Abstract
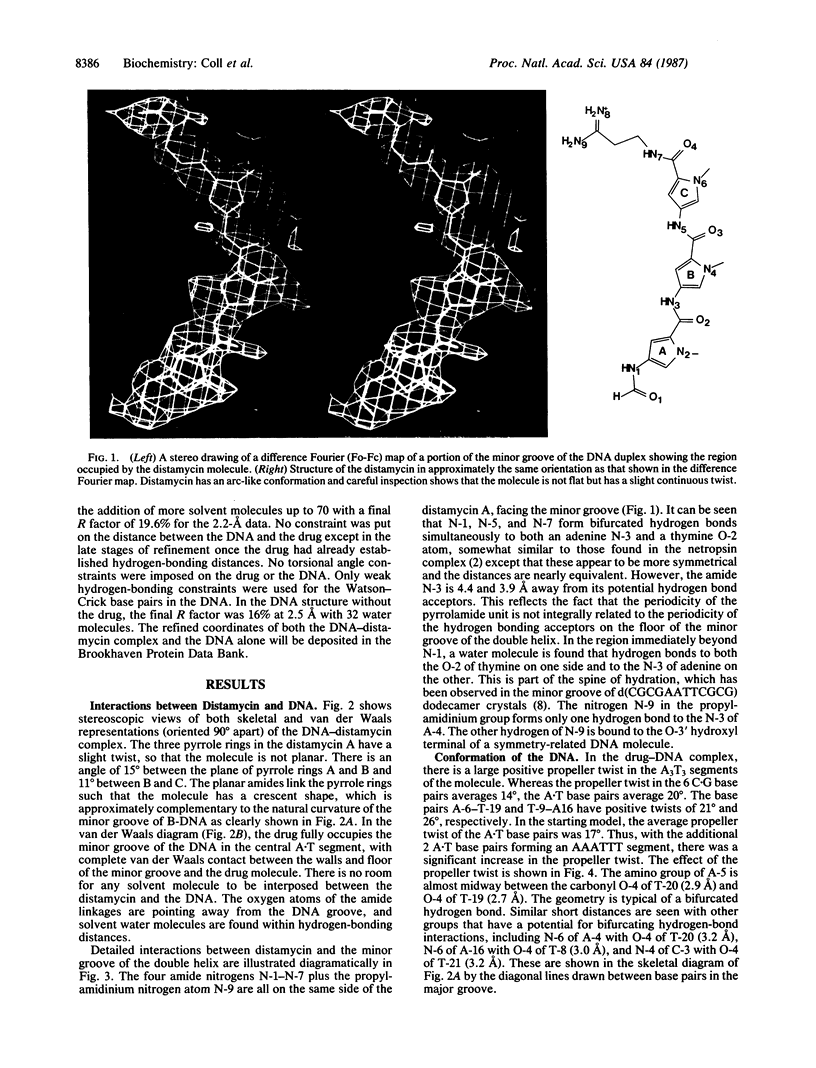
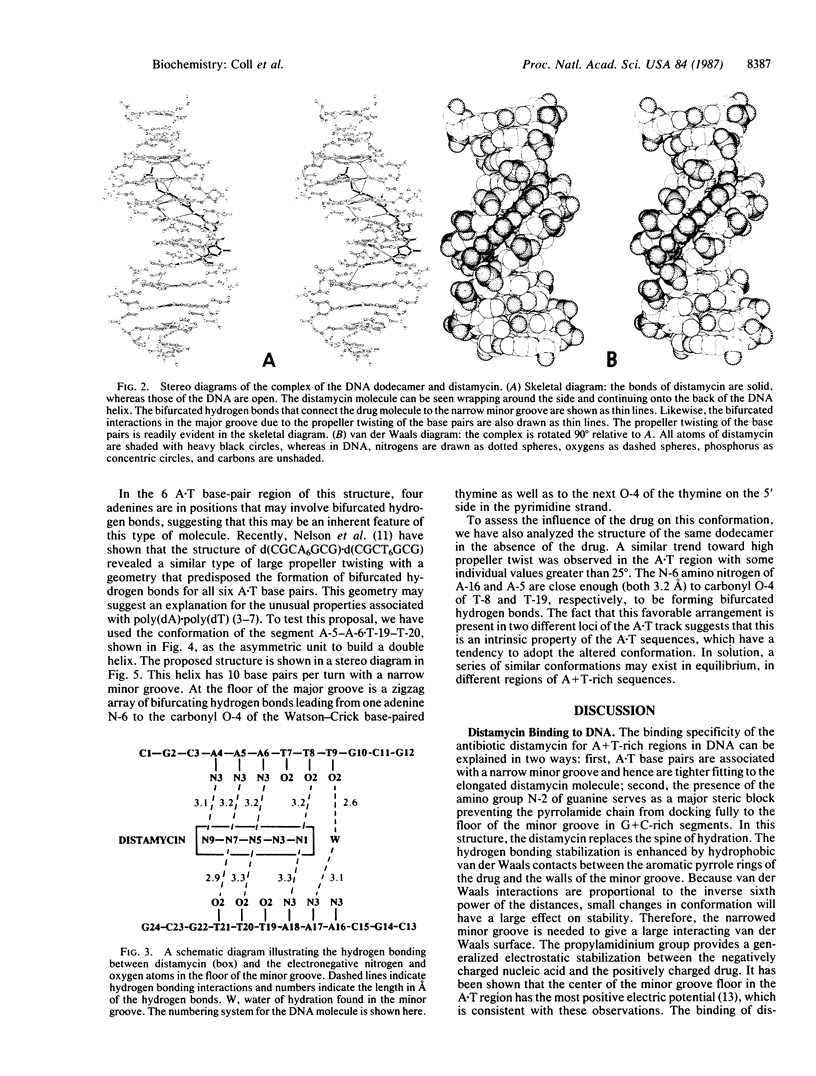
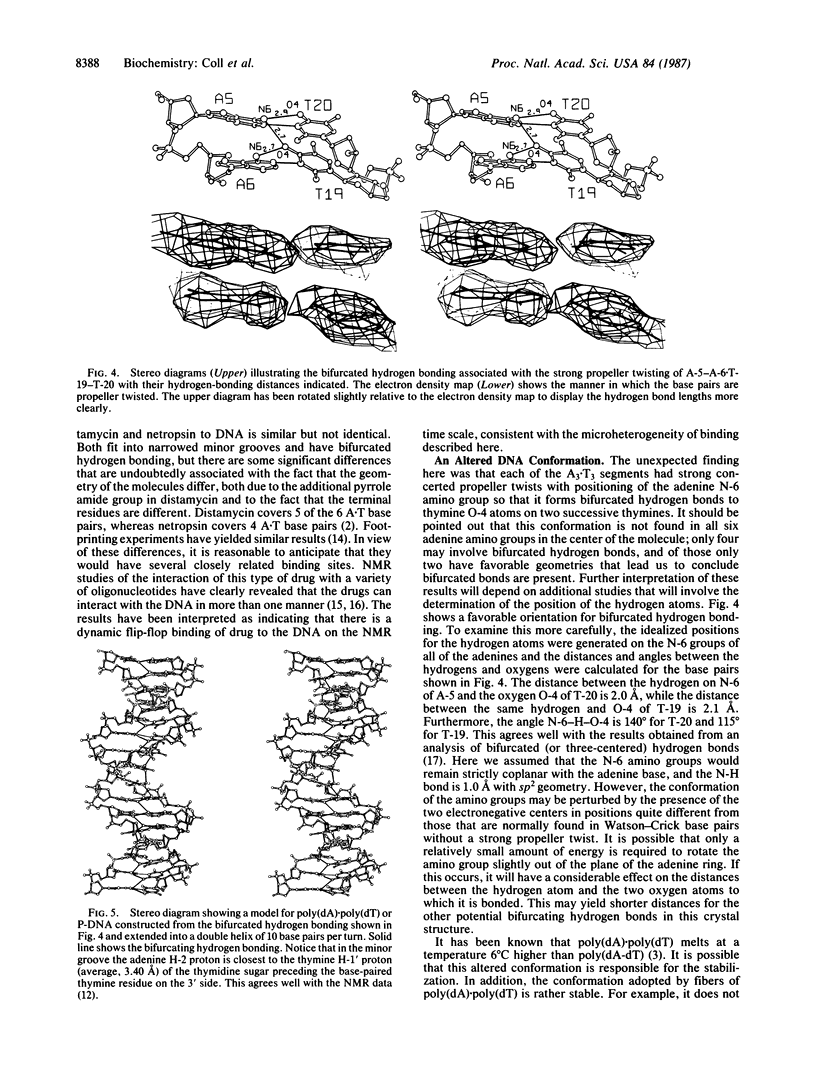
The crystal structures of d(CGCA3T3GCG) complex to the antitumor drug distamycin and the DNA fragment alone were solved by x-ray diffraction at 2.2 and 2.5 A resolution, respectively. The drug lies in the narrow minor groove near the center of the B-DNA fragment covering 5 of the 6 A.T base pairs. It is bound to the DNA by hydrogen bonding, van der Waals, and electrostatic interactions. In addition, the DNA was found to have an unusual conformation in the (dA)3.(dT)3 regions. These base pairs have a high positive propeller twist so that in the major groove the adenine amino group is located intermediate between the carbonyl O-4 groups of two adjacent thymines of the opposite strand, making bifurcated hydrogen bonds to the two thymine residues. This suggests a model to explain the unusual properties of poly-(dA).poly(dT) in which a modified B conformation is associated with a large propeller twist of the bases and a set of continuous bifurcating hydrogen bonds along the major groove, which may provide incremental stability to these segments. In addition, shorter segments of (dA)3-6.(dT)3-6 may have this conformation in the midst of B-DNA and stabilize bends in the DNA that may be associated with stacking on one of the high propeller-twisted bases at the ends of these segments.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexeev D. G., Lipanov A. A., Skuratovskii IYa Poly(dA).poly(dT) is a B-type double helix with a distinctively narrow minor groove. 