Abstract
Objective
To classify automatically lung tumor–node–metastases (TNM) cancer stages from free-text pathology reports using symbolic rule-based classification.
Design
By exploiting report substructure and the symbolic manipulation of systematized nomenclature of medicine–clinical terms (SNOMED CT) concepts in reports, statements in free text can be evaluated for relevance against factors relating to the staging guidelines. Post-coordinated SNOMED CT expressions based on templates were defined and populated by concepts in reports, and tested for subsumption by staging factors. The subsumption results were used to build logic according to the staging guidelines to calculate the TNM stage.
Measurements
The accuracy measure and confusion matrices were used to evaluate the TNM stages classified by the symbolic rule-based system. The system was evaluated against a database of multidisciplinary team staging decisions and a machine learning-based text classification system using support vector machines.
Results
Overall accuracy on a corpus of pathology reports for 718 lung cancer patients against a database of pathological TNM staging decisions were 72%, 78%, and 94% for T, N, and M staging, respectively. The system's performance was also comparable to support vector machine classification approaches.
Conclusion
A system to classify lung TNM stages from free-text pathology reports was developed, and it was verified that the symbolic rule-based approach using SNOMED CT can be used for the extraction of key lung cancer characteristics from free-text reports. Future work will investigate the applicability of using the proposed methodology for extracting other cancer characteristics and types.
Cancer stage is assigned according to standard criteria such as the tumor–node–metastases (TNM) staging standard.1 Cancer stage annotation could add significant value to existing incidence and mortality data collected by population-based cancer registries as it is both a basis for planning clinical management as well as the major prognostic factor required to allow analysis of outcomes across a population. Despite its importance, the quality of stage data collected in population-based datasets is often incomplete or inaccurate due to incomplete medical records, data entry errors, or staging misinterpretations.2 Currently no Australian state or territory captures perpetual population-based cancer stage data.3 4 The absence of stage data has been identified in the Australian National Cancer Data Strategy as a fundamental gap in population-based data collections.4
Multidisciplinary cancer teams attended by pathologists, radiologists, and cancer specialists assign consensus cancer stage by reference to tissue-specific staging systems providing a ‘gold standard’; however, such teams only operate in larger centers. Automatic classification of relevant clinical and pathological data to assign stage could potentially reduce reliance on expert clinical staff, reducing cost and improving efficiency and availability of cancer stage data.
To assist the collection of TNM stages, the College of American Pathologists (CAP) produced cancer case summaries as synoptic checklists containing tumor site-specific items including cancer staging information.5 6 The value of cancer stage along with other key characteristics in the CAP cancer checklists has been recognized by the American College of Surgeons Commission on Cancer, and documentation of checklist items in pathology reports is now mandated as a minimum requirement at Commission on Cancer-approved cancer programs.5 6 While this is a positive step toward standardizing the collection of TNM stages, data reliability remains dependent on the skill and experience of the individual clinician assigning and documenting the stage. As a result, stages documented by clinicians in reports are not used for population-based data collections. Despite this, TNM stages explicitly recorded in free text can be reliably extracted using regular expression pattern matching.7 8
Related work in extracting key cancer characteristics from free text can be used to build logic to derive a cancer stage. To achieve reliable population cancer stage data, reference to staging guideline criteria is required. The medical text analysis system/pathology system proposed by Coden et al9 automatically instantiates an extensible and modifiable cancer disease knowledge representation model (CDKRM) to capture cancer disease characteristics including cancer stage. Natural language processing (NLP), machine learning and rules were used to populate the CDKRM. Selected cancer characteristics were evaluated and showed promise when evaluated against a corpus of pathology reports for patients with colon cancer. Although cancer stage characteristics were proposed to be populated in the CDKRM, either by extracting explicitly mentioned stages within pathology reports or deriving it from other information such as primary tumor description, lymph node status, and the presence or absence of metastases, cancer stage was not ultimately included in the evaluation.
