Abstract
Background
During B-cell development, precursor B cells transiently express the pre-B-cell receptor composed of μ heavy chain complexed with VpreB and λ5 surrogate light chain polypeptides. Recent profiling studies unexpectedly revealed abundant transcripts of one member of the VpreB family, VpreB3, in a subset of mature B cells and Burkitt lymphoma.
Design and Methods
Here we used a novel antibody to investigate the normal expression pattern of VpreB3 protein in human hematopoietic and lymphoid tissues, and to determine whether VpreB3 could serve as a useful diagnostic biomarker for select B-cell lymphomas.
Results
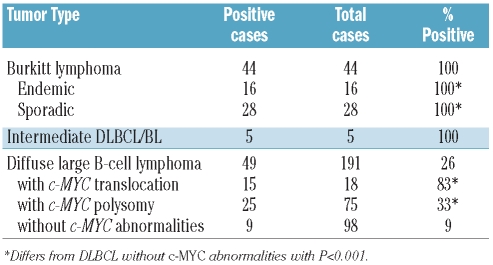
We found that VpreB3 protein is normally expressed by precursor B cells in bone marrow and by a subset of normal germinal center B cells in secondary lymphoid organs. Among lymphoid malignancies, we found an association between VpreB3 expression and B-cell tumors with c-MYC abnormalities. VpreB3 was highly expressed in all cases of Burkitt lymphoma, whether of endemic or sporadic origin (44/44 cases, 100%), all cases of B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma (5/5 cases, 100%), and the majority of diffuse large B-cell lymphomas harboring a c-MYC translocation (15/18 cases, 83%). The expression of VpreB3 in diffuse large B-cell lymphomas without a c-MYC translocation was associated with c-MYC polysomy in 25/75 cases (33%) but only rarely observed in diffuse large B-cell lymphomas lacking a c-MYC abnormality (9/98 cases, 9%).
Conclusions
We conclude that for B-cell tumors with features suggesting a possible c-MYC translocation, such as intermediate to large cell size and high proliferation rate, the presence of VpreB3 should prompt subsequent confirmatory genetic testing, whereas the absence of VpreB3 is virtually always associated with wild-type c-MYC alleles.
Keywords: VpreB3, pre-BCR, immunohistochemistry, lymphoma, Burkitt lymphoma, diffuse large B-cell lymphoma, c-Myc
Introduction
B-cell development in humans and mice begins in the bone marrow where it is intimately linked with the orderly rearrangement of variable (V), diversity (D) and joining (J) gene segments encoding the immunoglobulin heavy chain, and the V and J gene segments encoding the immunoglobulin light chain.1 Rearrangement of the heavy chain locus precedes rearrangement of the light chain locus, and a critical developmental step in B-cell ontology is the assembly of the nascent μ heavy chain polypeptides with the surrogate light chain proteins VpreB and λ5 within the pre-B-cell receptor (pre-BCR).2–4 Expression and signaling through the pre-BCR is necessary for further B-cell development and mice defective in many of the component genes encoding the pre-BCR, therefore, show a developmental arrest at the pre-B-cell stage.5–7
The human genome includes three VpreB genes. VpreB1 and VpreB2 appear to be the major family members associated with cell surface pre-BCR complexes.4 Very little is known about the expression and function of VpreB3 due, in part, to the lack of a VpreB3-deficient mouse.4,8,9 It has been discussed whether VpreB3 has a role in the chaperoning and/or assembly of pre-BCR complexes prior to surface expression since VpreB3 has been detected in association with μ heavy chain within the endoplasmic reticulum but not at the cell surface of pre-B-cell lines.10 Nevertheless, the major biological functions of VpreB3 during B-cell development remain to be established, and whether VpreB3 protein is expressed or has biological roles beyond early B-cell development is unknown.
Burkitt lymphoma (BL) is an aggressive B-cell tumor of germinal center cell origin. By definition BL harbors, usually as an isolated karyotypic abnormality, a chromosomal translocation involving the c-MYC locus that results in dysregulated expression of the c-Myc protein.11,12 In routine surgical pathology practice, the most common differential diagnosis for BL is diffuse large B-cell lymphoma (DLBCL) - a more frequent tumor of mature B cells that only rarely harbors a c-MYC translocation. Although the distinction between BL and DLBCL can often be made based on morphological and immunophenotypic features alone, no single phenotypic marker can uniformly distinguish these two tumor types and ambiguous cases are often encountered.13 Nevertheless, the proper classification of a tumor as BL or DLBCL is of paramount importance, as these tumors exhibit distinct biological behaviors and are treated with different chemotherapeutic regimens.11,14–16
Recently, it has become apparent that rare cases of DLBCL lacking the morphological and/or immunophenotypic features of BL can harbor a c-MYC translocation (MYC+ DLBCL).17 These tumors respond poorly to conventional, DLBCL-based chemotherapeutic regimens and might, therefore, be considered for BL-type regimens in the future.18 Taken together, these data raise the question of whether all B-cell lymphomas with intermediate to large cell morphology and a high proliferation fraction should be screened for a c-MYC abnormality, despite the rarity of the genetic lesion. The development of an immunohistochemical assay that is highly sensitive for tumors with a c-MYC translocation could prove a useful method to prevent unnecessary genetic testing for the majority of aggressive B-cell lymphomas.
