Abstract
Background
The RxNorm and NDF-RT (National Drug File Reference Terminology) are a suite of terminology standards for clinical drugs designated for use in the US federal government systems for electronic exchange of clinical health information. Analyzing how different drug products described in these terminologies are categorized into drug classes will help in their better organization and classification of pharmaceutical information.
Methods
Mappings between drug products in RxNorm and NDF-RT drug classes were extracted. Mappings were also extracted between drug products in RxNorm to five high-level NDF-RT categories: Chemical Structure; cellular or subcellular Mechanism of Action; organ-level or system-level Physiologic Effect; Therapeutic Intent; and Pharmacokinetics. Coverage for the mappings and the gaps were evaluated and analyzed algorithmically.
Results
Approximately 54% of RxNorm drug products (Semantic Clinical Drugs) were found not to have a correspondence in NDF-RT. Similarly, approximately 45% of drug products in NDF-RT are missing from RxNorm, most of which can be attributed to differences in dosage, strength, and route form. Approximately 81% of Chemical Structure classes, 42% of Mechanism of Action classes, 75% of Physiologic Effect classes, 76% of Therapeutic Intent classes, and 88% of Pharmacokinetics classes were also found not to have any RxNorm drug products classified under them. Finally, various issues regarding inconsistent mappings between drug concepts were identified in both terminologies.
Conclusion
This investigation identified potential limitations of the existing classification systems and various issues in specification of correspondences between the concepts in RxNorm and NDF-RT. These proposals and methods provide the preliminary steps in addressing some of the requirements.
Introduction
The complexity of patient data in electronic medical records, coupled with expectations that these data facilitate clinical decision making, healthcare cost effectiveness, medical error reduction, and evidence-based medicine, makes obvious the role of standardized terminologies as a foundation for comparable and consistent representation of patient information. Toward this end, the evolution of terminologies across the spectrum of detailed nomenclatures and sophisticated classifications has accelerated dramatically over this decade.
In the world of pharmaceutical drugs, the US National Library of Medicine and Department of Veteran Affairs have been involved in the development and maintenance of RxNorm1 and NDF-RT,2 respectively, for standardized nomenclature of clinical drugs. The goal of RxNorm is to facilitate various systems using different drug nomenclatures share and exchange data efficiently. It provides a way to link and map standard clinical drug names to many drug vocabularies commonly used in pharmacy management and drug interaction software, including First DataBank,3 Micromedex,4 Medi-Span,5 and Multum.6 The NDF-RT, on the other hand, uses a description logic-based7 reference model defining a set of abstractions for drug products along with a set of hierarchical and definitional relationships to capture the associated details. In particular, the model includes hierarchies of drug classes for Chemical Structure (eg, Acetic Acid), cellular or subcellular Mechanism of Action (eg, Hydrolases), organ-level or system-level Physiological Effect (eg, Cellular Motion Alteration), drug–disease relationship describing the Therapeutic Intent (eg, Bile Reflux), and Pharmacokinetics describing the mechanisms of absorption and distribution of an administered drug within a body (eg, Hepatic Excretion). Additionally, NDF-RT categorizes the drug products via a single-inheritance hierarchy of approximately 550 drug classes (eg, Antimicrobials).8
Various efforts in the recent past have focused on studying different aspects of the drug terminologies ranging from their coverage and adequacy of representation2 9 to categorization and classification,8 10–12 as well as their application for information exchange13 and linkage.14 The primary objective of this study is to investigate the categorization of the drug products in RxNorm and NDF-RT with respect to the drug classes in NDF-RT. In particular, our goal is to determine similarities, as well as dissimilarities, between the drug product-to-class mappings in RxNorm and NDF-RT and, where unavailable, propose an automatic technique to generate a candidate set of mappings for consideration.i Additionally, we will analyze the coverage of the NDF-RT drug class hierarchies for Chemical Structure, Mechanism of Action, Physiologic Effect, Therapeutic Intent and Pharmacokinetics as well as identify issues regarding inconsistent mappings and representation of incoherent information in the two terminology resources. The study concludes that significant additional effort is needed to bridge the ‘informational gap’ between the two drug terminologies to facilitate clinical and translational research activities, such as pharmacogenomics.
The rest of the article is organized as follows. We begin with a list of abbreviations used throughout (table 1). The next section provides background information about RxNorm and NDF-RT terminology resources. We then present the methods for analyzing the drug product-to-class mappings in RxNorm and NDF-RT and summarize our findings. Finally, we discuss the implications of our investigation along with strengths and limitations of our approach.
Table 1.
