments of the miRNA can contribute to recognition, seed pairing seems to be the dominant factor in determining regulation13. A very few extensive microRNA–target interactions can lead to mRNA cleavage in mammals14,15. However, none of these has yet been shown to be critical for target regulation16–18.
Despite the fact that animal microRNAs regulate targets without Ago-mediated cleavage, the Argonaute catalytic centre is deeply conserved. This consists of a catalytic DDH triad that serves as a metal coordinating site19. Examining the evolution of mammalian Ago proteins suggests a split into catalytic and non-catalytic family members occurring early in the vertebrate lineage (results not shown). Of the four Ago-clade proteins in mammals, only Ago2 has retained both the DDH motif and demonstrable endonuclease activity20–22. Ago1, Ago3 and Ago4 are linked within a single ~190 kilobase locus and have lost catalytic competence. An analysis of Ago2 mutant cells has indicated that proteins encoded by the Ago 1/3/4 locus can support miRNA-mediated silencing20. This leaves us without a clear explanation for the maintenance of a catalytically competent Ago family member, because miRNAs are the exclusive partners of these proteins in virtually all cell types20,23,24.
In mammals, endogenous small interfering RNAs have been detected in abundance only in oocytes and embryonic stem (ES) cells25,26 (and results not shown). In oocytes, these arise in part from double-stranded RNAs formed from the interaction of antisense pseudogene and sense protein-coding transcripts26,27. Loss of key components of small RNA biogenesis or effector pathways, Dicer, DGCR8 or Ago2, from growing oocytes indicates the importance of endo-siRNAs for gene regulation and ultimately for proper chromosome segregation in that cell type28–31. Although this phenomenon could provide one explanation for conservation of catalysis, Ago2-null mice are not simply female sterile. Instead, lethality is observed from early to mid gestation20,32,33. This strongly suggests unique functions for the single catalytic Ago protein outside the female germline. We envisioned that this could be due to Ago2 being the only family member expressed in a critical cell type or domain in the developing embryo or to a previously unsuspected and critical role for catalysis.
Ago2 is required for extra-embryonic development
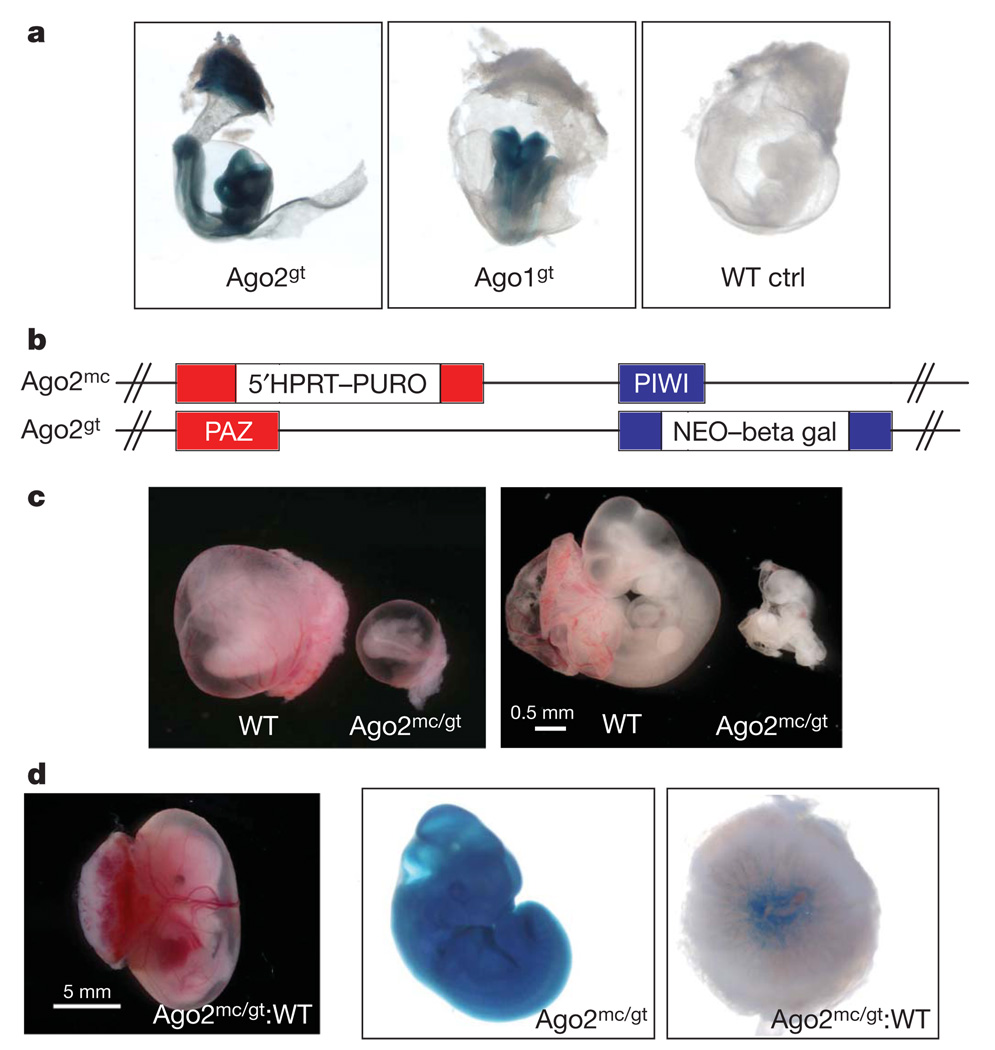
A number of studies have indicated that all four mammalian Ago proteins share broadly similar expression patterns and largely identical populations of bound small RNAs20,23,24,34. However, most of these were carried out by looking grossly at tissues of adult animals or by using continuous cell lines. We wished to probe the possibility that Ago2 occupies a unique expression domain during embryogenesis that might provide an explanation for the lethal phenotype of mutations in this gene. Towards this end, we took advantage of existing gene trap reporters, which bring lacZ under the control of the Ago1 or Ago2 promoters. Staining of mid-gestation embryos demonstrated broad expression of Ago2 in both the embryonic and extra-embryonic compartments. In contrast, Ago1 expression was restricted to the embryo (Fig. 1a). Our previous studies also demonstrated Ago3 expression within the developing embryo20.
