Abstract
Objective
To formulate a model for translating manual infection control surveillance methods to automated, algorithmic approaches.
Design
We propose a model for creating electronic surveillance algorithms by translating existing manual surveillance practices into automated electronic methods. Our model suggests that three dimensions of expert knowledge be consulted: clinical, surveillance, and informatics. Once collected, knowledge should be applied through a process of conceptualization, synthesis, programming, and testing.
Results
We applied our framework to central vascular catheter associated bloodstream infection surveillance, a major healthcare performance outcome measure. We found that despite major barriers such as differences in availability of structured data, in types of databases used and in semantic representation of clinical terms, bloodstream infection detection algorithms could be deployed at four very diverse medical centers.
Conclusions
We present a framework that translates existing practice—manual infection detection—to an automated process for surveillance. Our experience details barriers and solutions discovered during development of electronic surveillance for central vascular catheter associated bloodstream infections at four hospitals in a variety of data environments. Moving electronic surveillance to the next level—availability at a majority of acute care hospitals nationwide—would be hastened by the incorporation of necessary data elements, vocabularies and standards into commercially available electronic health records.
Keywords: Informatics, infection control, surveillance, bacteremia
Improving patient safety and healthcare quality has become a high-profile national goal.1 Public disclosure of the performance of hospitals has been promoted as integral to these efforts.2 Rates of hospital-acquired infections (HAIs)—in particular, bloodstream infections (BSIs)—are considered important measures for public reporting. HAIs are increasingly considered to be preventable through surveillance, adherence to infection control guidelines, and bundling of effective preventive practices.3 4 5 6 7 8
The Centers for Disease Control and Prevention (CDC) has conducted surveillance of HAIs since the 1970s; this relies almost exclusively on manual collection of data by infection preventionists (IPs) using HAI case definitions developed by the CDC's National Nosocomial Infections Surveillance System (NNIS) (currently the National Healthcare Safety Network or NHSN).9 Three barriers exist for implementation of surveillance of HAIs for performance measurement. First, surveillance of nosocomial infections is a time-consuming, labor-intensive process, requiring manual collection of clinical data from medical, laboratory, and pharmacy records.10 Second, proper manual surveillance requires that surveillance personnel have certain clinical and epidemiological skills. Third, because IPs must apply CDC case definitions to a broad range of clinical syndromes, clinical judgment is used, and despite training of surveillance staff, subjectivity or inconsistent classification may be introduced in the interpretation of surveillance definitions. Differences in surveillance practices by IPs within or between institutions can affect sensitivity of case-finding11 and introduce error in inter-hospital comparisons and ranking.12
With the increased public demand for reporting of performance measures that rank hospitals, we need measures that have good reliability for comparisons between hospitals. The automation of infection surveillance through use of information technology should increase reliability and allow for consistent application of rules in all institutions. An added benefit may be a more streamlined business process for detecting infections and a reduction in surveillance work, allowing IPs to spend more time on prevention efforts.13 14 Increased use of electronic data in surveillance has been emphasized in descriptions of the NHSN.15
Background
The work of automating infection control surveillance is in its infancy. While the value of repurposing electronic data for nosocomial infection surveillance is established16 17 18 19 and vendor solutions for infection surveillance exist, the knowledge bases that govern surveillance rules are often proprietary, and differences in local health record data make surveillance at each center a unique, non-scalable effort. A framework for developing electronic rules for infection detection would aid in the translation of manual surveillance approaches to those that are automated. In the development of such a framework, several questions should be addressed, including:
Are the necessary data sets and fields comprehensively and comprehensibly available in existing information systems?
Are standardized coding schemas used across multiple institutions in light of the semantic heterogeneity in local data repositories (eg, differences in bacterial nomenclature used between laboratory information systems)?
Do developed rules need to be faithful representations of current case definitions, or can simpler rules be used as a surrogate to achieve the aim of reproducible and accurate HAI detection? And as a corollary, is the goal to determine actual rates of infection or relative rates for ranking institutions?
Can the business logic of surveillance (ie, flowcharts, algorithms, or pseudocode describing the process) be deployed at individual hospitals and applied to individual data stores to produce accurate, reliable results?