We previously focussed on automating the assignment of TNM stages for lung cancer patients by an analysis of pathology and radiology reports2 10 using a system (cancer stage interpretation system; CSIS) predicated on the same guidelines used by clinicians in assigning stage based on pathology and radiology reports. An extract was produced consisting of sentences that were found to contribute to the final staging decision. These sentences from reports were evaluated against staging guideline criteria, herein termed ‘staging factors’. Machine learning techniques based on support vector machines were used to learn the associations between the set of sentences in the report and each of the staging factors. Classification accuracy was shown to be adequate for purposes of population-level research and for indicative staging before multidisciplinary team (MDT) meetings.
Although machine learning techniques show promise, extensibility and/or generalization to staging other types of cancer may be limited. Classification models not specific to lung cancer would have to be trained to relate new staging factors to sentences found in non-lung cancer pathology reports. We therefore hypothesized that the use of complex NLP principles in conjunction with medical ontologies such as the systematized nomenclature of medicine–clinical terms (SNOMED CT),11 herein termed ‘symbolic rule-based classification’, may improve the generalizability of classifiers to new cancer types or reporting modalities.
Here we parsed pathology reports using NLP to identify SNOMED CT concepts of relevance, and tested whether these concepts were subsumed by concepts relating to staging factors. References to staging factors in the free text were then used to build logic to derive the TNM cancer stage. Lung cancer TNM staging was used to illustrate the symbolic rule-based approach.
Methods
System description
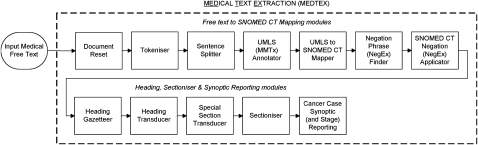
The symbolic rule-based cancer stage classification system proposed here was developed using General Architecture for Text Engineering (GATE),12 an open source architecture for NLP. GATE is a framework for the development and deployment of language engineering components and resources for natural language applications such as information extraction. The proposed pipeline application builds upon the medical text extraction (MEDTEX) system8 and is shown in figure 1. Here, the cancer case synoptic reporting module implements the relevant algorithms for extracting staging factors from free text and building the logic to calculate the TNM stage for each report.
Figure 1.
Medical text extraction (MEDTEX) pipeline application used to classify cancer stages.
The system comprises modules mapping free text to SNOMED CT (previously reported by Nguyen et al),8 along with further preprocessing to segment the free text into sections. Annotations generated from these steps were used for the extraction of staging factors and calculation of the TNM stage.
The free text to SNOMED CT mapping modules identify SNOMED CT concepts in medical free text. The tokeniser module splits the free text into tokens and also identifies length measurements and units, explicitly mentioned TNM cancer stages, and legacy SNOMED ID codes. In addition, lymph node station identifiers (eg, ‘No 5 lymph node’ and ‘No 7, 10, and 11 lymph nodes’) frequently encountered in free-text pathology reports were identified and are important for N staging.
The unified medical language system (UMLS) annotator and UMLS to SNOMED CT mapper maps strings in the free text to UMLS13 and SNOMED CT concepts, respectively. The negation phrase finder finds common medical negation (eg, ‘no evidence of’, ‘none of’, and ‘clear of’) and possibility (eg, ‘possible’ and ‘suspicious for’) phrases in the free text. Possibility phrases were included to minimize the number of false-positive findings and diseases that may be extracted. Negation phrases were associated with neighboring SNOMED CT findings and disease concepts in the SNOMED CT negation applicator module. An additional concept, ‘involved’ (248448006), was added to the list of terms considered for negation, as this concept was commonly used to refer to the involvement of tumors in body structures. Both negation phrase finder and SNOMED CT negation applicator modules were based on the NegEx algorithm.14 15
The heading, sectioniser and synoptic reporting modules were added to MEDTEX to allow for the filtering and processing of appropriate substructures (or sections) in the free text. The heading gazetteer searches for strings in the free text containing common pathology report headings, and the heading transducer refines the list of potential headings by applying regular expression templates to identify actual report headings. The sectioniser uses the report headings to segment the report into sections and annotates them with their corresponding section names. The optional special section transducer applied regular expression rules over self-contained parts of the document such as SNOMED ID and/or sections toward the end of the document containing identifier information such as names and facilities.