Figure 1. Ago2 is essential for extra-embryonic development.
a, LacZ whole-mount staining of embryonic day 9.5 embryos bearing the Ago2gt or Ago1gt gene trap beta-galactosidase reporter alleles and wild-type controls (WT ctrl). b, Allelic combination in Ago2 insertional mutant embryos, with the structure of each allele shown. HPRT, hypoxanthine-guanine phosphoribosyl transferase gene; NEO, neomycin resistance gene; PURO, puromycin resistance gene. c, Example wild-type (WT) and mutant (Ago2mmc/gt) embryos from heterozygous intercrosses. Left panel: embryonic day 10.5 embryos within their embryonic yolk sac and placentas. Right panel: embryos dissected from their extra-embryonic components. d, Representative embryonic day 12.5 chimaeric embryo developed from Ago2 null ES cells aggregated with wild-type tetraploid embryos. From left to right: whole chimaeric embryonic conceptus (Ago2mc/gt:WT), beta-galactosidase staining of the whole embryo showing contribution of Ago2 null ES cells (Ago2mc/gt), beta-galactosidase staining of the placenta showing contribution of the ES cells to the vasculature invading the placental labyrinth (Ago2mc/gt:WT).
Around embryonic day 8.5, the allantois grows out from the posterior mesoderm of the embryo to invade the chorion. This contact is essential for branching of the allantois structure and formation of a complex network of blood vessels that invade the trophoblast cells to form the labyrinthine layer. This is the site of nutrient, waste and gas exchange between the maternal blood islands and the embryo35.
We observed growth retardation, cardiac abnormalities and aberrant neural tube closure in Ago2-mutant embryos previously. On the basis of comparisons to other mutants with similar phenotypes, this seemed strongly indicative of a defect in extra-embryonic development36. An examination of this compartment revealed morphological abnormalities in implanted decidua and hypotrophic placentas in Ago2 mutants (Fig. 1b, c). Histological analysis of placental cross-sections showed a marked reduction in the thickness of the labyrinthine layer (Supplementary Fig. 1a). In contrast, the spongioblast and giant trophoblast layers seemed unaffected. This was consistent with detailed analysis of Ago2 gene trap animals, which strongly verified the presence of the reporter in the affected cell types (Supplementary Fig. 1b).
The malformation of the labyrinthine layer could be due either to a failure of the embryonic vasculature to invade the placenta, or an inherent defect in the cells derived from the trophoblast (extra-embryonic) lineage to support the embryonic vasculature. To distinguish between these two possibilities, we performed tetraploid complementation. Ago2-null ES cells were derived from mutant embryos (Supplementary Fig. 2) and aggregated with tetraploid cells derived from wild-type embryos. The latter can form extra-embryonic tissues but cannot contribute to the embryo proper. One Ago2 allele was marked by the gene-trap lacZ reporter, allowing us to visualize the contribution of null cells to the embryo. The presence of a wild-type extra-embryonic compartment was able to bypass the mid-gestational death of Ago2-null embryos (Fig. 1d). This verified an essential role for Ago2 in placental development. However, viable mice were not obtained, indicating additional critical Ago2 functions during later embryonic or perinatal development.
Ago2 catalytic activity is important for postnatal development
The requirement for Ago2 during embryonic development could be based solely on it being the only Argonaute protein abundantly expressed in critical extra-embryonic cell types or it could depend on Ago2 being catalytically competent. We therefore sought to generate a mouse in which Ago2 was still expressed but had been rendered catalytically inactive. We had previously shown that substitution of alanine for either of the two aspartate residues within the catalytic DDH motif disrupted RNA cleavage without impeding small RNA binding20. We therefore replaced the endogenous Ago2 allele with one carrying an ADH rather than a DDH triad (Supplementary Fig. 3).
We intercrossed heterozygous Ago2ADH mice and scored the ratios of each resulting genotype at various developmental time points (Supplementary Table 1). Expected Mendelian ratios were observed up until the point of weaning, at which time viable homozygotes were no longer present.