Here we used a novel anti-VpreB3 antibody to study the expression pattern of VpreB3 protein in normal lymphoid tissues and human B-cell malignancies.
Design and Methods
Antibodies, immunohistochemistry and evaluation
Three affinity-purified polyclonal antibodies raised against specific regions of the human VpreB3 protein were evaluated in frozen and formalin-fixed, paraffin-embedded tissue sections of human reactive tonsils. Only one antibody (raised against a protein sequence covering the immunoglobulin domain of VpreB3) was selected for this study based on its reactivity in paraffin-embedded tissue sections and background-free staining using both manual and automated immunohistochemistry protocols.22–24 Specificity of the antibody was confirmed by western blotting using protein lysates of the BL-derived cell lines Ramos and Daudi (Online Supplementary Figure S1E), previously shown to contain VpreB3 transcripts.8 Of the remaining two antibodies, one did not show tissue reactivity and the other labeled only plasma cells and was, therefore, excluded. Optimal immunohistochemical staining was obtained at 1:25 dilution and using a heat-induced epitope retrieval protocol with an EDTA-based solution. Details of the reagent used in this study are available on request to the corresponding author. Single and double immunohistochemistry were performed using protocols described previously.24
Cell lines and western blotting
Paraffin-embedded cell pellets of human lymphoma-derived cell lines were studied for VpreB3 expression by conventional immunohistochemistry following a protocol described elsewhere.22 The investigated cell lines included the DLBCL lines OCI-Ly3 and OCI-Ly10 (from Dr. R.E. Davis, Center for Cancer Research, NCI, Bethesda, USA); the follicular center lymphoma line FL-18 and the BL line Daudi (both provided by the Sir William Dunn School of Pathology, Oxford, UK); two other BL lines, Namalwa and Ramos, were obtained from the collection of the authors (JCP and TM), the T lymphoblastic leukemia line CCRF-CEM was a gift from Prof. E. Macintyre (Hôpital Necker-Enfants Malades, Paris, France). Protein extracts from some of the aforementioned cell lines were subjected to western blotting analysis following a protocol previously described.23 An anti-β-actin antibody (ab6276, Abcam, Cambridge, UK) was used as a control for protein loading.
Case selection
The samples of neoplastic lymphoid tissues comprised 635 B-, T- and Hodgkin’s lymphomas from the authors’ (SAP, LKT, MAP, TM) case files, the British Columbia Cancer Agency, and Brigham & Women’s Hospital and were analyzed with respective institutional review board approval. The original diagnoses were based on the combined morphological, phenotypic and cytogenetic features of the tumors and defined according to the World Health Organization (WHO) classification system.11,25
Patients with endemic BL (of Ugandan origin) ranged in age from 2 to 32 years with a median age of 8 years. Patients with sporadic BL ranged in age from 7 to 80 years with a median age of 42 years. All DLBCL samples were from adult patients. Among the cases analyzed, there are four that were originally classified as “atypical Burkitt lymphoma” and one case originally classified as “Burkitt-like lymphoma” according to the 2001 WHO criteria.25 Each of these cases were subsequently classified as “B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma” (intermediate DLBCL/BL) using the 2008 WHO criteria upon re-review of the morphological and phenotypic characteristics of these cases (Online Supplementary Table S1).11 All diagnoses were confirmed by one or more of this study’s authors. All stained cases were evaluated by at least two histopathologists (SR and TM), and considered positive for VpreB3 expression if 25% or more of the tumor cells stained positive. Approval for this study was obtained from the Oxford Research Ethics Committee B (Research Ethics Committee reference number: C02.162).