Storage of the rules governing automated surveillance processes in knowledge bases has been accomplished in a variety of settings, including the Health Evaluation through Logical Processing (HELP) system,16 as well as in vendor systems.20 Furthermore, standards for the sharing and generic application of knowledge bases and decision support have been developed. Examples include the Arden syntax,20 an American National Standards Institute standard that specifies Medical Logic Modules which contain information about both the logic and the context of a decision algorithm; the GuideLine Interchange Format,21 a methodology for the process of guideline development using flowcharts and a set of classes to support temporality of rules, ontology of concepts, and hierarchy of actions; and the GELLO (objective oriented guidelines) expression language,22 which is an object oriented language that represents algorithms used in decision support.
An excellent target for electronic surveillance is central vascular catheter (also referred to as central-line) associated bloodstream infections (CLABSIs),23 because CLABSIs are common and preventable adverse events in healthcare settings,1 24 25 and are a subset of a easily detectable electronic trigger, blood cultures with growth of bacteria. CDC's Healthcare Infection Control Practices Advisory Committee recommends ongoing surveillance for CLABSIs,24 and in many states CLABSI monitoring has been mandated for reporting to public agencies. Automated CLABSI detection has been achieved in single centers or in small populations of patients.26 27 28
As described by NNIS/NHSN rules, surveillance for CLABSIs entails several sequential events. The characteristics of these events make them amenable to automation (box 1). First, IPs review blood cultures sent to laboratories to identify those with microbial growth (ie, positive blood cultures) and that are hospital-associated (ie, occur after the incubation time period for community-acquisition, ie, ≥ second hospital day). Then, the number of positive blood cultures, clinical features in patients with the positive cultures and type and number of culture isolates are reviewed to determine whether the isolates are likely to represent true infection (ie, are not contaminants of the blood cultures). Finally, the clinical features and culture results from other body sites in patients with positive blood cultures are reviewed to classify positive cultures as due to a CLABSI or a ‘secondary’ bacteremia from another site (eg, pneumonia or pyelonephritis with associated bacteremia). Because a positive blood culture is the trigger for a surveillance action, these steps can be executed as an electronic rule.
Box 1. NNIS criteria for CLABSI surveillance (valid through January 1, 2008).
Nosocomial infection
There is no evidence that the infection was present or incubating at the time of hospital admission unless the infection was related to a previous NSHN patient admission to the current hospital.
Primary bloodstream infection criteria
Criterion 1
Patient has a recognized pathogen recovered from ≥1 blood culture
Organism cultured from blood is not related to an infection at another site.
Criterion 2a
Patient has ≥1 of the following signs or symptoms: fever (>38°), chills, or hypotension
Signs and symptoms and positive laboratory results are not related to an infection at another site
Common skin commensal (eg, diphtheroids, Bacillus sp., Propionibacterium sp., coagulase-negative staphylococci, or Micrococcus spp.) is cultured from ≥2 blood cultures drawn on separate occasions
Criterion 2b
Patient has ≥1 of the following signs or symptoms: fever (>38o), chills or hypotension
Signs and symptoms and positive laboratory results are not related to an infection at another site
Common skin contaminant (eg, diphtheroids, Bacillus spp., Propionibacterium spp., coagulase-negative staphylococci, or Micrococcus spp.) is cultured from at least one blood culture from a patient with an intravascular line, and the physician institutes appropriate antimicrobial therapy
Criteria for CVC associated
During 48 h before the first blood culture positive for this organism, did the patient have a vascular access device that terminated at or close to the heart or one of the great vessels (includes tunneled or non-tunneled catheters inserted into the subclavian, jugular or femoral veins; pulmonary artery (Swan–Ganz) catheters; hemodialysis catheter; totally implanted devices (ie, ports); peripherally inserted central catheters (PICCs)).
CLABSI, catheter associated bloodstream infection; NNIS, National Nosocomial Infections Surveillance System.
As a part of the CDC Prevention Epicenters Program, a multicenter collaborative group seeking to use information technology to prevent nosocomial infections, we conducted a multicenter project to build and apply a framework for developing electronic algorithms from manual surveillance efforts, using CLABSI surveillance as an application of this framework. This paper describes the framework developed, challenges encountered, and lessons learned in the development process. A planned evaluation of the performance characteristics of electronic CLABSI surveillance as compared with manual surveillance in the 20 enrolled units will be presented in a separate report. This evaluation is not complete, and awaits the expert review of patient charts with putative CLABSIs to determine the accuracy of manual (ie, IP determined) and automated (ie, algorithmically determined) surveillance.