List of abbreviations used in article
| Abbreviation | Full form |
| NDF-RT | National Drug File-Reference Terminology |
| UMLS | Unified Medical Language System |
| MeSH | Medical Subject Heading |
| SNOMED-CT | Systematized Nomenclature of Medicine-Clinical Terms |
| NDC | National drug code |
| RxCUI | RxNorm concept unique identifier |
| UMLSCUI | UMLS concept unique identifier |
| VUID | Veterans Health Administration (VHA) unique identifier |
Background
RxNorm
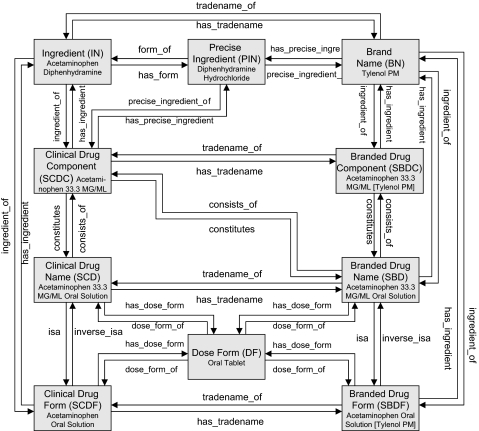
A nomenclature for clinical drugs, RxNorm is produced by the US National Library of Medicine.1 15 It contains the names of prescription and many nonprescription formulations approved for human use (primarily in the USA). An RxNorm clinical drug name reflects the active ingredients, strengths, and dose form comprising that drug. When any of these elements vary, a new RxNorm drug name is created as a separate concept identified by a concept unique identifier (RxCUI). Consequently, to distinguish between such drug entities, RxNorm uses ‘term types’ (TTYs) that represent categories for generic and branded drugs (figure 1). Specifically, RxNorm uses four categories for generic drugs: ingredient alone (Ingredient denoted by IN), ingredient plus strength (Clinical Drug Component denoted by SCDC), ingredient plus dose form (Clinical Drug Form denoted by SCDF) and ingredient plus strength and dose form (Clinical Drug denoted by SCD). Analogously, there are four categories for brand name drug concepts: brand name alone (Brand Name denoted by BN), brand name plus strength (Branded Drug Component denoted by SBDC), brand name plus dose form (Branded Drug Form denoted by SBDF), brand name plus strength and dose form (Branded Drug denoted by SBD). Furthermore, the RxNorm drug entities are related to each other by a well-defined set of named relationships that allows the traversal of the RxNorm graph and retrieval of information about different RxNorm drug entities. For example, as shown in figure 1, given the brand name Tylenol PM (RxCUI=220581), one can retrieve its ingredients Acetaminophen (RxCUI=161) and Diphenhydramine (RxCUI=3498) by traversing the direct path between BN and IN via the relationship tradename_of. In addition to the above, RxNorm contains mappings from its concepts to one or more concepts in external drug terminologies (or databases) including First DataBank, Micromedex, Medi-Span, Multum, and NDF-RT. For instance, to specify mappings between RxNorm and NDF-RT, an RxNorm drug product is assigned a Veterans Health Administration unique identifier (VUID) that corresponds to a uniquely identifiable concept code in NDF-RT. As an example, Acetaminophen 160 MG ORAL TABLET (RxCUI=282464) is assigned the VUID=4007166, which maps to NDF-RT code=C32112 (ACETAMINOPHEN 160MG TAB (VA Product)).
Figure 1.
Relationship between RxNorm Drug Entities (adapted from Peters L, Bodenreider O15).
NDF-RT
The NDF-RT is a drug information source produced by the US Department of Veteran Affairs that augments a ‘legacy’ classification system, called VA-NDF,12 via a description logic-based formal reference model that groups drug products into the high-level drug classes for Chemical Structure, cellular or subcellular Mechanism of Action, organ-level or system-level Physiologic Effect, drug–disease relationship describing the Therapeutic Intent, and Pharmacokinetics describing the mechanisms of absorption and distribution of an administered drug within the body. The model also represents a categorization of drug products based on a single-inheritance hierarchy of drug classes (VHA Drug Class).
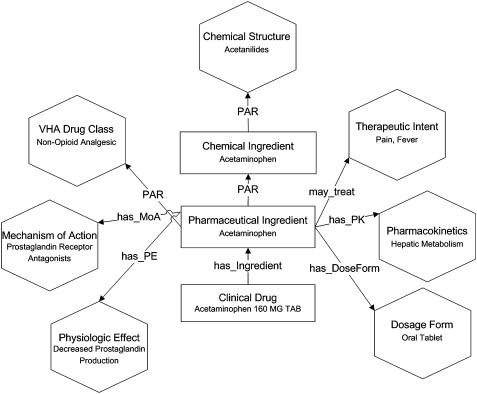
Figure 2 shows the structure of NDF-RT8: the hexagons represent multiple-inheritance reference hierarchies, whereas the rectangles are named sets of concepts each representing a level of abstraction used to describe medications. The hierarchies for Chemical Structure, Mechanism of Action, and Physiologic Effect were developed by matching ingredient names to the National Library of Medicine's Medical Subject Headings (MeSH16). For building the Therapeutic Intent hierarchy, automated algorithms were developed by Carter et al8 based on how a multitude of ingredients were used to treat diseases. Similarly, for developing the Pharmacokinetics hierarchy, techniques were proposed by Chute et al11. For example, Acetaminophen 160 MG Oral Tablet (RxCUI=282464), which has a correspondence to VUID=4007166, maps to NDF-RT code=C32112 (ACETAMINOPHEN 160MG TAB (VA Product)). This drug product is then categorized under the different NDF-RT hierarchies as follows (figure 2): Acetanilides (Chemical Structure), Prostaglandin Receptor Antagonists (Mechanism of Action), Decreased Prostaglandin Production (Physiologic Effect), Fever and Pain (Therapeutic Intent), Hepatic Metabolism (Pharmacokinetics), and Non-Opioid Analgesics (VHA Drug Class). Our objective is to study such a categorization of drug products within the different NDF-RT drug class hierarchies.
Figure 2.
NDF-RT Information Model (adapted from Carter JS, Brown SH, Erlbaum MS, et al8).