Classification and analyses of diffuse large B-cell lymphomas
DLBCL cases from Brigham & Women’s Hospital (n=50) were subclassified into cell-of-origin and comprehensive consensus cluster groups by gene expression profiling analysis as previously described.26,27 DLBCL cases from the British Columbia Cancer Agency (n=127) were subclassified into cell-of-origin groups using immunohistochemistry as previously described.28 Translocations and polysomy involving c-MYC were identified using a fluorescent in situ hybridization “break-apart” probe-set from Vysis/Abbott (Abbott Park, IL, USA). A subset of cases was screened for c-MYC polysomy by a chromogenic in situ hybridization technique in collaboration with Ventana Medical Systems (Roche Diagnostics). For each case at least 50 nuclei were counted and at least 5% of the nuclei had to show an abnormal hybridization signal to be considered positive for a c-MYC translocation or polysomy. There were no statistically significant differences in the overall proliferation rate (based on Ki67 staining) among DLBCL cases grouped according to c-MYC status (data not shown). Statistical analyses were performed using Student’s T-test and GraphPad software (La Jolla, CA, USA).
Results
Expression of VpreB3 in hematopoietic cell lines
Validation of the VpreB3 antibody was carried out in a selection of mature B-cell lymphoma-derived lines using immunohistochemistry and western blotting techniques. Immunostaining of cell pellets demonstrated strong VpreB3 expression in the BL line Ramos (Online Supplementary Figure S1A), and moderate expression in the BL lines Daudi and Namalwa (Online Supplementary Figure S1B-C). The DLBCL lines OCI-Ly3 and OCI-Ly10 (Online Supplementary Figure S1D), and the T-cell leukemia/lymphoma line CCRF-CEM, showed no VpreB3 expression. Western blotting of Ramos cell lysate (Online Supplementary Figure S1E) showed a strong band of the expected size of VpreB3 (13 kDa). OCI-Ly3, OCI-Ly10 and CCRF-CEM lymphoma-derived cell lines were negative. A weak band of the expected size was observed in the other BL-derived line Daudi. These findings show that the antibody recognizes a single protein of the expected size for VpreB3 in a subset of B-cell lymphoma-derived lines, including lines previously shown to express the VpreB3 transcript.8
Expression of VpreB3 in hematopoietic and lymphoid tissues
Immunohistochemical analysis of formalin-fixed, paraffin-embedded bone marrow trephine biopsies revealed only occasional VpreB3+ lymphoid cells (Figure 1A). Double immunostaining for VpreB3 and the B-lineage transcription factor PAX5 showed that virtually all VpreB3+ cells in the bone marrow were PAX5+ (data not shown). However, PAX5+/VpreB3− cells were also seen and may represent mature B cells, which account for approximately 1–3% of bone marrow cells under normal conditions.11 Staining for TdT, a marker of precursor B and T cells, demonstrated that many, but not all, VpreB3+ cells co-express TdT, consistent with lymphoblasts (Figure 1A, inset). Early and mature erythroid and myeloid cells and megakaryocytes were negative for VpreB3.
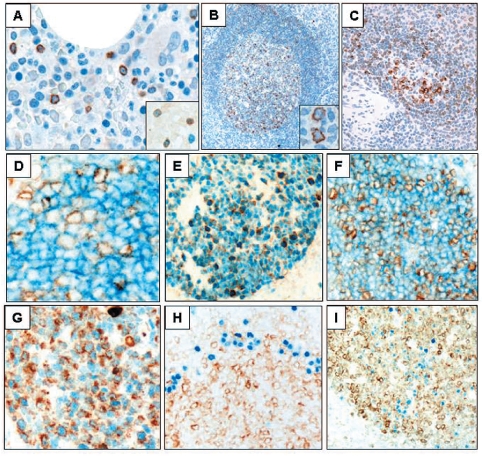
Figure 1.
The expression of VpreB3 (brown) in human hematopoietic and lymphoid tissues. Bone marrow biopsies (A) showing, scattered VpreB3 positive lymphoid cells (hematoxylin counterstain for nuclei, 400x), and (inset) partial co-localization of VpreB3 with the lymphoid immaturity marker TdT (dark blue nuclear, no hematoxylin counterstain, 1000x). Reactive tonsil (B) showing VpreB3 staining in a subset of lymphoid cells concentrated within germinal centers (hematoxylin counterstain, 100x, inset 1000x). Spleen (C) showing VpreB3 staining in a subset of lymphoid cells within the white pulp (hematoxylin counterstain, 100x). Tonsil (D-I) co-stained for VpreB3 (brown) and (D) CD10, (E) BCL6, (F) HGAL, (G) LMO2, (H) MUM1/IRF4, and (I) BLIMP1 (blue stains, no hematoxylin counter-stain, D and G at 1000x; E, F, H and I at 200x).