Formulation process
The value of a framework for electronic rule development is that it may reduce bias and misclassification introduced to electronic rules that results from the process of translating manual to automated surveillance methods. Surveillance systems seek to detect the absolute infection status of an individual (ie, either ‘infected’ or ‘not infected’). The action of classifying individuals as infected or not infected, however, is beset by problems of subjectivity and at times, a lack of sufficient data. Clinicians classify individuals as infected or not based often on subjective criteria, and often err on the side of presuming infection for treatment purposes; IP surveillance, on the other hand, uses specific criteria and case definitions in an attempt to reduce subjectivity. Despite the availability of surveillance criteria, the opportunity still exists for IPs to apply subjective interpretations of surveillance criteria which may introduce bias and reduce reproducibility. For example, IPs may subjectively apply surveillance criteria, may rely on clinician interpretations to aid determinations of infection status, may have access to clinical criteria that are not present in the electronic health record, or may have a culture of practice that is locally derived and differs substantially from other centers.
Automated surveillance, in contrast, provides for the strict and consistent application of surveillance definitions (with consequent elimination of clinical interpretations and judgment). However, such approaches risk misclassification errors when there are gaps in the availability of all necessary clinical information (eg, vital signs, patient appearance, unmeasured laboratory values, or evidence of infection elsewhere).
Automating CLABSI surveillance required that we formalize and operationalize the steps in translating existing manual surveillance criteria to algorithmic approaches. The NNIS (now called NHSN) definitions used to identify CLABSIs (box 1) are the standard criteria applied by IPs. We created algorithms to mimic these manual surveillance criteria; five rules were developed that ranged in complexity and in required data elements.
We sought to develop a model for rule development that would achieve the following aims:
Allow for code development by a single site with subsequent distribution to partners;
Use existing published knowledge about automating surveillance, while incorporating expert opinion;
Reduce bias that could result from a priori decisions made in the rule development process, or, in other words, implement the best practices for automated surveillance that are rooted in evidence and consensus, not opinion.
Our intent was to establish a formal process for developing automated surveillance criteria from existing manual processes. To allow for widespread distribution and adoption of the algorithms, standard table structures and vocabularies were used. To develop algorithms that synthesized existing definitions and incorporated clinical and informatics knowledge, we used a multidisciplinary approach. To reduce bias in the electronic rules, we followed an orderly development process composed of conceptualization, rule development, programming, and local testing and troubleshooting.
Model description
Conceptualization
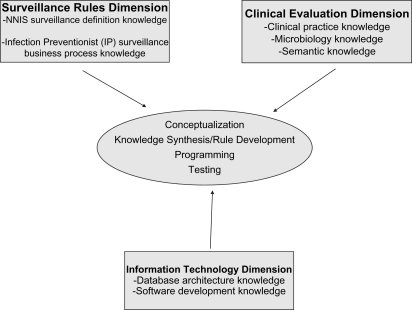
Due to a dearth of comparative trials of different applications of CLABSI rules, reliance on expert opinion for a priori assumptions in code development was necessary. When creating electronic rules, we identified three dimensions of expert knowledge that govern the transition from manual to automated surveillance: (1) the surveillance rules dimension; (2) the clinical evaluation dimension; and (3) the information technology dimension (figure 1).
Figure 1.
Knowledge dimensions required for electronic surveillance algorithm development.
The surveillance rules dimension relates to current practice in infection surveillance and incorporates existing surveillance methods into the approach for electronic rule creation. In the case of CLABSI surveillance, this dimension includes both the NNIS and NHSN rules for CLABSI surveillance and the IP implementation of these rules. We used our knowledge of the IP approach for assessing individual patients to develop this dimension of knowledge. For example, IPs may use clinical criteria to aid in classification of infected versus not infected or catheter-related versus secondary (ie, from an alternate site, such as the urinary tract) BSI.
The clinical evaluation dimension describes the knowledge necessary to relate healthcare events to the clinical significance of those events. For CLABSI surveillance, this dimension describes whether, for a given episode of positive blood cultures, clinicians will classify the positive cultures as due to true infections and the data needed to make that determination. Examples of information contained within this dimension include the importance of potential (especially uncommon) skin commensal organisms as true pathogens, the value of differing patterns of culture positivity with skin commensals in classifying culture results as due to contaminants or CLABSI episodes, the association of specific microbes recovered with secondary BSIs and the value of positive cultures from other body sites in determining that positive blood cultures likely represent seeding of the blood from those sites (ie, secondary infections).
The information technology dimension describes knowledge of the contents of typical data repositories and standard vocabularies with respect to the inclusion of surveillance logic in the resulting code development. It encompasses the knowledge of available data elements, the construct of local databases and the methods required to translate surveillance criteria into business logic applicable to databases. The implementation of CLABSI surveillance requires microbiology data to identify positive blood cultures as well as patient location data to detect location of origin of infections.