Related work
In the recent past, numerous researchers have studied the issue of content coverage and adequacy of representation in drug terminologies. Brown et al2 investigated the coverage of medication list phrases extracted from dictated patient notes using NDF-RT. The authors used a health-vocabulary-indexing tool to preprocess the medication list phrases and to algorithmically map them to NDF-RT codes. The preliminary mappings were used to populate evaluation forms for human expert review, and the results indicated that NDF-RT covered 97.8% of the medication list phrases. This group of researchers also studied the representation adequacy of the Physiologic Effects hierarchy in NDF-RT for commonly prescribed medications.9 In this work, 10 physician reviewers classified the physiologic effects of 10 drugs, and rated the accuracy of the selected terms. The results were analyzed by generating descriptive statistics of the number of Physiologic Effect classes identified for each drug, the number of raters who assigned each class to the drugs, and the mean confidence score of the reviewers for the accuracy of the assigned classes. Overall, reviewers provided 308 drug classifications using 127 unique classes. The numbers of classes assigned to a drug ranged from two for Omeprazole to 34 for Triamcinolone. Arguably, the physiologic effects modeled became more disperse with drugs having and inducing multiple physiologic processes. The study concluded that the Physiologic Effects reference hierarchy in NDF-RT is appropriate for modeling the physiologic effects of medications. Fung et al17 created an interface terminology called RxTerms derived from RxNorm. The RxTerms provides drug name information intended for use with electronic prescribing and medication histories as needed in personal health records. It excludes drugs from RxNorm that are obsolete or unavailable in the USA, and has commonly used synonyms and abbreviations (eg, HCTZ for Hydrochlorothiazide). Additional synonyms from other sources further enhance the user-friendliness. An initial study for the top 200 prescriptions for 2003 by number of US prescriptions dispensed indicated that RxTerms covered 99% of generic and branded drug names.
Various studies have also been carried out that have focused on the categorization and classification of drug products, primarily involving NDF-RT. Carter et al8 evaluated a UMLS metathesaurus co-occurrence mining algorithm to connect medications and diseases that may indicate a treatment relationship in NDF-RT. The study was based on 16 years of co-occurrence data for drug–disease pairs, out of which, the mining algorithm generated 977 candidate drug–disease pairs for approximately 100 ingredients present in those drugs. Individual physician reviewers rated 80% of those candidates as ‘appropriate’, which were then used to initialize the hierarchy for Therapeutic Intent in NDF-RT. A similar study was carried out by Chute et al11 to create a hierarchy for capturing information about Pharmacokinetics in NDF-RT. Orthogonal to such work, in a separate effort, Carter and his colleagues10 examined the hierarchies for Chemical Structure, Mechanism of Action, Physiologic Effect, and Therapeutic Intent from three different source terminologies to better understand their information content and evaluate NDF-RT's semantic coverage. The study revealed that NDF-RT's categorical reference model accommodates more than 76% of the information identified in drug class names, although a new NDF-RT reference axis of drug formulations could improve NDF-RT's coverage to 85%.
Our work in this report is also aims to evaluate the categorization and classification of drug products in RxNorm with respect to the classes in NDF-RT. Specifically, we will study how RxNorm drug products are categorized under different single- and multiple-inheritance NDF-RT drug classes, as well as investigate the coverage of the NDF-RT drug class hierarchies. Furthermore, we will discuss issues regarding inconsistent mapping, as well as representation of incoherent information in both RxNorm and NDF-RT.
Materials
The following materials were used in this study:
RxNorm November 17, 2008 full update release data consistent with the 2008AB version of the UMLS, and accessible via http://download.nlm.nih.gov/umls/kss/rxnorm/RxNorm_full_11172008.zip. This dataset included 4112 ingredients, 100 dose forms, 13 923 clinical drug components, 8180 clinical drug forms, 18 228 clinical drugs, 10 029 brand names, 14 154 branded drug components, 11 643 branded drug forms, 14 891 branded drugs, 288 branded packs, and 224 generic packs. Furthermore, the dataset had over 500 000 relationships between these RxNorm entities. The numbers quoted above do not include the RxNorm entities that are ‘obsolete’ (ie, tagged ‘O’ (for obsolete) in the rich release format (RRF) tables).
NDF-RT March 11, 2008 public inferred edition released with 2008AB version of the UMLS, and accessible via http://evs.nci.nih.gov/ftp1/NDF-RT/Archive/NDF-RT_XML_Inferred%202008-03-11.zip. This dataset included 24 671 drug concepts which could be of types ‘Ingredient’ (6675), ‘VA Class’ (572), ‘VA Product’ (17 424), and so on. From this, 13 128 mapped to RxNorm CUIs, and 12 724 mapped to MeSH unique identifiers. Additionally, the dataset had over 300 000 relationships between NDF-RT entities.
Methods
Classification of RxNorm drug products using NDF-RT single-inheritance drug class hierarchy
We evaluated three different approaches to assigning NDF-RT single-inheritance drug classes (VHA Drug Class) to RxNorm drug products, and describe them below.