We next examined VpreB3 expression in secondary lymphoid organs. In the tonsil, VpreB3 positivity was observed in cells populating both the light and dark zones of germinal centers (Figure 1B), although the greatest number of positive cells were concentrated in the proliferative dark zone corresponding to approximately 50% of all cells. Morphologically, the VpreB3+ cells comprised both centroblasts and centrocytes (Figure 1B, inset). In the mantle zones, only occasional lymphoid cells showed weakly positive staining; furthermore, only rare positively-stained lymphocytes were found in the T-cell-rich interfollicular areas.
In the spleen, VpreB3 expression was predominantly confined to the white pulp, labeling germinal center B cells and scattered cells in the mantle and marginal zones (Figure 1C). Little to no staining was found in the red pulp.
Taken together, these data indicate that VpreB3 is expressed by immature lymphoid cells of B lineage within the bone marrow and, unexpectedly, by a subset of mature lymphocytes, most frequently those within reactive germinal centers of secondary lymphoid organs.
VpreB3 is expressed by mature germinal center B cells
We next examined the phenotype of VpreB3-expressing cells localized to germinal centers. Double immunolabeling showed that a subpopulation of VpreB3+ cells in germinal centers co-express the germinal center markers CD10, BCL6, HGAL, LMO2 (Figure 1D–G, respectively), LRMP1/Jaw1 and GCET1 (data not shown).23,29–31 VpreB3+ cells rarely co-localized with the post-germinal center cell markers MUM1/IRF4 and BLIMP1 (Figure 1H–I).32 These findings demonstrate that VpreB3 expression is characteristic of a subset of mature, germinal center B-cells.
VpreB3 expression in Burkitt lymphoma and diffuse large B-cell lymphoma with c-MYC translocations
Data from gene expression profiling (GEP) studies have shown that high levels of VpreB3 transcript are characteristic of tumors carrying the pathological diagnosis of BL and bearing an IgH-cMYC fusion (Online Supplementary Figure S2).19,20 This finding prompted us to examine VpreB3 protein expression in a cohort of BL cases. VpreB3 expression was 100% sensitive for BL cases, being found in all 44 screened BL cases (of both endemic and sporadic origin) (Figure 2A–B, Table 1). In addition, we found that all five cases with morphological and/or immunophenotypic features intermediate between BL and DLBCL (intermediate DLBCL/BL), but bearing a c-MYC translocation (Online Supplementary Table S1) showed robust staining for VpreB3 (Table 1). We conclude that VpreB3 is an excellent marker of c-MYC translocations being universally expressed by BL and intermediate DLBCL/BL.
Figure 2.
Expression of VpreB3 in B-cell tumors. Representative staining for VpreB3 (brown) in (A) endemic BL, (B) sporadic BL, and (C) DLBCL with c-MYC translocation. (D) Chromogenic in situ hybridization staining for c-MYC (blue dots) and chromosome 8 (red dots) in a case of DLBCL showing extra copies of c-MYC (>2 dots per cell) in a subset of cells, and (E) staining for VpreB3 in the same case. (F) Positive staining for VpreB3 in a case of B lymphoblastic leukemia. BL: Burkitt lymphoma, DLBCL: diffuse large B-cell lymphoma.
Table 1.
Association of VpreB3 expression with c-MYC abnormalities.
Approximately 5–10% of cases of DLBCL harbor a c-MYC translocation.19 Currently there are no known morphological or phenotypic characteristics that can be reliably used to distinguish MYC+ DLBCL from MYC− DLBCL and establishing these diagnoses requires genetic testing.17 Of 18 cases of MYC+ DLBCL, we found that 15 (83%) expressed VpreB3 protein (Figure 2C, Table 1). In contrast, among 173 cases of MYC− DLBCL, 34 cases (20%) showed VpreB3 protein expression. The difference in VpreB3 expression between these two groups is highly significant (P<0.01) and suggests either a direct or indirect a role for c-Myc in the expression of VpreB3 in this tumor type.
Correlation between VpreB3 expression and c-MYC polysomy in diffuse large B-cell lymphoma
We next explored whether the 34 VpreB3+ DLBCL that lacked a c-MYC translocation shared any genetic or phenotypic features that would distinguish them from VpreB3− DLBCL. Intriguingly, 25 of the 34 cases (74%) showed polysomy for the c-MYC locus as determined by fluorescent or chromogenic in situ hybridization (Table 1, Figure 2D–E). These VpreB3+ cases represented 33% of all DLBCL with more than two copies of c-MYC (Table 1). In the vast majority, c-MYC was present in three or four copies per cell. In contrast, among cases without a c-MYC abnormality, only 9% (9/98 cases) expressed VpreB3, a difference that was statistically significant (P<0.01). We, therefore, conclude that, in addition to its association with a c-MYC translocation, VpreB3 expression is associated with increased c-MYC copy number in a minority of cases.