Knowledge synthesis and rule development
As noted in figure 2, NNIS surveillance rules were implemented as algorithms for CLABSI surveillance. Because the rules were planned for use at multiple institutions, a standard table schema was constructed (available at http://bsi.cchil.org (accessed 1 Oct 2009)). The databases required for full implementation of the electronic algorithms were: (1) microbiology data, including all positive and negative culture results from both blood and other body sites, but excluding catheter tips or surveillance (ie, non-clinical) cultures; (2) admission, discharge and transfer data, using patient admission and transfer events to assign bed locations; and (3) pharmacy dispensing or ordering data, to assess if vancomycin (a common treatment for CLABSIs) prescriptions had occurred. Catheter tips were not included since it was believed that centers differed in the frequency that catheter tip cultures were obtained when CLABSIs were suspected. The code was developed based on a plan for collaborating hospitals to extract relevant data, harmonize semantic differences using standard vocabularies and translation tables and fit the data to the standard table structure.
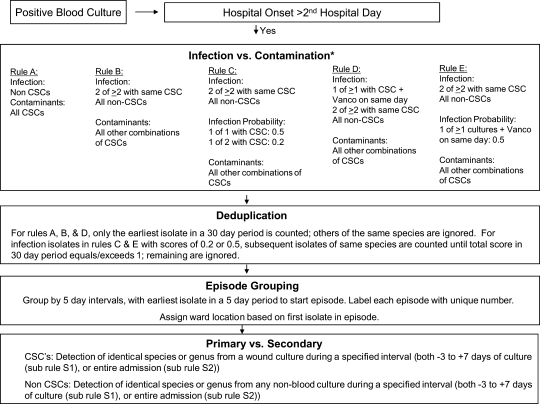
Figure 2.
Algorithm for electronic BSI surveillance. BSI, bloodstream infection; CSC, common skin commensals.
Programming
The business logic for CLABSI rules is shown in figure 2. This figure was used by programmers to develop SQL code. Code development required 80 h of one programmer. The final SQL code is available at http://bsi.cchil.org.
Since the algorithms were to be applied locally at each site, generic code was developed to accommodate differences in local databases. In addition, to promote use of standards, accommodate nomenclature differences between local hospital data sets and enhance interoperability, standard table structures for input data and standard vocabularies (ie, SNOMED and LOINC29) were used, and a structured query language pseudocode was developed in T/SQL to allow transparency of rule implementation to investigators. Other sites were allowed to adapt and test the code for use in their data warehouse environments.
Testing
The CLABSI algorithms were tested at the development site in several steps. First, a random sample of isolates was queried from the local database at the development site and the paper charts were reviewed for these episodes. To test the fidelity of the algorithm, an investigator who was a clinical knowledge expert (ML) audited the clinical charts of patients using a convenience sample (n=30) of consecutive isolates evaluated by the algorithm, and made assessments of the classification status of the isolates (ie, true infection vs contaminant, CLABSI vs secondary). Discrepancies between manual and electronic algorithm determination were closely examined. Discrepancies had two causes: (1) missing electronic data that changed the algorithm's interpretation; and (2) incorrect representation of the algorithm business logic in SQL, leading to misclassification by the electronic algorithm. Missing data and errors in code creation were resolved locally at this point, and we performed iterative retesting using both already-sampled and newly-sampled isolates.
Validation through example
The final CLABSI surveillance rules are shown in figure 2. Each episode, or BSI event, begins with a blood culture with microbial growth (ie, a positive blood culture). Each episode is defined as all unique positive blood cultures found in a patient identified during a five calendar-day period. The collection day of the positive blood culture isolate is assessed; isolates obtained before the third hospital day are considered community-onset and disregarded. Isolates obtained on or after the third hospital day are considered hospital-onset in origin and are classified as infections or contaminants. The classification of common skin commensals (CSCs)—organisms that commonly grow on skin and can contaminate blood cultures as they are being obtained or processed (ie, coagulase-negative staphylococci, Corynebacterium spp., Micrococcus spp., Bacillus spp., and Propionibacterium spp.)—differs among the five algorithms; for all rules, isolates that are not CSCs are considered true infections.