Classification using the RxNorm database
The RxNorm data are distributed in metathesaurus relational or rich release format (RRF) tables. These tables contain information about concepts that appears in RxNorm such as the concept identifier (RxCUI), concept name, or correspondences to other drug vocabularies. We first extracted all the RxNorm concepts with TTY=SCD (Semantic Clinical Drug) from the RRF tables, as SCD is essentially the most complete generic name for a drug, giving ingredients, their strengths, and a dose form. Then, for all extracted concepts, we queried for two types of attribute-value information: VAC and VA_CLASS_NAME representing the VHA Drug Class identifier and its name, respectively. Together, these two attributes allow us to determine which VHA Drug Class is assigned to a given RxNorm drug product. As an example, Acetaminophen 160 MG Oral Tablet is assigned a VAC=C8838 and VA_CLASS_NAME=Non-Opioid Analgesics. Hence, we classify Acetaminophen 160 MG Oral Tablet under the VHA Drug Class Non-Opioid Analgesics.
Classification using mappings between RxNorm and NDF-RT drug products
As described above, RxNorm contains mappings from its concepts to one or more concepts in external drug terminologies. In the case of mappings to NDF-RT, the RxCUI for an RxNorm concept is mapped to one or more VUIDs of NDF-RT concepts, which are subsequently classified under a VHA Drug Class. For example, Cetirizine 5 MG Oral Tablet is mapped to VUID=4012844 (CETIRIZINE HCL 5MG TAB) which, in turn, is assigned to the VHA Drug Class Antihistamines, Piperazine. Consequently, we leveraged this mapping between RxCUIs and VUIDs to assign Cetirizine 5 MG Oral Tablet under the VHA Drug Class Antihistamines, Piperazine.
Classification using mappings between RxNorm and NDF-RT ingredients
Although our methods described above enable assignment of RxNorm drug products to VHA Drug Classes, many RxNorm drug products could not be classified under any NDF-RT drug class for two main reasons.
Approximately 52% (9535 of 18 228) of RxNorm drug product concepts (ie, SCDs) did not have the VAC and VA_CLASS_NAME information in the RRF tables (eg, Diazoxide 50 MG Oral Tablet). Consequently, classification using the RxNorm database could not assign a VHA Drug Class to such RxNorm drug products.
Approximately 53% (9660 of 18 228) of RxNorm drug product concepts (ie, SCDs) did not have correspondence to a NDF-RT clinical drug concept (eg, Sodium Bicarbonate 500 MG Oral Capsule). As a result, classification using mappings between RxNorm and NDF-RT drug products could not assign a VHA Drug Class to such RxNorm drug products.
To address these limitations, we explored leveraging chemical ingredients present in a particular drug product for assignment of drug classes. In particular, for a given drug product in RxNorm, our algorithm first identifies all the RxNorm and NDF-RT ingredient concepts for the drug. The method then determines the drug product(s) in NDF-RT that contain only those NDF-RT ingredient concepts identified from the first step, and extracts the corresponding VHA Drug Classes. For example, Cimetidine 2 MG/ML Oral Solution contains the ingredient Cimetidine. This ingredient is, in turn, present in seven different drug products in NDF-RT, all of which are categorized under the VHA Drug Class, Histamine Antagonists. As a result, our method assigns Cimetidine 2 MG/ML Oral Solution to the VHA Drug Class, Histamine Antagonists.
Although in most cases, this method will assign only one VHA Drug Class to a particular RxNorm drug product, there are two exceptions.
In some cases, the NDF-RT drug products having the exact same chemical ingredient may be categorized under unique and different VHA Drug Classes. For instance, Diazoxide 50 MG Oral Tablet contains the ingredient Diazoxide, which is categorized under the VHA Drug Classes Antihypertensives, Other, and Antihypoglycemics. Consequently, our method assigns Antihypertensives, Other, and Antihypoglycemics as the drug class for the RxNorm drug product Diazoxide 50 MG Oral Tablet.
In some cases, a particular drug product might comprise more than one ingredient, as a result of which it might be categorized under multiple VHA Drug Classes. For example, Ethinyl Estradiol 0.0025 MG/Norethindrone 0.5 MG Oral Tablet is composed of two ingredient concepts: Ethinyl Estradiol and Norethindrone. Incidentally, NDF-RT contains two clinical drug products, Femhrt 1/5 Tab and Necon 0.5/35 Tab, that comprise both these ingredients, and are categorized under different VHA Drug Classes, Hormones/Synthetics/Modifiers, Other and Contraceptives, Systemic. Consequently, our technique categorizes the RxNorm drug product Ethinyl Estradiol 0.0025 MG/Norethindrone 0.5 MG Oral Tablet under both these drug classes.
Classification of RxNorm drug products using NDF-RT multiaxial class hierarchies
As illustrated above, in addition to the single-inheritance hierarchy for VHA Drug Classes, NDF-RT groups drug products into the high-level classes of Chemical Structure, Mechanism of Action, Physiologic Effect, Therapeutic Intent, and Pharmacokinetics. We evaluated the categorization of RxNorm drug products for these drug classes by traversing the relationships between corresponding NDF-RT drug products with the multiaxial class hierarchies (table 2 and figure 2). For example, by traversing the has_PE association in NDF-RT, we could assign the Physiologic Effect classes Decreased Dopamine Activity and Decreased Norepinephrine Activity to the RxNorm drug Chlorprothixene 50 MG Oral Tablet.
Table 2.