This demonstrates that Ago2 catalysis is not required for extra-embryonic development and strongly indicates that it is simply the presence of Ago2 as the dominant microRNA partner in this tissue that leads to its loss-of-function phenotype. We have identified many abundant placental microRNAs that could underlie the observed effects via their association with Ago2 (Supplementary Fig. 4). Catalytic competence is, however, required for viability. Mice lacking a catalytic Ago2 became pale after birth and generally died within a few hours (Fig. 2a). Histological examination revealed no gross morphological defects. However, the appearance of these animals was strongly indicative of anaemia.
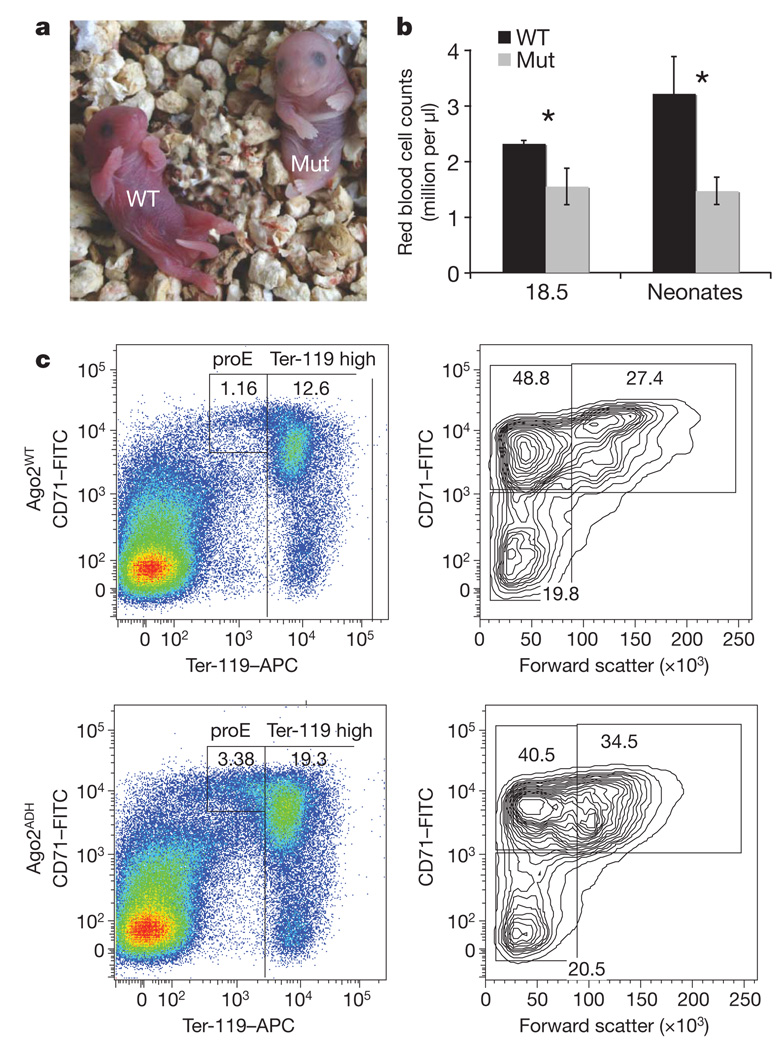
Figure 2. Ago2 catalysis is essential for development.
a, Representative neonates from Ago2ADH heterozygous inter-crosses. WT, wild type; Mut, Ago2 homozygous mutant. b, Peripheral blood count of litter mates from WT and Mut. Data are the mean ± s.d. *t-test (unequal variance for embryonic day 18.5 time point P = 0.035, equal variance for the birth time point P = 1.95 × 1027). c, Representative FACS analysis using CD71/Ter-119 erythroid population marking of individual bone marrow samples of mutant versus wild-type litter mates. Three independent pairs from three different litters showed virtually identical profiles. APC, allophycocyanin; FITC, fluorescein isothiocyanate; FACS, fluorescence-activated cell sorting.
To address this possibility, we performed a complete blood count. By embryonic day 18.5, red blood cell (RBC) counts in mutants begin to deviate from those of control animals (Fig. 2b). This was even more marked at birth, with mutants showing a roughly 50% reduction in peripheral RBCs. This seems specific to erythroid cells, because no alteration in the abundance of other cell types within the haematopoietic compartment was observed.
Loss of circulating RBCs could result from haemolysis, haemorrhage, or a defect in erythropoiesis. We therefore assessed RBC production in the haematopoietic sites of neonates using Ter-119 and CD71 double marking of differentiating erythroblast cell populations (Fig. 2c). We detected a two- to threefold proportional increase in proerythroblasts (Pro-E) in liver, bone marrow and spleen in the AgoADH homozygotes. These results indicated that Ago2 catalysis might be important for erythroid maturation during the transition from pro-E to basophilic erythroblasts.