Following the classification of an isolate as a contaminant or a true infection, all additional blood isolates of the same species are ignored for the subsequent 30 days and are considered duplicates. Isolates obtained within a 5 day period are grouped together into an episode. Episodes are characterized as primary BSIs (ie, an intravascular infection) or secondary (ie, related to infection at another site, such as the respiratory or urinary tract infection). Cultures from body sites other than blood are compared to blood culture isolates (all sites for non-CSCs, only wound sites for CSCs); if any blood isolate in an episode is deemed secondary (ie, due to match with culture results from a non-bloodstream site), then the entire episode is classified as a secondary BSI episode. Two rules are used for the assessment of secondary BSIs: in the first (S1), only non-bloodstream isolates obtained within 3 days before, to 7 days after a bloodstream isolate are considered, while the second (S2) uses all non-bloodstream isolates obtained during the same hospital admission as a bloodstream isolate. The ward location of BSI episodes is assigned based on the patient location at the presumed time of incubation of infection, judged based on the location of the patient 2 days prior to collection date of the first positive blood culture in an episode. Prior admissions are used in the deduplication process, in that the 30 day rule for deduplication can cross admissions, but prior admission information is not used in the assignment of community—or nosocomial—onset for BSIs. Clinical information such as the presence of hypotension or fever is not used in the algorithms. Future iterations of the algorithms, with more robust data requirements, may include these data elements and might augment the performance characteristics of rules particularly in the setting of CSCs.
Primary BSI episodes identified by the electronic rules are further classified as CLABSIs if a central venous catheter was present at the time of the episode. Where these data are available electronically (eg, nursing documentation on a structured form), this determination can be automated. If the presence of a central vascular catheter is not determinable electronically, manual assessments of the presence of central lines must be made by chart review. Similarly, determination of the denominator—central-catheter days for the studied unit—relies on either manual data collection or through electronic documentation.
Distribution and implementation of CLABSI surveillance code
Following central testing of the algorithm and correct classification of CLABSI episodes as compared to human review of sampled episodes, code was distributed to partner institutions with the aim of rule implementation and validation at each site. The code was developed at Hospital A, which used Microsoft SQL Server 2000, with microbiology and bed information tables for patient location. Of the three partner hospitals that adopted the CLABSI algorithms, Hospital B did not have a data warehouse incorporating laboratory and other clinical data; Hospital C used SYBASE for its clinical data warehouse and had stored microbiology, bed information and central-line utilization tables; and Hospital D used an ORACLE database platform for its clinical data warehouse.
Several challenges for code implementation were noted—issues in data structure and format and in compatibility of SQL code (table 1). Because code was redeveloped at each center as needed, custom solutions were developed at each center. Two problems were notable. First, microbiology data exhibited significant semantic differences between centers, and in two centers were not available as discrete data, but rather, in free text format. Second, differences in local SQL languages required redevelopment time at each center. Specific examples of issues unique to each hospital will now be described.
Table 1.
Hospitals, database systems and challenges encountered in automated bloodstream infection surveillance
| Hospital | Database system | Challenges | Solutions |
| Hospital A | Microsoft SQL Server 2000 | Leveraging knowledge dimensions | Consultation among knowledge experts |
| Hospital B | None—extract of data given to Hospital A | Free text, unstructured microbiology data | Use of templates to import data to flat files (Datawatch Monarch) |
| Ad hoc requests needed for individual data extracts | Python script to standardize free text data | ||
| Bed information obtained from billing data warehouse | |||
| Hospital C | SYBASE | Negative cultures not available | Could not run 1 of 5 rules (Rule C) |
| Hospital D (30) | Oracle | Free text, unstructured microbiology data | Natural language processing |
| Review of business processes of microbiology reporting | |||
| SQL language version incompatibility (ie, T-SQL vs PL/SQL) | Algorithm rewritten from conceptual flowchart |
Because Hospital B did not have an accessible data warehouse at the time of the project, a data extract based on an existing report found in the microbiology system, printed to a text file, was imported into the SQL server database used at Hospital A. The data were unstructured, and converted to a flat table using customized templates developed using Monarch (Datawatch Corporation, Chelmsford, MA, USA). The resulting report contained organism names that were reported in free text fields, while specimen source information was discrete and mapped to standard nomenclature. Organism names were further refined using python scripts and validated to ensure accuracy.
Hospital C, which had an internally developed and validated data warehouse, was able to use the SQL-coded algorithms with minor modification, that is, correction of capitalization and formatting of SQL and adaptation of code to local data structures. This was possible because of the similarities in implementation of SQL between Microsoft SQL Server and SYBASE (both use T/SQL).