Correlation between VpreB3 expression and the cell of origin in diffuse large B-cell lymphomas
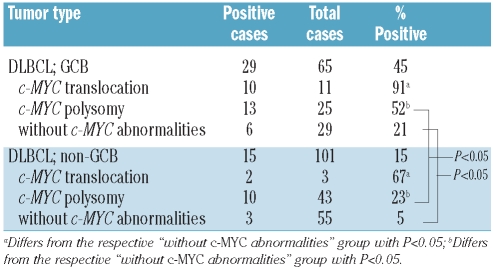
DLBCL may be subdivided by transcriptional profiling according to their cell of origin into germinal center B-cell (GCB) or non-GCB types.26 The expression of VpreB3 in a subset of normal GCB led us to investigate whether or not VpreB3+ DLBCL would preferentially fall into the GCB category. By sorting the cohort of DLBCL cases according to their cell of origin, we found increased VpreB3 expression among tumors classified as GCB (29/65 cases, 45%, Table 2) relative to non-GCB (15/101 cases, 15%, Table 2) - a difference of statistical significance.
Table 2.
Association of VpreB3 expression with cell of origin.
This analysis was, however, complicated by the observation that DLBCL cases with a c-MYC translocation or with c-MYC polysomy were more frequently of the GCB subtype (Table 2). Nevertheless, among the DLBCL of GCB type, VpreB3 was expressed by 91% of cases with a c-MYC translocation, 52% with c-MYC polysomy, and 21% without a c-MYC abnormality (Table 2). These differences are statistically significant and indicate that VpreB3 expression correlates with c-MYC status among DLBCL of the GCB subtype. Similarly among the non-GCB DLBCL, VpreB3 was expressed by 67% of cases with a c-MYC translocation, 23% with c-MYC polysomy and 5% without a c-MYC abnormality (Table 2). These differences are also statistically significant (P<0.05) and indicate that VpreB3 expression correlates with c-MYC status among DLBCL of non-GCB type. Given that we found only three non-GCB DLBCL with a c-MYC translocation (Table 2), some of these results will need to be validated in additional, larger cohorts. We did not find an association between VpreB3 expression and the subclassification of DLBCL by the comprehensive consensus cluster scheme (data not shown).27 The above data suggest that, independently of c-MYC status, the differentiation state of the DLBCL can contribute to VpreB3 expression in a minority of cases.
Correlation between VpreB3 expression and c-Myc transcript abundance
Our analyses (Table 1), coupled with published GEP data on B-cell lymphomas bearing a c-MYC translocation (Online Supplementary Figure S2), indicate a close association between VpreB3 expression and c-Myc transcript abundance in tumors with genetic abnormalities in c-MYC. We, therefore, investigated whether VpreB3 protein expression could serve as a marker of elevated c-Myc transcripts in DLBCL cases lacking a c-MYC translocation. Among 45 cases of MYC− DLBCL for which we have GEP data (12 with c-MYC polysomy; 33 without a c-MYC abnormality), we did not find a statistical association between VpreB3 and c-Myc transcript abundance (Online Supplementary Figure S3, Pearson correlation coefficient= 0.198). By this assay, we also failed to find a statistical association between c-MYC polysomy and c-Myc transcript abundance (Online Supplementary Figure S3; data not shown). The lack of correlation between VpreB3 expression, determined by immunohistochemistry, and c-Myc transcript abundance, determined by GEP, in our cohort of DLBCL might be due to small differences in gene expression levels within this group, which are not optimally quantified by GEP, as well as to the limited sample size. This analysis contrasts with the markedly elevated c-Myc transcript levels that are consistently seen in BL when large cohorts of BL are directly compared to DLBCL by GEP (Online Supplementary Figure S2). Thus, to the limits of the sensitivity of our assay, we did not find a correlation between VpreB3 and c-Myc transcript levels which is independent of the tumors’ genetic status.
VpreB3 expression and clinical outcome in patients with diffuse large B-cell lymphoma
The potential prognostic value of VpreB3 was analyzed in DLBCL. For this analysis we included a separate cohort of cases (different from those studied by GEP) but for which clinical outcome data were available. No significant difference in overall survival was found between patients with VpreB3+ and VpreB3− tumors treated with a standard chemotherapeutic regimen (R-CHOP, Online Supplementary Figure S4).