Non-canonical biogenesis of an erythropoietic miRNA
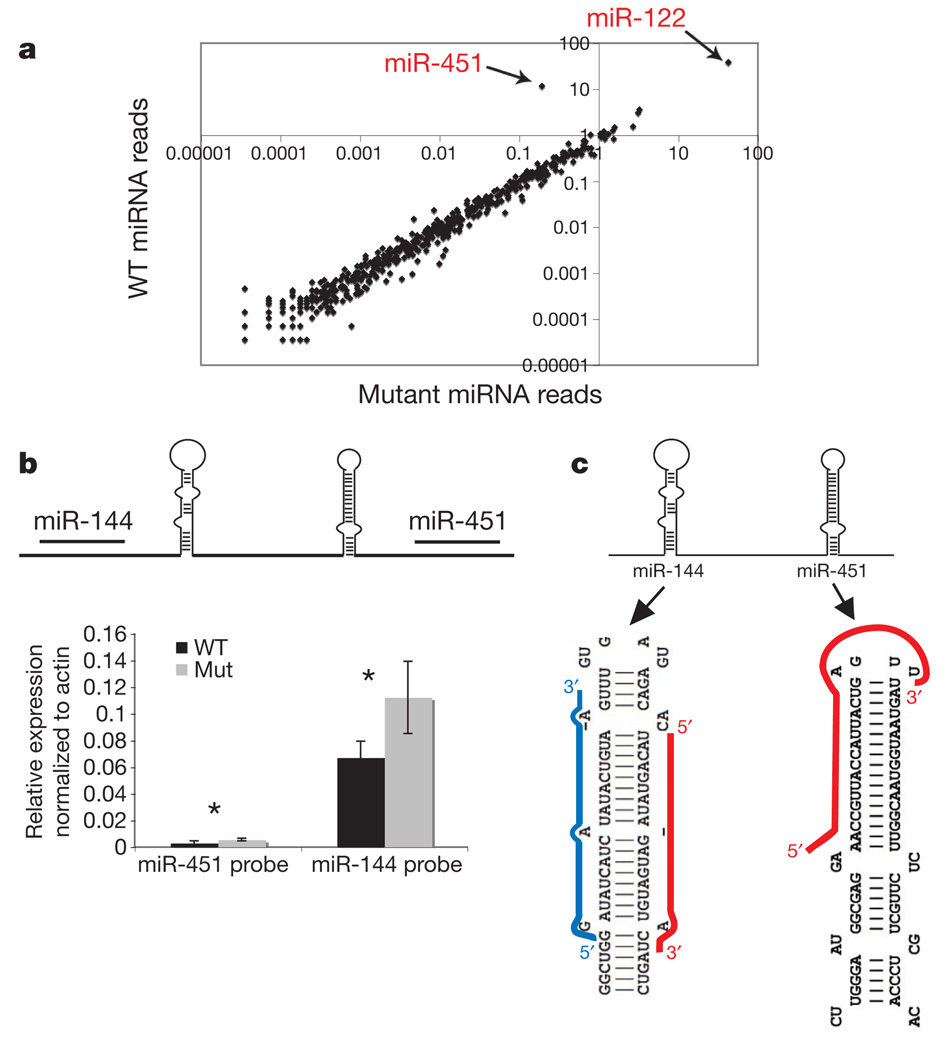
Our results indicated that miRNA-directed target cleavage might prove important for erythrocyte maturation. As a step towards identifying such a target, we profiled miRNAs expressed in the liver, one of the fetal haematopoietic sites. Deep sequencing from wild-type animals and Ago2ADH homozygotes showed that virtually all microRNAs were present at nearly identical levels. One miRNA, miR-451, represented 11% of all miRNA reads in normal fetal liver but was markedly reduced in the mutants (Fig. 3a).
Figure 3. Mature miR-451 expression depends on Ago2 catalysis.
a, Scatter plot of miRNA reads in wild-type versus mutant fetal liver. b, Quantitative RT–PCR of primary transcript levels of miR-144 and miR-451 in wild-type and mutant liver samples. Data are the mean of three biological replicates ± s.d. *t-test with equal variance P > 0.05. c, The unique structure of the miR-451 hairpin compared to miR-144 with the miRBase annotation of mature miR-451 and miR-144 mapped to the predicted secondary hairpin structure shown. Guide strand in red and passenger strand in blue.
Previous studies have demonstrated a strong dependency of the development of pro-E into basophilic erythroblasts on the expression of miR-451 (ref. 37). Together, miR-451 and miR-144 form a microRNA cluster with robust expression in erythroid cells. This pattern can be explained in part on the basis of the presence of regulatory sites for the GATA-1 zinc finger transcription factor, which acts as a master regulator of erythroid differentiation38. The regulatory circuit seems to be intact in Ago2ADH animals, because we observe no changes in the levels of precursor pre-miR-144-451 in homozygous mutants (Fig. 3b). This strongly pointed to an impact of catalysis on miR-451 maturation rather than miR-451 expression.
MicroRNA biogenesis occurs via a two-step processing pathway wherein Drosha initially cleaves the primary microRNA transcript to liberate a hairpin pre-miRNA39. This is exported to the cytoplasm and recognized and cleaved by Dicer to yield the mature duplex, which is loaded into Ago. The passenger strand is removed through unknown mechanisms to yield a complex ready for target recognition.