Finally, the cancer case synoptic reporting module extracts staging factors and computes TNM stage. The SNOMED CT encoded version of the CAP cancer checklist (version 1.5)5 relating to lung cancer resections (based on the AJCC 6th edition)1 was used to identify staging factors for inclusion into the module. Extraction of checklist items from free text relevant to staging was implemented using the subsumption querying protocol discussed in the next section. Logic based on the staging guidelines was built from the staging factor subsumption results to calculate the TNM stage.
Subsumption querying protocol
The subsumption querying protocol uses the subsumption (IS_A) relationships in SNOMED CT. It allows for the testing of specializations (subtypes) or generalizations (supertypes) of expressions. Expressions consist of single concepts to more complex expressions that include multiple concepts and refinements. Refinements were defined using attribute name–value relationships. Over 50 defining attributes (or concept model attributes) can be used to model concepts. Valid attributes used to define concepts can be obtained from the SNOMED CT user guide.9 Multiple attributes can be grouped together (into role groups) to avoid ambiguity in concept definitions when, for example, multiple anatomical structures are involved. Expressions can be constructed to represent clinical concepts precisely when there is no suitable predefined concept. This process is termed post-coordination. An example notation of an expression with a single ‘focus’ concept, ungrouped attribute and grouped attribute is as follows:
<focus concept>:
<ungrouped attribute name>=<ungrouped attribute value>
{<grouped attribute name>=<grouped attribute value>}
Multiple focus concepts are separated by a plus sign (‘+’), while multiple attributes are separated by commas (‘,’).
For subsumption querying, expressions are transformed into a common normalized form16 from which rules can be used to test subsumption between expressions. In any subsumption test, a candidate expression is tested for subsumption by a predicate expression. In other words, the candidate expression is being tested to see if it is subsumed by the predicate expression (specialization), or conversely the predicate expression is being tested to see if it subsumes the candidate expression (generalization). Piecewise subsumption queries were implemented whereby concepts and attribute names and values from predicate and candidate expressions were individually tested for subsumption using rules detailed in the SNOMED CT transforming expressions to normal forms publication.16
Two methods for generating expressions were employed. When concepts are fully modeled, expressions were obtained from the concept's normal form. For partly modeled concepts, either a new SNOMED CT extension was created to provide sufficient characterization,17 or in situations in which attributes were insufficient to model the concept, ‘special case’ subsumption queries were performed using ad hoc concepts to test for subsumption.
An example predicate and candidate expression for the direct tumor extension staging factor value of ‘chest wall involved by direct extension of malignant neoplasm’ (384963009) is shown in table 1.
Table 1.
An example predicate and candidate expression for subsumption querying
| Concept | 384963009 | chest wall involved by direct extension of malignant neoplasm | |
| Normal form | 384963009 | chest wall involved by direct extension of malignant neoplasm |: 116676008 | associated morphology | = 49755003 | morphologically abnormal structure | |
| Predicate expression (SNOMED CT extension) | 385413003 | tumor extension finding |: 363714003 | interprets | = 370052007 | status of invasion by tumor | {116676008 | associated morphology | = 49755003 | morphologically abnormal structure |, 363698007 | finding site | = 78904004 | chest wall structure | } |
| Candidate expression template | 385413003 | tumor extension finding |: 363714003 | interprets | = <invasion> {116676008 | associated morphology | = <morphology>, 363698007 | finding site | = <topology> } |
where <invasion>, <morphology> and <topology> are invasion, morphology and topology concepts from the free text used to populate the post-coordinated candidate expression template.