VpreB3 expression in additional lymphoma types
Staining for VpreB3 was extended to a broader survey of non-Hodgkin and Hodgkin lymphomas. Among small B-cell lymphomas the tumor with the highest percentage (40%) of VpreB3 expression was B-cell acute lymphoblastic leukemia (4/10 cases, Figure 2F; Online Supplementary Table S2). The findings suggest that although VpreB3 is expressed by immature B cells (Figure 1), VpreB3 is not a universal marker of B-cell acute lymphoblastic leukemia (at least to the limits of the sensitivity of our antibody). VpreB3 expression was also observed in a minority of other mature B-cell neoplasms including primary mediastinal large B-cell lymphoma, mantle cell lymphoma, and chronic lymphocytic leukemia (Online Supplementary Table S2). An analysis of VpreB3 and c-Myc transcript levels in primary mediastinal large B-cell lymphoma did not reveal coordinate regulation of these genes in this tumor type (Pearson’s correlation coefficient=0.164, sig.=0.354; Online Supplementary Figure S3B). Similarly, an examination of the published mantle cell lymphoma and chronic lymphocytic leukemia GEP data failed to show an overt correlation between VpreB3 and c-Myc transcript abundance in these tumor types (data not shown).
Discussion
The expression and biological roles of VpreB3 in normal B-cell development remain poorly characterized. Initial data indicated that VpreB3 is an important member of the pre-BCR complex in precursor B cells.8,10 However, recent expression profiling studies detected VpreB3 transcripts in mature B cells and derived tumors.6,19,20 In this study we show that VpreB3 is expressed by a subset of GCB as also demonstrated by its co-expression with germinal center-associated molecules i.e. BCL6, GCET1, HGAL, LMO2, and LRMP1/Jaw1 and absent in post-germinal center cells (VpreB3+ cells were MUM1−/IRF4− and BLIMP1−). These results raise the question as to whether VpreB3 serves biological roles beyond those in early B-cell development. One possibility, currently under investigation, is whether in mature B cells, VpreB3 coincides with AID expression or whether VpreB3 interacts with nascent somatically mutated or class-switched Ig during the germinal center reaction.
The evidence that VpreB3 transcripts were detected in BL (Online Supplementary Figure S1)19,20 prompted us to study the diagnostic usefulness of VpreB3 protein expression in BL. VpreB3 was universally expressed by BL of both endemic and sporadic origin indicating that its immunohistochemical detection serves as a relevant marker of BL. Moreover, our observation that all five cases classified as intermediate DLBCL/BL and harboring a c-MYC translocation were positive for VpreB3, points to VpreB3 as being a useful molecule for identifying this type of tumor.
Unexpectedly, we found that 26% of DLBCL express VpreB3 (Table 1). Further analyses of this heterogenous group of tumors revealed that VpreB3 identifies the majority of cases of DLBCL harboring a c-MYC translocation as well as a minority of DLBCL with polysomy for c-MYC and a minority of DLBCL of GCB origin (Table 2). Despite these associations, we did not find a general correlation between VpreB3 and c-Myc transcript levels in DLBCL or other B-cell tumors by GEP which was independent of an underlying genetic abnormality in c-MYC (Online Supplementary Figure S3).
Collectively, our findings have several implications. First, the presence of VpreB3 in cases classified as intermediate DLBCL/BL and in the majority of DLBCL harboring a c-MYC translocation suggests that at molecular level these tumors resemble BL.19,20 Second, the association between VpreB3 expression and a subset of DLBCL with c-MYC polysomy raises the possibility that, in a minority of cases, increased c-MYC copy number may partially recapitulate the genetic program of tumors with a c-MYC translocation.33 Third, our inability to find a direct correlation between VpreB3 and c-Myc transcript abundance in tumors lacking c-MYC genetic abnormalities (Online Supplementary Figure S3) suggests that the expression of VpreB3 may not be directly regulated by the c-Myc protein itself but the result of a broader dysfunctional genetic program in tumors harboring a c-MYC aberration. Further studies are needed to delineate the molecular mechanisms underlying the expression of VpreB3 in these tumors.