An examination of the miR-451 precursor and its mature strand revealed an unusual feature. As annotated, the six terminal nucleotides of the 23-nucleotide-long mature miR-451 span the loop region and extend into the complementary strand of the hairpin precursor. This arrangement seems incompatible with the well-studied enzymatic activities of Drosha and Dicer, which would normally liberate the mature microRNA mapping to the stem only (Fig. 3c). We therefore explored the possibility that miR-451 might adopt an unusual mode of biogenesis.
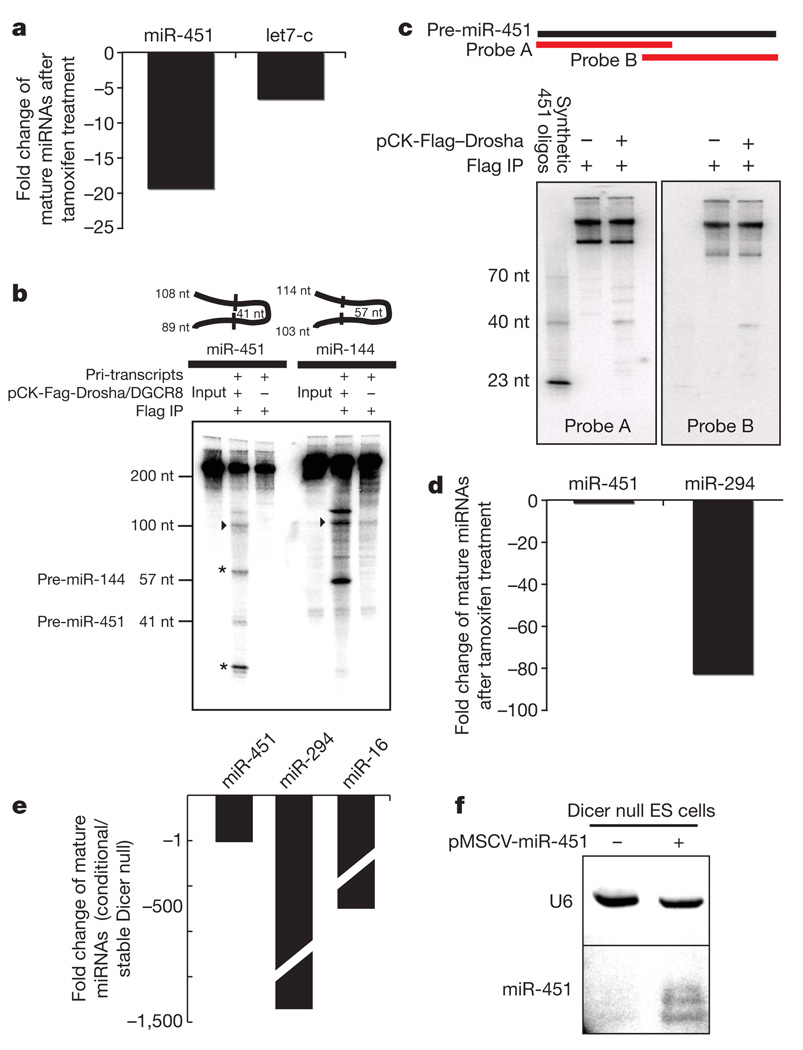
We began by assessing the dependency of miR-451 on Drosha. We created a construct that drives the expression of the miR-144/451 precursor from a strong viral promoter and introduced this into mouse embryonic fibroblasts (MEFs) homozygous for a conditional Drosha allele. After activation of Cre-ER and Drosha loss of function, we noted a 20-fold reduction in levels of mature miR-451. This was even more marked than the effect on a miRNA, let-7c, with a well-established dependency on canonical processing factors (Fig. 4a).We also assessed the ability of Drosha to liberate pre-miR-451 in vitro. Drosha complexes were affinity-purified from human 293T cells and mixed with in vitro-synthesized fragments of primary pri-miR-451 or pri-miR-144. In both cases, bands of the appropriate size for the pre-miRNA were observed (Fig. 4b). In the case of pri-miR-451 processing the 5′ flank of the transcript folds into an additional hairpin, which may be released by Drosha to give additional fragments. As a result, only one flank was observed. The identities of pre-miRNA bands were confirmed by northern blotting with oligonucleotide probes corresponding to the predicted species (Fig. 4c, not shown). Considered together, these experiments provide both genetic and biochemical support for Drosha catalysing the excision of pre-miR-451 from its primary transcript.
Figure 4. Non-canonical biogenesis of miR-451.
a, Effect on mature miRNA levels of Drosha conditional ablation in Drosha flox/flox Cre-ER MEFs. b, In vitro processing of miR-451 and miR-144 primary transcripts by Drosha immunoprecipitates (IP). Pre-miR-144 and pre-miR-451 are indicated with their corresponding expected sizes. nt, nucleotides. Additional fragments released by possible Drosha processing of the 5′ miR-451 flank are indicated with asterisks. Flanks are indicated with arrowheads. c, Northern blots for confirmation of in vitro Drosha processing assays. d, Effect on mature miRNA levels of Dicer conditional ablation in Dicer flox/flox Cre-ER ES cells. e, Effect on mature miRNA levels in Dicer null stable ES cells. f, Northern blot of mature miR-451 expression in Dicer null stable ES cells. U6 is used as a loading control. MSCV, murine stem cell virus.