SNOMED CT, systematized nomenclature of medicine–clinical terms.
The search for concepts in the free text to populate the post-coordinated expression templates has a limited scope: six terms bounded by conjunction phrases and the start and end of a sentence within relevant sections of the free text. The scoping criteria are similar to those used to identify negated terms in the NegEx algorithm.14 15 In the event that four or more concepts were tested for subsumption in a single expression, then the six-term proximity scope restriction was removed.
Concepts from the free text were symbolically manipulated to populate the post-coordinated expression templates that were defined for each staging factor. The subsumption querying protocol was used to test for subsumption against staging factor concepts from the checklist, including other factors relevant to staging but were not separately itemized in the checklist (eg, regional lymph node and distant metastasis staging factors).
The staging factor subsumption results were then used to build the logic based on the staging guidelines to calculate the TNM stage. In brief, T[X,is,0–4], N[X,0–3] and M[X,1] stage assignments were assigned by searching for staging factors that relate to the most advanced stage (ie, T4, N3 and M1 for T, N and M stage, respectively), and then working down toward the least. This iterative search process terminates when a positive finding for a staging factor was found. For more information on the individual post-coordinated expression templates for each staging factor and the logic used to generate the final TNM stage, see the data supplement available online only.
Corpus description
A corpus of 1205 de-identified pathology reports for 1054 lung cancer patients extracted from a statewide pathology information system (AUSLAB) within Queensland Health was obtained with research ethics approval from the Queensland Health Research Ethics Committee. A development (or training) set of 114 reports pertaining to 100 random lung cancer patients from the corpus was used to identify (or annotate) staging factor findings. These findings were subsequently used to define post-coordinated expressions and/or rules for each staging factor.
For system evaluation, a corresponding database of pathological TNM staging decisions18 containing ‘patient-level’ pathological TNM stages collected by MDT over a 5-year period ending in December 2005 was used as the gold standard. All MDT staging decisions were based on the same reports from AUSLAB, and the same staging criteria used for system development (ie, AJCC 6th edition).1 The database had corresponding TNM staging decisions for 718 of the 1054 lung cancer patients in AUSLAB. One problem was that M0 stage (no distant metastases) was recorded in the database, but was not reportable by the lung cancer resection checklist (because the absence of distant metastases could not be ascertained by pathological examination limited to resected lung). Therefore M0 (database) was assigned as equivalent to MX (distant metastasis status unable to be assessed) in resection reports. The proposed system had potential to classify all stages in the database, except for T0 (no evidence of primary tumor), which was assumed to be TX (primary tumor not assessable) by the system. As the database was not used for system development, the entire 718 patient cases were used to evaluate the cancer stage classified by the system. Database statistics are given in table 2.
Table 2.
Key statistics for the database of pathological TNM staging decisions
| Data | Cases | Stage breakdown | Reports | |||||
| Pathology reports with TNM stage decision | 718 | TX | 3 | NX | 59 | MX | 695 | 828 |
| Tis | 0 | N0 | 437 | M1 | 23 | |||
| T0 | 5 | N1 | 150 | |||||
| T1 | 204 | N2 | 72 | |||||
| T2 | 405 | N3 | 0 | |||||
| T3 | 52 | |||||||
| T4 | 49 | |||||||
TNM, tumor–node–metastases.
Results
Using the symbolic rule-based classification system described, staging factors were automatically extracted to calculate the TNM stage for each report. Reports not relevant to lung cancer staging were assigned TX, NX, and MX by the system, indicating that the respective stage cannot be assessed. Post-processing of the system's ‘report level’ stages were performed such that the most advanced T, N or M stage output from reports for the same patient is output as the final ‘patient level’ TNM stage.
Overall system TNM stage accuracy with respect to the database of staging decisions is shown in table 3. The breakdown of cases by stage is shown in the confusion matrix in table 4.
Table 3.