Relationships between NDF-RT drug products and multiaxial class hierarchies
| Class type | No of NDF-RT classes | NDF-RT relationship |
| Chemical Structure | 8339 | has_Ingredient |
| Mechanism of Action | 346 | has_MoA |
| Physiologic Effect | 1758 | has_PE |
| Therapeutic Intent | 4188 | may_treat |
| Pharmacokinetics | 58 | has_PK |
Results
Classification of RxNorm drug products using NDF-RT single-inheritance drug class hierarchy
Coverage
Table 3 shows the number of RxNorm drug products (TTY=SCD) that were not assigned a VHA Drug Class using the methods described above: Direct-Map (under Classification using the RxNorm database), Drug-Level-Map (under Classification using mappings between RxNorm and NDF-RT drug products), and Ingredient-Level-Map (under Classification using mappings between RxNorm and NDF-RT ingredients).
Table 3.
RxNorm drug products (not mutually exclusive) without VHA Drug Class assignments
| Mapping type | % of RxNorm drugs without a VHA Drug Class assignment (n=18228) |
| Direct-Map | 53% (9711) |
| Drug-Level-Map | 55% (10060) |
| Ingredient-Level-Map | 49% (8893) |
For approximately 53% (9711 of 18 228) of RxNorm drug products that could not be mapped to VHA Drug Classes via the Direct-Map approach, 9604 (out of 9711) did not have any mapping specified to a NDF-RT drug entity in the RRF tables (and hence the attributes VAC and VA_CLASS_NAME were missing), and, for the remaining 107 (out of 9711) RxNorm drug products with corresponding NDF-RT entries in the RRF tables, the attributes VAC and VA_CLASS_NAME were missing. Examples of the latter include Nifedipine 60 MG Extended Release Tablet and Verdenafil 10 MG Oral Tablet.
For approximately 55% (10 060 of 18 228) of RxNorm drug products that could not be mapped to VHA Drug Classes via the Drug-Level-Map approach, 9621 (out of 10 060) did not have a corresponding VUID entry for NDF-RT concepts. Consequently, such RxNorm drug products could not be assigned any VHA Drug Class. For the remaining 439 (out of 10 060) uncategorized drug products, their VUIDs from the RRF tables did not match any corresponding NDF-RT entity. As an example, Nebivolol 5 MG Oral Tablet had VUIDs 4027134 and 4027137 listed in the RRF tables, although both 4027134 and 4027137 were missing from NDF-RT (ie, there were no NDF-RT entities that corresponded to these VUIDs).
For the Ingredient-Level-Map approach, 49% (8893 of 18 228) of the RxNorm drug products could not be mapped to VHA Drug Classes. Most of these cases were because either ingredient VUIDs between RxNorm and NDF-RT did not match or RxNorm ingredients were mapped to multiple VUIDs of NDF-RT ingredient concepts, and there were no NDF-RT drug products containing only those ingredients. For example, Glucose 500 MG/ML Oral Solution contains the RxNorm ingredient Glucose, which maps to NDF-RT ingredient VUIDs 4019541 (Glucose) and 4017760 (Dextrose). However, there are no NDF-RT drug products with both Glucose and Dextrose as the ‘sole ingredients’, and as a consequence no VHA Drug Class was assigned to Glucose 500 MG/ML Oral Solution.
Similarities
Table 4 shows the ‘similarity’ (or overlap) of the drug class assignment for the methods described above.ii Comparing the Direct-Map and Drug-Level-Map approaches, 44% (8801 of 18 228) of RxNorm drug products were assigned the same VHA Drug Class.
Table 4.
RxNorm drug products (not mutually exclusive) with similar VHA Drug Class assignments
| Mapping type | % of RxNorm drugs with similar VHA Drug Class assignment (n=18228) |
| Direct-Map vs Drug-Level-Map | 44% (8001) |
| Drug-Level-Map vs Ingredient-Level-Map | 24% (4379) |
| Direct-Map vs Ingredient-Level-Map | 25% (4473) |
Similarly, comparing the Drug-Level-Map and Ingredient-Level-Map approaches, approximately 24% (4379 of 18 228) of RxNorm drug products were assigned the same VHA Drug Class, whereas, for the Direct-Map versus the Ingredient-Level-Map approaches, 25% (4473 of 18 228) of RxNorm drug products had the same VHA Drug Class.
Dissimilarities
We also analyzed the differences in class assignments for the three approaches as shown in table 5. Comparing the Direct-Map and Drug-Level-Map approaches, approximately 3% (562 of 18 228) of RxNorm drug products were assigned different VHA Drug Classes. For instance, 24 HR Verapamil 180 MG Extended Release Capsule was assigned to the classes Calcium Channel Blockers and Antiarrhythmics by Direct-Map and Drug-Level-Map, respectively. Furthermore, in 525 (out of 562) cases, drug products were assigned classes by only one approach: Tramadol 200 MG Extended Release Tablet was assigned to the class Non-Opioid Analgesics by the Direct-Map approach, whereas no class was assigned by the Drug-Level-Map approach.
Table 5.
RxNorm drug products (not mutually exclusive) with dissimilar VHA Drug Class assignments
| Mapping type | % of RxNorm drugs with dissimilar VHA Drug Class assignment (n=18228) |
| Direct-Map vs Drug-Level-Map | 3% (562) |
| Drug-Level-Map vs Ingredient-Level-Map | 53% (9679) |
| Direct-Map vs Ingredient-Level-Map | 54% (9828) |
Comparing the Drug-Level-Map and Ingredient-Level-Map approaches, approximately 53% (9679 of 18 228) of RxNorm drug products were assigned different VHA Drug Classes, whereas 54% (9828 of 18 228) of drugs were assigned different VHA Drug Classes when comparing the Direct-Map and Ingredient-Level-Map approaches. As an example, both Direct-Map and Drug-Level-Map approaches assign Acetaminophen 24 MG/ML Oral Solution to the VHA Drug Class Non-Opioid Analgesics. However, the Ingredient-Level-Map technique assigned this drug to three additional classes: Cold Remedies, Other Pharmaceutical Aids/Reagents, and Antimigraine Agents.