Pre-miR-451 has an unusually short, 17-nucleotide stem region. Previous studies indicate that this is too short to be efficiently recognized and processed by Dicer40. We therefore examined the role of Dicer in miR-451 maturation. We introduced the pri-miR-451 expression vector into ES cells that are homozygous for Dicer conditional alleles and express tamoxifen-inducible oestrogen receptor–Cre fusion protein (Cre-ER). Whereas acute Dicer loss caused a roughly 80-fold reduction in a control ES cell microRNA (miR-294), miR-451 levels did not change (Fig. 4d). A pure population of continuous Dicer-null ES cells showed a more than 500-fold reduction in conventional microRNA, whereas levels of miR-451 were unaffected (Fig. 4e). We also confirmed these results using northern blot analysis of Dicer-null ES cells transiently expressing the miR-451 precursor (Fig. 4f). Finally, we incubated synthetic miR-451 pre-miRNA with recombinant Dicer and observed no mature cleavage products, although pre-let-7c was efficiently processed (not shown). Thus, conversion of pre-miR-451 into a mature miRNA proceeds independently of Dicer. We therefore strove to identify an alternative maturation pathway.
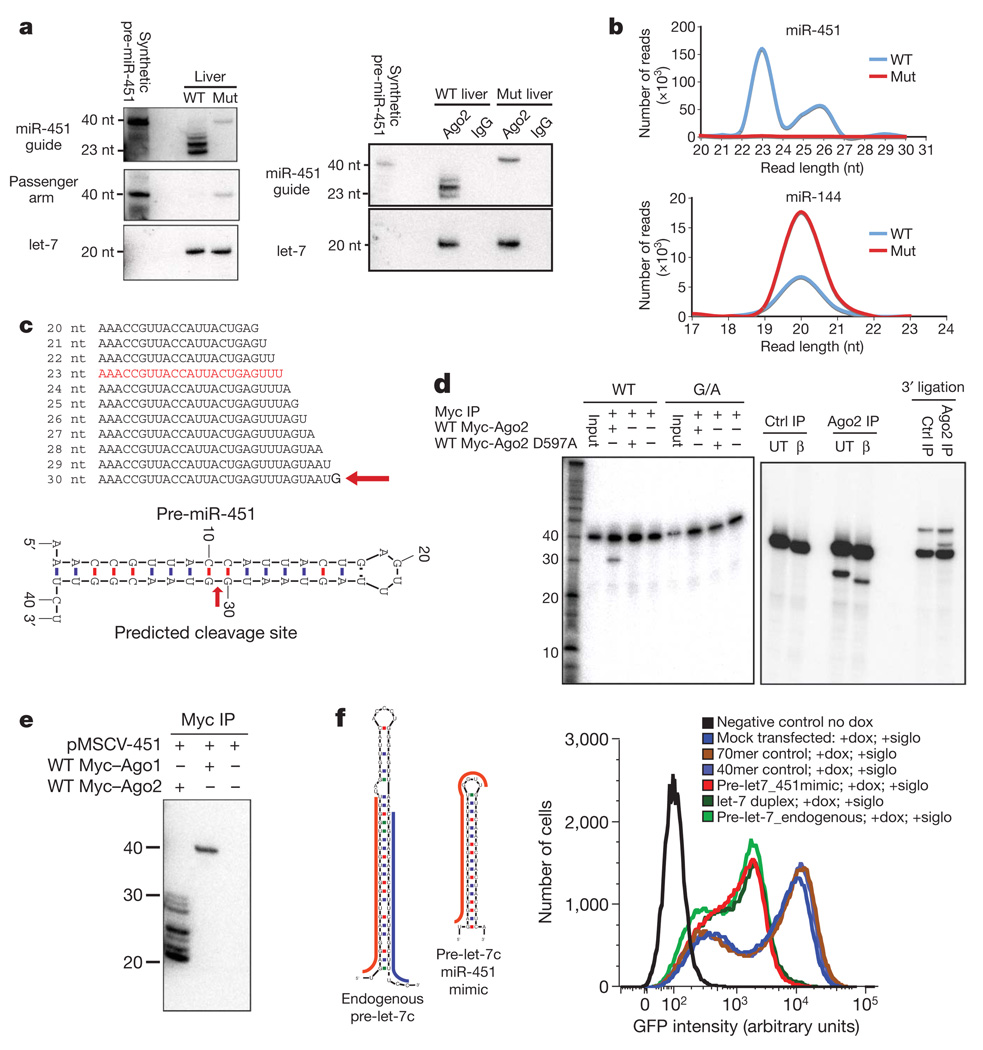
We examined miR-451 species in wild-type and Ago2ADH mutant livers by northern blotting. This confirmed loss of the mature miRNA in the mutant animals. However, we noted the appearance of a ~40- nucleotide band that co-migrated with a synthetic pre-miR-451 and hybridized to probes to its 5′ and 3′ arms (Fig. 5a). This indicated accumulation of the Drosha cleavage product in mutant animals. Notably, the same bands seen in total RNA were also detected in Ago2 immunoprecipitates (Fig. 5a). This demonstrated the direct loading of the pre-miRNA-451 into Ago2 and raised the possibility that the Ago2 catalytic centre might help to catalyse the maturation of this microRNA.