Accuracy of system with respect to database of pathological staging decisions
| Stage | Cases | Accuracy % (95% CI) |
| T | 718 | 72 (69 to 75) |
| N | 718 | 78 (75 to 81) |
| M | 718 | 94 (92 to 96) |
Table 4.
Confusion matrices for system with respect to database of pathological staging decisions
| System | System | System | ||||||||||||||
| TX | Tis | T0 | T1 | T2 | T3 | T4 | NX | N0 | N1 | N2 | N3 | MX | M1 | |||
| Database | Database | Database | ||||||||||||||
| TX | 2 | 0 | 0 | 1 | 0 | 0 | 0 | NX | 51 | 1 | 7 | 0 | 0 | MX | 670 | 25 |
| Tis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | N0 | 48 | 334 | 54 | 1 | 0 | M1 | 18 | 5 |
| T0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | N1 | 4 | 5 | 138 | 3 | 0 | |||
| T1 | 5 | 4 | 0 | 159 | 32 | 0 | 4 | N2 | 3 | 2 | 28 | 39 | 0 | |||
| T2 | 10 | 0 | 0 | 54 | 327 | 0 | 14 | N3 | 0 | 0 | 0 | 0 | 0 | |||
| T3 | 0 | 0 | 0 | 3 | 23 | 20 | 6 | |||||||||
| T4 | 2 | 0 | 0 | 4 | 30 | 3 | 10 | |||||||||
To allow direct comparison between the proposed symbolic rule-based system and the predecessor machine learning-based lung cancer staging system (CSIS),2 post-processing of the proposed system's output stages was required. The database of pathological staging decisions and the same corpus of pathology reports were used as the development set in CSIS. Due to limited numbers of cases of TX, T0, Tis, and N3 assignments, these stages were not implemented in CSIS and pathological M stage was also omitted for the same reason. The CSIS was therefore developed using the database of staging data and pathology reports for the remaining 710 lung cancer patients. Unbiased classifier results were achieved using N-fold cross-validation. In contrast, the proposed system used a fraction of the available reports for development (see Corpus description).
To compare the results from the proposed system directly with CSIS, the same 710 cases were compared. Post-processing of the proposed system's outputs was required for stages not implemented in CSIS. As TX stages in CSIS were implicitly assigned using the T and N relevance classifiers and no T0 or N3 stages were output from the proposed system, no TX, T0 or N3 adjustments were required in the proposed system. However, the four Tis cases output by the proposed system were assigned as equivalent to a T1 stage (indicating that the tumor is at least assessable). Overall TNM stage accuracy with respect to the database of pathological staging decisions for both the proposed system and CSIS are shown in table 5.
Table 5.
Accuracy of proposed symbolic rule-based and machine learning-based system with respect to database of pathological staging decision*
| Stage | Cases | Accuracy % (95% CI) | |
| Proposed system (symbolic rule-based) | CSIS (machine learning) | ||
| T | 710 | 73 (70 to 76) | 78 (74 to 81) |
| N | 710 | 79 (75 to 81) | 82 (79 to 85) |
| M | 710 | 94 (92 to 96) | N/A |
Results for the cancer stage interpretation system (CSIS) are based on unbiased classifier outputs obtain by using N-fold cross-validation from the 710 case development set. The proposed system used a fraction of the available reports for system development.
Discussion
Overall TNM stage accuracy with respect to the database of pathological staging decisions (table 3) and against a machine learning-based approach (table 5) were very encouraging. In particular, the proposed methodology performs close to state-of-the-art statistical machine learning approaches. Given that the machine learning-based approach learned examples from the entire corpus to build its classifiers (although active learning may be investigated to reduce the amount of training data required), the small difference in performance improvement was not expected. The machine learning approach was able to learn the variations in reporting styles at both an individual and institutional level. In contrast, the proposed system only used a fraction of the corpus for system development, which substantially reduces the amount of annotated training data but at the expense of additional expert resources to define post-coordinated expressions and/or rules.