Classification of RxNorm drug products using NDF-RT multiaxial class hierarchies
For multiaxial NDF-RT drug class hierarchies, we focused on the coverage of RxNorm drug products under these classes (table 6) and also investigated classes that were not assigned any drug products (table 7).
Table 6.
RxNorm drug products (not mutually exclusive) without NDF-RT multiaxial drug class assignment
| Drug class type | % of RxNorm drugs without multiaxial drug class assignment (n=18228) |
| Chemical Structure | 65% (11871) |
| Mechanism of Action | 74% (13483) |
| Physiologic Effect | 74% (13525) |
| Therapeutic Intent | 74% (13443) |
| Pharmacokinetics | 98% (18012) |
Table 7.
NDF-RT multiaxial drug classes (not mutually exclusive) without any drug product classified
| Drug class type | % of NDF-RT drug classes without drug product classified |
| Chemical Structure | 81% (6810/8339) |
| Mechanism of Action | 42% (147/346) |
| Physiologic Effect | 75% (1312/1758) |
| Therapeutic Intent | 76% (3207/4188) |
| Pharmacokinetics | 88% (51/58) |
Chemical Structure
Approximately 65% (11 871 of 18 228) of RxNorm drug products were not assigned a Chemical Structure class. The majority (11 035 of 11 871) did not have a correspondence to a NDF-RT code, and hence were not classified. For the remainder (836 of 11 871 having a correspondence to a NDF-RT code), the has_Ingredient association was not specified. Furthermore, 81% (6810 of 8339) of Chemical Structure classes did not have any drug product categorized under them.
Mechanism of Action
Approximately 74% (13 483 of 18 228) of RxNorm drug products were not assigned a Mechanism of Action class, of which, 11 043 did not have a correspondence to a NDF-RT code. For the remainder (2448 RxNorm drug products having correspondence to a NDF-RT code), has_MoA association was not specified. Additionally, 42% (147 of 346) of Mechanism of Action classes did not have any drug product categorized under them.
Physiologic Effect
Around 74% (13 525 of 18 228) of RxNorm drug products were not assigned a Physiologic Effect class, of which, 11 035 did not have a correspondence to a NDF-RT code, and the remainder (2490) did not have a has_PE association specified in NDF-RT. Additionally, 75% (1312 of 1758) of Physiologic Effect classes did not have any drug product categorized under them.
Therapeutic Intent
Approximately 74% (13 443 of 18 228) of RxNorm drug products were not classified under a Therapeutic Intent drug class. A total of 11 035 drugs from this list did not have a correspondence to a NDF-RT code, and for the remaining 2408, the may_treat association in NDF-RT was not specified. Also, around 76% (3207 of 4188) of Therapeutic Intent classes did not have any drug products classified under them.
Pharmacokinetics
A very large proportion (98% (18 012 of 18 228)) of RxNorm drug products were not classified under a Pharmacokinetics drug class. Of these, 11 035 did not have a corresponding code in NDF-RT, and for the remaining 6977 drugs, no has_PK association was specified in NDF-RT. Furthermore, 88% (51 of 58) of Pharmacokinetics drug classes did not have a drug product classified under them.
Discussion
Significance
The overarching goal of this study was to analyze the utility of multiaxial NDF-RT drug classes for supporting aggregation and analysis of generic and branded drug products coded in RxNorm. Several studies18–21 have previously reported the use of RxNorm for coding clinical medication data and its application in clinical research, although the ability to aggregate and classify medication data has always been a common problem for any kind of clinical and epidemiological research, particularly research on large populations. Such issues become particularly relevant in the context of building a national health information network where medical institutions and organizations will be required to comply with national terminology standards for achieving interoperability, and the ability to classify standards-based medication data using drug classes and multiple ingredient generics will be essential in several research areas (eg, studying drug allergies, pharmacy prescriptions, adverse event reactions, pharmacogenomics, and clinical decision support). Our study investigates this problem on the basis of RxNorm and NDF-RT mappings, and proposes methods for determining new, as well as validating existing, ways of classifying drugs using NDF-RT drug classes. Our findings not only illustrate the applicability of RxNorm and NDF-RT as standards to coding medication data for building and deploying interoperable electronic health record systems, but also identify multiple issues in drug classification that can be improved to facilitate the use of RxNorm and NDF-RT mappings for virtually all clinical research.
In particular, we investigated several interesting and relevant discussion points. Firstly, as is evident from our findings, approximately 54% of RxNorm drug products (TTY=SCD) do not have a correspondence in NDF-RT. Similarly, approximately 45% of drug products in NDF-RT are missing from RxNorm. Although this can mostly be attributed to differences in drug strength, dose form, route, and reaction time (12HR, 24HR, etc), it presents a significant ‘gulf’ between the two drug terminologies, as this is one of the major factors contributing to a small percentage of RxNorm drug products being classified in NDF-RT multiaxial drug classes (and vice versa).