At Hospital D, reuse of the SQL code was abandoned due to differences between implementation of SQL in ORACLE (used at Hospital D) and Microsoft SQL Server (used at Hospital A) (ie, PL/SQL vs T-SQL). Therefore, the algorithms and flowchart were used to develop a database agnostic, generic code.30 Microbiology data at Hospital D, like Hospital B, were limited by the use of free text fields in organism name fields, prompting standardization of the organism name list and a review of the business process of microbiology reporting of organism names. Natural language processing methods were used to translate free text organism name reports to a set of discrete organisms.
Development of validation data sets
To verify that code redevelopment at each partner hospital faithfully represented the original code, each hospital tested its algorithms against a specially developed test data set (available at http://bsi.cchil.org). Derived from cleaned Hospital A data, the data set contained de-identified, linked results from microbiology, pharmacy and bed information databases. Five test tables were developed representing a 2 year period: (1) a microbiology data set for all positive and negative blood cultures; (2) a microbiology data set of positive non-blood site cultures; (3) patient location information, based on admission-discharge-transfer transactions; (4) a pharmacy data set of Vancomycin prescriptions; and (5) a SNOMED mapping table, with organism names mapped to SNOMED codes. In addition, sample outputs, with raw counts and aggregated counts for each of the five rules, were provided. Using the example of the test data set, Hospitals B–D compared their rule-generated BSI counts to aggregated monthly data and patient level counts. When discrepancies were noted, algorithms were reviewed and improved iteratively until observed results matched expected results for the test data set. This process continued at each site until an exact match occurred. The time to achieve an exact match varied between sites, occurring within 3–4 weeks at one site (Hospital C) and requiring several months for the entire process at another (Hospital D).
Discussion
Using a planning and development process that included coordination of informaticians, programmers, clinicians and surveillance experts, we developed and deployed algorithms for the automatic surveillance of CLABSIs. These algorithms allowed for the comparison of several methods of CLABSI classification and consistent surveillance rule application across large disparate academic medical centers. Despite differences in data availability, structure and SQL language type, we were able to adapt the business logic such that it could be universally applied. Our framework should assist others in developing and distributing electronic algorithms that approximate existing surveillance definitions, which often rely on local institutional practices and subjective interpretation of clinical findings in patients. As more data are available in electronic health records (eg, vital signs and clinical impressions), opportunity for augmentation of surveillance algorithms with these clinical data will occur. Consumers of the results of surveillance algorithms, for example, CDC, could play an integral role in ensuring that algorithm development uses a standard approach, in determining that developed algorithms generate expected results when using test data sets and in disseminating validated knowledge bases and algorithms using standards based approaches like Arden syntax.
Major barriers in this process were identified and will need to be addressed for national automated CLABSI surveillance. First, the process of translating current surveillance definitions which use manual processes into electronic rules may lead to algorithms that vary among institutions. As a result, unless a standard approach to develop rules and validate rates produced by the resulting automated surveillance tools is developed, rates generated by these rules may vary due to differing rule development methods and coding. Such discrepancies may affect inter-hospital comparisons. The use of standards-based approaches to rendering generic knowledge bases20 21 22 31 may be a method to disseminate guidelines and algorithms to multiple centers. Test data sets, as we used, could be a way to test the implementation of generic knowledge bases at centers. Second, substantial differences in the electronic availability and format of data elements stored at each hospital provided challenges for the implementation of rules. For example, we encountered the use of free text descriptions of microbiology specimens and the absence of negative cultures in some data sets in this project. Given that the data flow of microbiology results begins in instruments, moves to a laboratory information system and often is transferred to a full electronic health record, devising workflows to capture discrete data instead of free text would seem to be a desirable, and achievable, goal. In addition, incorporating fields in laboratory information systems that have discrete interpretations of free text reports would aid this process. Pattern matching and natural language processing, while promising techniques, require evaluation to determine whether sufficient sensitivity and specificity exist in interpretation of reports. Third, differences in database types make the use of a database agnostic algorithm a necessary goal. A major challenge was the heterogeneity of local nomenclatures and lack of standard vocabulary usage for describing microbiology data in most centers. Therefore, exposing data using standards (ie, in vocabulary and database schema) will minimize development time required at individual centers. Use of standards in code sets and knowledge bases may have limited utility unless the difficult task of harmonizing data sets is accomplished first.