Symbolic rule-based classification systems allow semantics and relationships between symbols (or concepts) to be taken into account and provide a flexible and extensible framework for clinical information extraction. Symbolic approaches allow a greater coverage of stages, for example, Tis, N3, and M1, which have very low prevalence and are generally not suitable for training using machine learning approaches. Finer grain extractions were also able to be achieved to highlight phrases (bounded by concepts) that refer to staging factors rather than at a sentence or document level as typically achieved by machine learning approaches.
The confusion matrices presented in table 4 show the most common stage confusions for the symbolic rule-based system against the database of pathological staging decisions. For T staging, the most common confusions were between T1 and T2, and T2 and T4 stages, with a number of cases falsely called T2 when the assignments should have been T3. For N staging, the most common confusions were between N0 and N1, and N1 and N2. For M staging, high accuracy is gained due to a high prevalence of the MX stage, but there were a large number of incorrectly assigned M1 stages.
Examination of misclassifications revealed that the sources of errors were largely attributable to terms referring to the proximity of tumors to certain body structures (eg, ‘near’ and ‘abuts’), and possibility phrases (eg, ‘most likely’) found in the reports but not part of the list of negation or possibility phrases used for negation detection. These false-positive findings consequently incorrectly assign a more advanced stage. Other sources of errors include staging factors in guidelines that had no corresponding item in the cancer checklist (eg, invasion of trachea, and separate tumors in same or different lobes). These staging factors were not implemented, and as a result were overlooked by the system and thus a less advanced stage would be output. Another source of error relates to the parsing of lymph nodes in which a combination of numbers and words were used to represent lymph nodes. For example, the phrase ‘No 5 and peribronchial lymph node metastases’ was unable to output a higher N stage (ie, N2) for the ‘No 5 lymph node metastasis’. Having knowledge of examples of false positives and negatives allows for potential further refinement of the free-text extraction algorithms (including the NLP components) to improve system accuracy, however, possibly at the expense of increased complexity.
In addition to the errors above, the confusion matrix shows that many reports were incorrectly assigned either a TX or NX stage. This suggests that no relevant staging information from these patients' reports were extracted. For T staging, the minimum requirement for a non-TX stage in the system was that the histologic type was non-default. This was observed to be not sufficient and did not capture all specializations of ‘carcinoma’ (68453008). An additional subsumption query on ‘carcinoma’ would upgrade many of the false-positive TX stages. For N staging, many of the NX false-positive cases were biopsy or wedge resection reports, which generally do not have any regional lymph node information relevant for staging. It is conjectured that in these cases, the ground truth N stages were acquired from data sources beyond those available in this study.
Other limitations and considerations
A symbolic rule-based clinical information extraction system based on SNOMED CT has several limitations. First, there are gaps in the SNOMED CT ontology where concepts were not fully modeled and may have missing or no defining relationships. This was overcome by creating SNOMED CT extensions,17 in which valid concept model attributes were used to model the concepts. In some cases, no concept model attributes were available to define the concept and special case (ad hoc) subsumption queries on free-text concepts were necessary.
Another limitation of the system is that piecewise subsumption queries were performed rather than direct expression-based subsumption querying. Implementations of expression-based subsumption querying were not available at the time of system development and as a result piecewise subsumption queries on concepts and attribute names and values were performed to achieve an equivalent result. Expression-based subsumption querying is currently under development and will be available in the near future to replace the piecewise subsumption queries in the proposed system. This will provide a unified framework for subsumption querying and decreases the amount of concept manipulation that is required for piecewise subsumptions.
Furthermore, TNM stages extracted or derived from pathology reports may not accurately reflect a patient's cancer stage, particularly for the M component in which the required information may not be available from pathology reports. Supplementing M staging results from pathology reports with those found in radiology reports would potentially reduce the number of false negatives and improve the M staging results.