Figure 5. Ago2 catalysis is required for miR-451 biogenesis.
a, Left panels, northern blot on total RNA from WT and mutant livers probing for the miR-451 guide strand and the passenger arm of the hairpin (indicated). Let-7c is used as a loading control. Right panels, northern blots of Ago2 and IgG control immunoprecipitates from WT and mutant liver extracts with the indicated probes. b, miRNA read length distribution for the indicated miRNA from deep sequencing of WT and mutant livers. c, prediction of a miR-451 Ago2 cleavage site. Top, miR-451 3′ end heterogeneity. Bottom, predicted cleavage site at the thirtieth phosphodiester bond of pre-miR-451. d, Left gel, in vitro cleavage assay of pre-miR-451 by an Ago2 immunoprecipitate. Right gel, confirmation of the 3′ end character of the Ago2 cleavage product using beta elimination and 3′ end ligation reactions. β, beta-eliminated; UT, untreated. e, Immunoprecipitation-northern blot indicating presence of the mature form of miR-451 in Ago2 complexes and loading of pre-miR-451 without processing in Ago1 complexes. f, Left, schematic depictions of the pre-let-7c-miR-451 mimic hairpin compared to the native pre-let-7c. Guide strand in red and passenger strand in blue. Right, FACS analysis for GFP in the indicated samples. Cells were co-transfected with phycoerythrin (PE)–siRNA control. PE-positive cells (105) were gated and analysed for GFP expression. siglo is a fluorescently labelled siRNA used as a transfection control and dox is doxycycline.
The well-established biochemical properties of Ago2 predict that it would cleave a loaded pre-miR-451 after its thirtieth base. We searched for evidence of such an intermediate in fetal liver small RNA libraries encompassing an expanded size range. Plotting a size distribution of reads corresponding to a conventional miRNA, miR-144, gave the expected pattern, a sharp peak at ~20 nucleotides. In contrast, miR-451 showed a heterogeneous size distribution, exclusively because of variation at its 3′ end. One abundant species corresponded precisely to the predicted Ago cleavage product (Fig. 5b, c).
We confirmed the capacity of Ago2 to load and cleave pre-miR-451 using in vitro assays (Fig. 5d). Wild-type or catalytically inactive Ago2 complexes (Fig. 5d) were affinity-purified from 293T cells and mixed with 5′ end-labelled pre-miR-451. Only wild-type Ago2 produced the expected product, and this depended on the presence of Mg2+ (not shown). No product was produced if we provided a mutant version of the precursor in which a single point mutation disrupted pairing at the cleavage site. Beta elimination and ligation reactions confirmed that cleavage left a free 3′ OH terminus as expected of Argonaute proteins. These data strongly support a role for the Ago2 catalytic centre in miR-451 maturation. This is perhaps akin to the proposed role of passenger strand cleavage in the maturation of RISC (RNA-induced silencing complex). Ago1 could load pre-miR-451 but was unable to process it to its mature form (Fig. 5e)
To investigate sequence versus structural requirements for entry into the alternative miRNA biogenesis pathway, we created a structural mimic of miR-451 that might instead produce let-7c. At the concentration tested, this was as potent as the native pre-let-7c in suppressing a green fluorescent protein (GFP) or luciferase reporter containing perfect let-7c complementary sites (Fig. 5f and Supplementary Fig. 5a). The miR-451 precursor could also be remodelled to express a short hairpin RNA that efficiently represses p53 (also known as Trp53; Supplementary Fig. 5b). Given the unique ability of Ago2 to productively process miR-451, these data could have practical implications in the experimental use of shRNAs.
Considered together, our results suggest a model (Supplementary Fig. 6) in which miR-451 enters RISC through an alternative biogenesis pathway. Although Drosha cleavage proceeds normally, the Dicer step is skipped and the pre-miRNA is loaded directly into Argonaute. This is surprising, considering earlier studies indicating a coupling of Dicer cleavage and RISC loading41,42. Such a complex would also lack interactions between the PAZ domain and the 3′ end of a conventional Dicer product43,44. A previous report indicated the ability of RISC to accommodate such species and posited a potential for Ago cleavage in the maturation of canonical microRNAs45. However, no physiological role for such an activity was demonstrated, and we detect no measurable defects in the processing of canonical miRNAs in Ago2ADH mutants. miR-451 maturation proceeds with Ago-mediated cleavage, producing an intermediate that is further trimmed. Although this could occur via either endo- or exonucleolytic digestion, the observed distribution of 3′ ends, many bearing single non-templated U residues (Supplementary Figs 6, 7), seems more consistent with the latter model. Although the precise enzymology of this step remains obscure, preliminary studies fail to support roles for Eri-1 (also known as Eri1) or the exosome complex (not shown).