Our experience mimics that of Kahn et al,19 in their description of the GermWatcher expert system, in that the data required are only loosely coupled with a center's electronic medical record databases. Our attempt to develop a detection algorithm knowledge base which is modular and shareable is similar to other knowledge base methods for decision support.16 20 Despite the 20 year history in the literature of knowledge base development for nosocomial infection surveillance and the great potential for healthcare quality improvement with use of these methods, our experience suggests that nationwide deployment of electronic infection surveillance faces significant barriers due to the lack of harmonization between electronic health records.
Conclusion
The CDC has a stated goal to streamline surveillance practices, and the automation of infection detection would achieve this aim.15 For automated surveillance, IT infrastructure is essential. Despite the call for a paperless medical record by the year 2001 by the Institute Of Medicine, many hospital systems continue to use paper reports for laboratory values and lack the capability to share results electronically.32 In some settings, electronic clinical data may be stored in data warehouses, but there are no national assessments of how these data are structured, how semantically distinct they are or how accessible they are for surveillance activities.
We present a framework that translates existing practice—manual infection detection—to an automated process for surveillance. We believe that a multidisciplinary approach with active consultation among informaticians, surveillance experts, and clinicians will be required to allow interpretation and structuring of data in data warehouses to enable enhanced electronic surveillance activities. In addition, clear steps for validating the results of automation are essential. Our experience details barriers and solutions discovered during development of electronic surveillance for CLABSIs at four hospitals in a variety of data environments (table 1). Moving electronic surveillance to the next level—availability at a majority of acute care hospitals nationwide—would be hastened by incorporation of necessary data elements, vocabularies and systems into commercially available electronic health records. Such dissemination of standards may be achieved by increasing visibility of existing deficiencies, by using national mandates or by improving financial support for hospitals to deploy electronic health records.
Footnotes
Funding: This study was provided by Grants U01 CI 000327-01, U01 C1 000333 and U01 CI000328-03 from the Centers for Disease Control and Prevention.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Kohn LT, Corrigan JM, Donaldson MS, eds. To err is human: building a safer health system. Committee on quality of health care in America. Institute of Medicine report. Washington: National Academy of Press, 2000 [PubMed] [Google Scholar]
- 2.Lansky D. Improving quality through public disclosure of performance information. Health Aff (Millwood) 2002;21:52–62 [DOI] [PubMed] [Google Scholar]
- 3.Pronovost P, Needham D, Berenholtz S, et al. An intervention to decrease catheter-related bloodstream infections in the ICU. N Engl J Med 2006;355:2725–32 [DOI] [PubMed] [Google Scholar]
- 4.Bleasdale SC, Trick WE, Gonzalez IM, et al. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med 2007;167:2073–9 [DOI] [PubMed] [Google Scholar]
- 5.Vernon MO, Hayden MK, Trick WE, et al. Chlorhexidine gluconate to cleanse patients in a medical intensive care unit: the effectiveness of source control to reduce the bioburden of vancomycin-resistant enterococci. Arch Intern Med 2006;166:306–12 [DOI] [PubMed] [Google Scholar]
- 6.Evans B. Best-practice protocols: VAP prevention. Nurs Manage 2005;36:10, 2, 4 passim. [DOI] [PubMed] [Google Scholar]
- 7.Haley RW, Culver DH, White JW, et al. The efficacy of infection surveillance and control programs in preventing nosocomial infections in US hospitals. Am J Epidemiol 1985;121:182–205 [DOI] [PubMed] [Google Scholar]
- 8.Public health focus: surveillance, prevention, and control of nosocomial infections. MMWR Morb Mortal Wkly Rep 1992;41:783–7 [PubMed] [Google Scholar]
- 9.Garner JS, Jarvis WR, Emori TG, et al. CDC definitions for nosocomial infections, 1988. Am J Infect Control 1988;16:128–40 [DOI] [PubMed] [Google Scholar]
- 10.Tokars JI, Richards C, Andrus M, et al. The changing face of surveillance for health care-associated infections. Clin Infect Dis 2004;39:1347–52 [DOI] [PubMed] [Google Scholar]