1987 Feb 26-Mar 4Nature. 325(6107):821–823. doi: 10.1038/325821a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Behling R. W., Kearns D. R. 1H two-dimensional nuclear Overhauser effect and relaxation studies of poly(dA).poly(dT) Biochemistry. 1986 Jun 3;25(11):3335–3346. doi: 10.1021/bi00359a037. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Wemmer D. E., Reid B. R. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986 Jun 3;25(11):3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J. Netropsin . dG-dG-dA-dA-dT-dT-dC-dC complex. Antibiotic binding at adenine . thymine base pairs in the minor groove of the self-complementary octanucleotide duplex. Eur J Biochem. 1979 Sep;99(2):369–378. doi: 10.1111/j.1432-1033.1979.tb13265.x. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6(5):1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P. G., Dervan P. B. Distamycin and penta-N-methylpyrrolecarboxamide binding sites on native DNA. A comparison of methidiumpropyl-EDTA-Fe(II) footprinting and DNA affinity cleaving. J Biomol Struct Dyn. 1984 Mar;1(5):1133–1147. doi: 10.1080/07391102.1984.10507508. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Zakrzewska K., Lavery R., Pullman B. Theoretical studies of the selective binding to DNA of two non-intercalating ligands: netropsin and SN 18071. Nucleic Acids Res. 1983 Dec 20;11(24):8825–8839. doi: 10.1093/nar/11.24.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]