A previous report noted severe defects in erythropoiesis in Ago2-null cells46. In that case, the phenotype could be rescued by transplantation of adult haematopoietic stem cells expressing a catalytically inactive Ago2 allele. Although the two studies may seem at odds, we feel that the differences simply reflect the quantitative nature of our respective phenotypes and the fact that the strong reduction in miR-451 that we observe results in only a partial loss of RBCs. Anaemia in the Ago2 mutants could be a complex phenotype, of which miR-451 loss is only one component. Finally, in the earlier study, a different inactive Ago2 allele was expressed in adult haematopoietic stem cells from a strong viral promoter, whereas in the work presented here, a knock-in allele had its impact on the haematopoietic niche as a whole during fetal development.
Although the anaemia of the Ago2ADH animals is profound, it is unclear whether this alone is sufficient to cause the perinatal death of mutant animals. Although no other well-defined pathologies were observed, we did find haemorrhages in the lungs and intra-abdominal bleeding in some animals. Moreover, miR-451 is expressed in the gastric epithelium, where its loss could have impacts that we have as yet failed to detect47. We leave open the possibility that additional microRNAs might rely on the alternative biogenesis pathway for maturation, although none presently annotated in miRBase share the usual structure or 3′ heterogeneity of miR-451. Additional defects could also arise from a role of Ago2-mediated target mRNA cleavage. Thus far, miR-196 and microRNAs from the imprinted Rtl1 cluster have been ascribed this property; however, none of the phenotypes that we observe can be explained by defects in these particular species16–18.
The structure of the miR-451 precursor and the extension of its mature sequence around the loop and into the complementary strand of the precursor are present throughout vertebrate evolution (Supplementary Fig. 8). Thus, a conserved pathway of miR-451 maturation may provide at least some of the evolutionary pressure to maintain a catalytic Argonaute protein in animals.
METHODS SUMMARY
Mouse strains were generated as described in the Methods. Embryos were collected at embryonic day 3.5 for ES cells derivation, at midgestation for embryo phenotyping and beta-galactosidase reporter analysis, and at embryonic day 12.5 for the analysis of chimaeras. Neonates were dissected to collect peripheral blood and haematopoietic organs (liver, bone marrow and spleen) to perform a complete blood count and erythroid FACS analysis, respectively. Small RNA fractions isolated from wild-type and mutant livers were cloned and deeply sequenced as described previously48. Gene expression analysis was done by quantitative PCR with reverse transcription (RT–PCR) using custom primers or specific TaqMan probes. Immunoprecipitations were performed as described in the Methods. Northern blotting of small RNAs was performed using miRNA-specific DNA probes. In vitro Drosha, Dicer and Ago2 processing experiments were carried out as described previously or as described in the Methods20,49. The substrates for the enzymatic reactions were either in vitro [α32P] UTP-labelled transcripts or [γ32P]ATP end-labelled synthetic RNAs. The conditional cell lines were treated with tamoxifen to induce gene deletion or doxycycline to induce gene expression. All cell lines were transfected as described in the Methods and collected for gene expression analysis or reporter assays.
Supplementary Material
Acknowledgements
We thank M. Mosquera, S. Y. Kim, D. Frendewey and A. Economides for help with generating mutants and caring for animals. A. Nagy, M. Shen, J. Murn, V. Vagin and A. Aravin provided discussion. N. Kim, D. Littman, I. Ibarra and E. Wagenblast provided critical reagents, and O. Tam, R. Sachidanandam, Z. Xuan, D. McCombie and M. Rooks provided support for generation and analysis of deep sequencing data. This work was supported by grants from the NIH and by a gift from K. W. Davis.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions S.C. and G.J.H. planned experiments, interpreted data and wrote the paper. S.C. and C.O.D.S. performed studies, and M.M.W.C. provided critical unpublished reagents and discussion.
Author Information Sequencing data has been deposited in GEO and assigned accession number GSE21370. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 2.Joshua-Tor L. The Argonautes. Cold Spring Harb. Symp. Quant. Biol. 2006;71:67–72. doi: 10.1101/sqb.2006.71.048. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan YR, et al. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Bohmert K, et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 15.Davis E, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 16.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl Acad. Sci. USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekita Y, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the fetomaternal interface of mouse placenta. Nature Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- 18.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 19.Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nature Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 21.Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 22.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 23.Azuma-Mukai A, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl Acad. Sci. USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ender C, et al. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 28.Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, et al. MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh N, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alisch RS, Jin P, Epstein M, Caspary T, Warren ST. Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet. 2007;3:e227. doi: 10.1371/journal.pgen.0030227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita S, et al. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 35.Rossant J, Cross JC. In: Mouse Development: Pattering, Morphogenesis and Organogenesis. Rossant J, Tam PPL, editors. Academic; 2002. pp. 155–180. [Google Scholar]
- 36.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nature Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 37.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2009;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 38.Dore LC, et al. AGATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 40.Siolas D, et al. Synthetic shRNAs as potent RNAi triggers. Nature Biotechnol. 2004;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 41.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang HW, et al. Structural insights into RNA processing by the human RISC-loading complex. Nature Struct. Mol. Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diederichs S, Haber DA. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 46.O’Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandres E, et al. MicroRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 48.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.