Further, as illustrated in table 7, 81% (6810 of 8339) of Chemical Structure classes, 42% (147 of 346) of Mechanism of Action classes, 75% (1312 of 1758) of Physiologic Effect classes, 76% (3207 of 4188) of Therapeutic Intent classes, and 88% (51 of 58) of Pharmacokinetics classes did not have any drug products classified under them. This presents an important lack-of-coverage problem in NDF-RT, and addressing this may provide significant benefits for analyzing and utilizing the drug classification hierarchies.
Our techniques also discovered multiple issues in mappings between RxNorm and NDF-RT concepts. In general, RxNorm contains mappings from its concepts to one or more concepts in external drug terminologies, and in the case of mappings to NDF-RT, the RxCUI for an RxNorm concept is mapped to one or more VUIDs of NDF-RT concepts. However, in many cases, the VUIDs present in the RxNorm RRF tables did not match the RxCUI entry in NDF-RT. As an example, Clemastine 0.1 MG/ML Oral Solution (RxCUI=755824) has a VUID=4006429. However, this VUID in NDF-RT maps to RxCUI=197513, which, according to RxNorm RRF tables, has been archived on Febraury 6, 2008, and subsequently merged with RxCUI=755824. Similarly, Polyvinyl Alcohol 0.014 ML/ML Ophthalmic Solution (RxCUI=142004) maps to two VUIDs in RxNorm: 4009166 and 4009167. In NDF-RT, both these VUIDs correspond to RxCUI=312485, which was archived on September 9, 2008 and merged with RxCUI=142004. Such housekeeping and maintenance issues can lead to erroneous mappings and classifications, and hence should be addressed by the terminology curators.
Along the same lines, we also found that, in some cases, a particular RxNorm ingredient concept may map to more than one NDF-RT ingredient concept. For example, Beta Carotene maps to two different VUIDs, 4017793 and 4020599, representing Beta Carotene and Carotene, Beta, respectively. In this case, the RxCUI has not been archived in RxNorm; however, the NDF-RT VUIDs seem to be redundant.
For multiaxial drug classes in NDF-RT, a drug product can be categorized under multiple classes. However, this is not applicable in the case of single-inheritance class hierarchy for VHA Drug Classes. To quote from previous work10: ‘This single-class structure has obvious limitations: it is impossible to categorize a drug as both an Anti-hypertensive and a Beta-Blocker. Although NDF allows products to be placed in up to two classes, in practice all products belong to a single class’. In concordance with Carter et al,10 we believe that this limitation needs to be addressed, since the VHA Drug Classes, although derived from the legacy VA-NDF, have significant clinical implications for drug classifications.
Specifically, we hypothesized that our Ingredient-Level-Map technique for drug product categorization provided the initial steps to address this requirement. This technique leveraged chemical ingredients present in a particular drug product that gives the drug its distinctive clinical properties to determine which VHA Drug Class would be the most ‘appropriate’ assignment. For instance, while Urea 200 MG/ML Topical Lotion, which was assigned to only a single VHA Drug Class (Emollients) originally, was assigned to three additional classes by our technique: Oxytocics, Diuretics, Other and Pharmaceutical Aids/Reagents. While a formal evaluation of the accuracy and clinical significance of such class assignments is forthcoming, the Ingredient-Level-Map technique provides preliminary insights to allow drug products to be assigned to multiple VHA Drug Classes. Furthermore, this technique may also prove useful in assigning new VHA Drug Classes to RxNorm drug products that were originally never assigned (ie, both Direct-Map and Drug-Level-Map methods did not find a class association).
Limitations and future work
In this study, we focused on only two drug terminologies, RxNorm and NDF-RT. Realizing that many clinical and healthcare practices use various other drug databases (eg, Micromedex, First DataBank), we believe that additional investigations between other publicly available drug terminology resources/databases will be beneficial in identifying and addressing issues similar to those studied here. Furthermore, conceptually, our approach relates to the problem of ontology and terminology alignment: the process of finding correspondence between concepts in two or more terminologies. Numerous approaches (see Choi et al22 and Kalfoglou and Schorlemmer23 for surveys) have been proposed over the last few years for addressing this problem, which apply a range of lexical and semantic matching techniques for finding correspondences. In general, mappings are created semiautomatically, with the user working directly with a tool or manipulating the output generated by a mapping tool. This is often an iterative process where the user approves or rejects the proposed correspondences, and that information is used by an automated procedure to make further suggestions and refinements. Although in this work our primary goal was to evaluate existing correspondences, in future we plan to investigate several state-of-the-art ontology alignment techniques for creating correspondences between RxNorm and NDF-RT drug entities. In particular, we plan to investigate CogZ24—a suite of tools that provide cognitive support and visualization for human-guided ontology alignment. Based on the Protégé Prompt mapping framework,25 CogZ supports visual construction of user-defined, as well as automatic, generation of candidate mappings. A user can apply a range of filters to the automatically generated mappings to limit the complexity of the correspondences, and review and verify the appropriate ones. For automatic detection of candidate mappings, CogZ uses linguistic similarity (based on lexical distance) between the terms and concepts of the source and target ontologies to find similar matches. The tool also allows easy extension for adding more sophisticated term-comparison algorithms, such as WordNet26 lookup for detecting synonymy or concept negation.