- 11.Emori TG, Edwards JR, Culver DH, et al. Accuracy of reporting nosocomial infections in intensive-care-unit patients to the National Nosocomial Infections Surveillance System: a pilot study. Infect Control Hosp Epidemiol 1998;19:308–16 [DOI] [PubMed] [Google Scholar]
- 12.Nosocomial infection rates for interhospital comparison: limitations and possible solutions. A Report from the National Nosocomial Infections Surveillance (NNIS) System. Infect Control Hosp Epidemiol 1991;12:609–21 [PubMed] [Google Scholar]
- 13.Panackal AA, M'Ikanatha NM, Tsui FC, et al. Automatic electronic laboratory-based reporting of notifiable infectious diseases at a large health system. Emerg Infect Dis 2002;8:685–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Effler P, Ching-Lee M, Bogard A, et al. Statewide system of electronic notifiable disease reporting from clinical laboratories: comparing automated reporting with conventional methods. JAMA 1999;282:1845–50 [DOI] [PubMed] [Google Scholar]
- 15.Tokars JI. Predictive value of blood cultures positive for coagulase-negative staphylococci: implications for patient care and health care quality assurance. Clin Infect Dis 2004;39:333–41 Epub 12 Jul 2004. [DOI] [PubMed] [Google Scholar]
- 16.Evans RS, Gardner RM, Bush AR, et al. Development of a computerized infectious disease monitor (CIDM). Comput Biomed Res 1985;18:103–13 [DOI] [PubMed] [Google Scholar]
- 17.Kahn MG, Steib SA, Spitznagel EL, et al. Improvement in user performance following development and routine use of an expert system. Medinfo 1995;8(Pt 2):1064–7 [PubMed] [Google Scholar]
- 18.Evans RS, Larsen RA, Burke JP, et al. Computer surveillance of hospital-acquired infections and antibiotic use. JAMA 1986;256:1007–11 [PubMed] [Google Scholar]
- 19.Kahn MG, Steib SA, Fraser VJ, et al. An expert system for culture-based infection control surveillance. Proc Annu Symp Comput Appl Med Care 1993;171–5 [PMC free article] [PubMed] [Google Scholar]
- 20.Hripcsak G, Ludemann P, Pryor TA, et al. Rationale for the Arden Syntax. Comput Biomed Res 1994;27:291–324 [DOI] [PubMed] [Google Scholar]
- 21.Peleg M, Boxwala AA, Bernstam E, et al. Sharable representation of clinical guidelines in GLIF: relationship to the Arden Syntax. J Biomed Inform 2001;34:170–81 [DOI] [PubMed] [Google Scholar]
- 22.Sordo M, Boxwala AA, Ogunyemi O, et al. Description and status update on GELLO: a proposed standardized object-oriented expression language for clinical decision support. Stud Health Technol Inform 2004;107(Pt 1):164–8 [PubMed] [Google Scholar]
- 23.McKibben L, Horan T, Tokars JI, et al. Guidance on public reporting of healthcare-associated infections: recommendations of the Healthcare Infection Control Practices Advisory Committee. Am J Infect Control 2005;33:217–26 [DOI] [PubMed] [Google Scholar]
- 24.O'Grady NP, Alexander M, Dellinger EP, et al. Guidelines for the prevention of intravascular catheter-related infections. Centers for Disease Control and Prevention. MMWR Recomm Rep 2002;51(RR-10):1–29 [PubMed] [Google Scholar]
- 25.Wenzel RP, Edmond MB. The impact of hospital-acquired bloodstream infections. Emerg Infect Dis 2001;7:174–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trick WE, Zagorski BM, Tokars JI, et al. Computer algorithms to detect bloodstream infections. Emerg Infect Dis 2004;10:1612–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoe DS, Anderson J, Chambers R, et al. Simplified surveillance for nosocomial bloodstream infections. Infect Control Hosp Epidemiol 1998;19:657–60 [DOI] [PubMed] [Google Scholar]
- 28.Bellini C, Petignat C, Francioli P, et al. Comparison of automated strategies for surveillance of nosocomial bacteremia. Infect Control Hosp Epidemiol 2007;28:1030–5 [DOI] [PubMed] [Google Scholar]
- 29.Wurtz R, Cameron BJ. Electronic laboratory reporting for the infectious diseases physician and clinical microbiologist. Clin Infect Dis 2005;40:1638–43 [DOI] [PubMed] [Google Scholar]
- 30.Borlawsky T, Hota B, Lin MY, et al. Development of a reference information model and knowledgebase for electronic bloodstream infection detection. AMIA Annu Symp Proc 2008:56–60 [PMC free article] [PubMed] [Google Scholar]
- 31.Hripcsak G. Arden Syntax for medical logic modules. MD Comput 1991;8:76, 78. [PubMed] [Google Scholar]
- 32.Institutes of Medicine Key capabilities of an electronic health record system: letter report. National Academic Press, 2003. http://www.nap.edu/catalog/10781.html (accessed May 29, 2008). [PubMed] [Google Scholar]