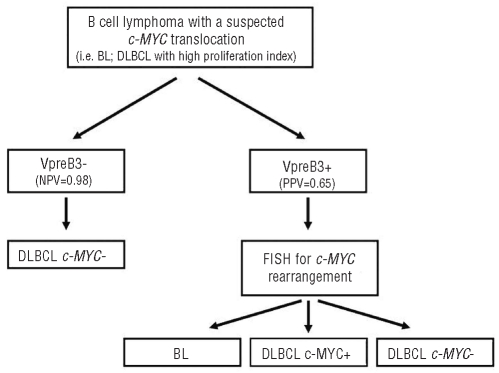
We did not find VpreB3 expression to be a prognostic marker among patients with DLBCL treated with R-CHOP. This result can be explained by the modest specificity of VpreB3 as a marker of an underlying c-MYC translocation (specificity=80%; Table 3). Rather, the utility of VpreB3 as a novel biomarker is suggested by its extremely high negative predictive value (0.98) for an underlying c-MYC translocation in aggressive B-cell tumors, and advocates VpreB3 staining as a very useful screening test applicable to the clinical setting (Figure 3). BL, intermediate DLBCL/BL, and DLBCL with a c-MYC translocation are all rare tumors (each <3% of all non-Hodgkin lymphomas)11,18 and thus by limiting genetic testing to only those cases that show VpreB3 expression (as revealed by immunohistochemistry), unnecessary and costly genetic analysis can be avoided (Figure 3). This is especially true for c-MYC+ DLBCL, which can be morphologically and phenotypically indistinguishable from c-MYC− DLBCL, yet show an inferior response to conventional chemotherapy.17 Finally, due to the expression of VpreB3 in a minority of low-grade B-cell lymphomas that usually do not harbor c-MYC translocations, the diagnostic utility of this marker is best considered only in the context of aggressive intermediate- and large-sized B-cell lymphomas in which an underlying c-MYC abnormality might occur.
Figure 3.
Proposed algorithm for evaluating a B-cell lymphoma with a suspected c-MYC translocation using immunohistochemistry for VpreB3. NPV: negative predictive value; PPV: positive predictive value.
The power of expression profiling studies in human tumors lies in the identification of genes that are associated with their stage of development.21 The biological roles of many of these gene products in influencing tumor growth and survival remain unknown. We show here that VpreB3 expression by precursor B cells and B-cell acute lymphoblastic leukemia is in keeping with its postulated role in B-cell development. However, the novel aspect of our findings that opens the possibility for further molecular studies is the observation of VpreB3 expression by a subset of GCB and some mature B-cell neoplasms, but most strikingly its expression in BL. The detailed examination of VpreB3+ DLBCL suggests that VpreB3 expression is driven, if indirectly, by c-MYC dysregulation. In conclusion, the immunohistochemical detection of VpreB3 can represent a useful screening test for identifying aggressive B-cell malignancies that potentially harbor a c-MYC translocation and therefore warrant additional genetic testing.
Footnotes
Funding: this work was supported by a Project Grant (N. 0382) from Leukaemia Research.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 2.Melchers F, ten Boekel E, Seidl T, Kong XC, Yamagami T, Onishi K, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev. 2000;175:33–46. [PubMed] [Google Scholar]
- 3.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5(7):578–84. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 4.Martensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19(2):137–42. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. A critical role of lambda 5 protein in B cell development. Cell. 1992;69(5):823–31. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 6.Minegishi Y, Coustan-Smith E, Wang YH, Cooper MD, Campana D, Conley ME. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187(1):71–7. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mundt C, Licence S, Shimizu T, Melchers F, Martensson IL. Loss of precursor B cell expansion but not allelic exclusion in VpreB1/VpreB2 double-deficient mice. J Exp Med. 2001;193(4):435–45. doi: 10.1084/jem.193.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosnet O, Mattei MG, Delattre O, Schiff C. VPREB3: cDNA characterization and expression in human and chromosome mapping in human and mouse. Cytogenet Cell Genet. 1999;87(3–4):205–8. doi: 10.1159/000015468. [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi K, Takemori T. Molecular components and assembly of mu.surrogate light chain complexes in pre-B cell lines. J Biol Chem. 1994;269(45):28347–53. [PubMed] [Google Scholar]
- 10.Rosnet O, Blanco-Betancourt C, Grivel K, Richter K, Schiff C. Binding of free immunoglobulin light chains to VpreB3 inhibits their maturation and secretion in chicken B cells. J Biol Chem. 2004;279(11):10228–36. doi: 10.1074/jbc.M312169-A200. [DOI] [PubMed] [Google Scholar]
- 11.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon: IARC; 2008. [Google Scholar]
- 12.Hecht JL, Aster JC. Molecular biology of Burkitt’s lymphoma. J Clin Oncol. 2000;18(21):3707–21. doi: 10.1200/JCO.2000.18.21.3707. [DOI] [PubMed] [Google Scholar]