Conclusion
The RxNorm and NDF-RT are publicly available and widely used drug terminologies in the US healthcare system. In this work, we studied how drug products in RxNorm are classified under drug classes in NDF-RT. Our investigation identified potential limitations of the existing classification system, as well as various issues in specification of correspondences between the concepts in RxNorm and NDF-RT. Our proposals and methods provide the preliminary steps to addressing some of the requirements.
Acknowledgments
This research was supported in part by NHGRI grant U01HG04599 (eMERGE network). We also thank Olivier Bodenreider and Mark Siska for numerous fruitful discussions leading to this work.
Footnotes
Funding: This research was supported in part by the NHGRI grant U01HG04599 (eMERGE network).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Note that in RxNorm, drug products are only mapped to the NDF-RT single hierarchy of ∼550 drug classes.
References
- 1.Liu S, Ma W, Moore R, et al. RxNorm: prescription for electronic drug information exchange, IT Professional 2005;7:17–23, ISSN 1520–9202. [Google Scholar]
- 2.Brown SH, Elkin PL, Rosenbloom T, et al. VA national drug file reference terminology: a cross-institutional content coverage study, in: Medinfo: studies in health technology and informatics, IOS Publications, 2004:477–81 [PubMed] [Google Scholar]
- 3.First DataBank http://www.firstdatabank.com (accessed 10 Sep 2009).
- 4.Micromedex http://www.micromedex.com (accessed 10 Sep 2009).
- 5.Medispan http://www.medispan.com (accessed 10 Sep 2009).
- 6.Multum http://www.multum.com (accessed 10 Sep 2009).
- 7.Baader F, Calvanese D, McGuinness D, et al. The description logic handbook, Cambridge University Press, 2003 [Google Scholar]
- 8.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA Medication Reference Terminology Using UMLS Metathesaurus Co-Occurrences. AMIA Annu Symp Proc 2002:116–20 [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbloom ST, Awad J, Speroff T, et al. Adequacy of representation of the national drug file reference terminology physiologic effects reference hierarchy for commonly prescribed medications. AMIA Annu Symp Proc 2003:569–73 [PMC free article] [PubMed] [Google Scholar]
- 10.Carter J, Brown S, Bauer BA, et al. Categorical information in pharmaceutical terminologies. AMIA Annu Symp Proc 2006:116–20 [PMC free article] [PubMed] [Google Scholar]
- 11.Chute CG, Carter J, Tuttle M, et al. Integrating pharmacokinetics knowledge into a drug ontology as an extension to support pharmacogenomics. AMIA Annu Symp Proc 2003:170–4 [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson SJ, Brown SH, Erlbaum MS, et al. A semantic normal form for clinical drugs in the UMLS: early experiences with the VANDF. AMIA Annu Symp Proc 2002;557–61 [PMC free article] [PubMed] [Google Scholar]
- 13.Bouhaddou O, Warnekar P, Parrish F, et al. Exchange of computable patient data between the Department of Veterans Affairs (VA) and the Department of Defense (DoD): Terminology Mediation Strategy. J Am Med Inform Assoc 2008;15:174–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton MM, Simonaitis L, Schadow G, et al. Medication and indication linkage: a practical therapy for the problem list? AMIA Annu Symp Proc 2008:86–90 [PMC free article] [PubMed] [Google Scholar]
- 15.Peters L, Bodenreider O. Using the RxNorm web services API for quality assurance purposes. AMIA Annu Symp Proc 2008:591–5 [PMC free article] [PubMed] [Google Scholar]
- 16.Medical Subjects Heading http://www.nlm.nih.gov/mesh (accessed 10 Sep 2009).
- 17.Fung KW, McDonald C, Bray BE. RxTerms: a drug interface terminology derived from RxNorm. AMIA Annu Symp Proc 2008:227–31 [PMC free article] [PubMed] [Google Scholar]
- 18.Richesson R, Krischer J. Data standards in clinical research: gaps, overlaps, challenges and future directions. J Am Med Inform Assoc 2007;14:687–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richesson R, Smith S, Malloy J, et al. Achieving Standardized medication data in clinical research studies: two approaches and applications for implementing RxNorm. J Med Syst 2009. doi:10.1007/s10916-009-9278-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnekar P, Bouhaddou O, Parrish F, et al. Use of RxNorm to exchange codified drug allergy information between Department of Veterans Affairs (VA) and Department of Defense (DoD). AMIA Annu Symp Proc 2007:781–5 [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez P, Podchiyska T, Weber S, et al. Automated mapping of pharmacy orders from two electronic health record systems to RxNorm within the STRIDE Clinical Data Warehouse. AMIA Annu Symp Proc 2009:244–8 [PMC free article] [PubMed] [Google Scholar]
- 22.Choi N, Song I, Han H. A survey on ontology mapping. ACM SIGMOD Record 2006;35:34–41 [Google Scholar]
- 23.Kalfoglou Y, Schorlemmer M. Ontology mapping: the state of the art. The Knowledge Engineering Review 18:1–31 [Google Scholar]
- 24.Falconer S. Cognitive support for semi-automatic ontology mapping. PhD Doctoral Thesis, Department of Computer Science, University of Victoria, 2009 [Google Scholar]
- 25.Noy N, Musen M. The PROMPT Suite: interactive tools for ontology merging and mapping. International Journal of Human-Computer Studies 19:983–1024 [Google Scholar]
- 26.WordNet A lexical database for English. http://wordnet.princeton.edu/ (accessed 22 Apr 2010).