- 13.Rodig SJ, Vergilio JA, Shahsafaei A, Dorfman DM. Characteristic expression patterns of TCL1, CD38, and CD44 identify aggressive lymphomas harboring a MYC translocation. Am J Surg Pathol. 2008;32(1):113–22. doi: 10.1097/PAS.0b013e3180959e09. [DOI] [PubMed] [Google Scholar]
- 14.Magrath IT, Janus C, Edwards BK, Spiegel R, Jaffe ES, Berard CW, et al. An effective therapy for both undifferentiated (including Burkitt’s) lymphomas and lymphoblastic lymphomas in children and young adults. Blood. 1984;63(5):1102–11. [PubMed] [Google Scholar]
- 15.Patte C, Auperin A, Gerrard M, Michon J, Pinkerton R, Sposto R, et al. Results of the randomized international FAB/LMB96 trial for intermediate risk B-cell non-Hodgkin lymphoma in children and adolescents: it is possible to reduce treatment for the early responding patients. Blood. 2007;109(7):2773–80. doi: 10.1182/blood-2006-07-036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossafa H, Damotte D, Jenabian A, Delarue R, Vincenneau A, Amouroux I, et al. Non-Hodgkin’s lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leuk Lymphoma. 2006;47(9):1885–93. doi: 10.1080/10428190600687547. [DOI] [PubMed] [Google Scholar]
- 17.Cook JR, Tubbs RR, Goldman B, LeBlanc M, Rimsza LM, Fisher RI, et al. Diffuse large B-cell lymphomas with high grade morphologic features and/or MYC translocations lack distinctive clinicopathologic features at presentation: a SWOG S9704 study. USCAP annual meeting, 2010. 2010 Abstract #1308. [Google Scholar]
- 18.Savage KJ, Johnson NA, Ben-Neriah S, Connors JM, Sehn LH, Farinha P, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114(17):3533–7. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 19.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354(23):2431–42. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 20.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–30. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 21.Staudt LM. Molecular diagnosis of the hematologic cancers. N Engl J Med. 2003;348(18):1777–85. doi: 10.1056/NEJMra020067. [DOI] [PubMed] [Google Scholar]
- 22.Marafioti T, Pozzobon M, Hansmann ML, Delsol G, Pileri SA, Mason DY. Expression of intracellular signaling molecules in classical and lymphocyte predominance Hodgkin disease. Blood. 2004;103(1):188–93. doi: 10.1182/blood-2003-05-1487. [DOI] [PubMed] [Google Scholar]
- 23.Marafioti T, Pozzobon M, Hansmann ML, Ventura R, Pileri SA, Roberton H, et al. The NFATc1 transcription factor is widely expressed in white cells and translocates from the cytoplasm to the nucleus in a subset of human lymphomas. Br J Haematol. 2005;128(3):333–42. doi: 10.1111/j.1365-2141.2004.05313.x. [DOI] [PubMed] [Google Scholar]
- 24.Paterson JC, Ballabio E, Mattsson G, Turner SH, Mason DY, Marafioti T. Labeling of multiple cell markers and mRNA using automated apparatus. Appl Immunohistochem Mol Morphol. 2008;16(4):371–81. doi: 10.1097/PAI.0b013e318164fc63. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe ES, Harris NL, Stein H, Vardiman JW. Tumours of the Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2001. [Google Scholar]
- 26.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 27.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8(1):68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 28.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 29.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109(4):1636–42. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedoldi S, Paterson JC, Cordell J, Tan SY, Jones M, Manek S, et al. Jaw1/LRMP, a germinal centre-associated marker for the immunohistological study of B-cell lymphomas. J Pathol. 2006;209(4):454–63. doi: 10.1002/path.2002. [DOI] [PubMed] [Google Scholar]
- 31.Montes-Moreno S, Roncador G, Maestre L, Martinez N, Sanchez-Verde L, Camacho FI, et al. Gcet1 (centerin), a highly restricted marker for a subset of germinal center-derived lymphomas. Blood. 2008;111 (1):351–8. doi: 10.1182/blood-2007-06-094151. [DOI] [PubMed] [Google Scholar]
- 32.Cattoretti G, Angelin-Duclos C, Shaknovich R, Zhou H, Wang D, Alobeid B. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol. 2005;206(1):76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- 33.Stasik CJ, Nitta H, Zhang W, Mosher CH, Cook JR, Tubbs RR, et al. Increased MYC gene copy number correlates with increased mRNA levels in diffuse large B-cell lymphoma. Haematologica. 2010;95(4):597–603. doi: 10.3324/haematol.2009.012864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Copie-Bergman C, Gaulard P, Leroy K, Briere J, Baia M, Jais JP, et al. Immuno-fluorescence in situ hybridization index predicts survival in patients with diffuse large B-cell lymphoma treated with R-CHOP: a GELA study. J Clin Oncol. 2009;27(33):5573–9. doi: 10.1200/JCO.2009.22.7058. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102(12):3871–9. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]