The proposed symbolic rule-based approach using SNOMED CT is flexible and robust and provides a framework for clinical information extraction from free text. Although staging lung cancers may be more straightforward than other cancers from pathology reports due to the detailed relevant information contained in these reports, future work will investigate the applicability of pathology reports (and other report types) for staging other cancer types. Indeed, if detailed summaries, synoptic reports or even explicitly mentioned stages are available, then this may make determining stage easier for all cancers.
Conclusion
A symbolic rule-based MEDTEX system using the SNOMED CT ontology and its semantics was proposed to extract lung TNM staging factors automatically from free-text pathology reports. The extraction methodology used was based on the subsumption querying of concepts in free text using post-coordinated SNOMED CT expression templates. Lung TNM stages were classified by building logic from the relevant staging factors. Although lung cancer was used as a case study, future work will investigate the applicability of the proposed symbolic rule-based approach for extracting other cancer characteristics and types.
Supplementary Material
Acknowledgments
The research in this article was conducted in partnership between the Australian e-Health Research Centre (AEHRC) within CSIRO and the Queensland Cancer Control Analysis Team (QCCAT) within Queensland Health. The authors acknowledge Iain McCowan and Darren Moore from CSIRO, and Mary-Jane Courage, Hazel Harden, Julie Moore and Steven Armstrong from QCCAT; Jaccalyne Brady and Donna Fry from the Queensland Integrated Lung Cancer Outcomes Project; and Wayne Watson from the AUSLAB Support Group.
Footnotes
Competing interests: None.
Ethics approval: This study was conducted with the approval of the Queensland Health Research Ethics Committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Greene FL, Page DL, Fleming ID, et al., eds. AJCC cancer staging manual. 6th edn New York: Springer-Verlag, 2002 [Google Scholar]
- 2.McCowan IA, Moore DC, Nguyen AN, et al. Collection of cancer stage data by classifying free-text medical reports. J Am Med Inform Assoc 2007;14:736–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Threlfall T, Wittorff J, Boutdara P, et al. Collection of population-based cancer staging information in Western Australia—a feasibility study. Melbourne, Australia: National Cancer Control Initiative, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Australia A national cancer data strategy for Australia. 2008. http://www.canceraustralia.gov.au (accessed Nov 2009).
- 5.College of American Pathologists SNOMED CT–Encoded CAP cancer checklist (version 1.5). 2006. http://www.cap.org/ (accessed Jun 2006). [Google Scholar]
- 6.College of American Pathologists An overview of the College of American Pathologists cancer checklists. 2009. http://www.cap.org/ (accessed Mar 2009).
- 7.D'Avolio LW, Litwin MS, Rogers SO, et al. Facilitating clinical outcomes assessment through the automated identification of quality measures for prostate cancer surgery. J Am Med Inform Assoc 2008;15:341–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen AN, Lawley MJ, Hansen DP, et al. A simple pipeline application for identifying and negating SNOMED clinical terminology in free text. Proceedings of the Health Informatics Conference; August 2009, Canberra, Australia; 2009:188–93 [Google Scholar]
- 9.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a disease knowledge representation model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 10.Nguyen AN, Lawley M, Hansen D, et al. Automated metastasis stage classification for lung cancer patients using free text radiology reports. Proceedings of the American Medical Informatics Association Annual Symposium; November 2009, San Francisco, CA; 2009:474 [Google Scholar]
- 11.International Health Terminology Standards Development Organisation SNOMED Clinical Terms® User Guide. 2008. http://www.ihtsdo.org (accessed Sep 2008).
- 12.Cunningham H, Maynard D, Bontcheva K, et al. GATE: a framework and graphical development environment for robust NLP tools and applications. Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics; July 2002, Philadelphia; 2002 [Google Scholar]
- 13.U.S. National Library of Medicine Unified medical language system (UMLS). http://www.nlm.nih.gov/research/umls/ (accessed Feb 2008).
- 14.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 15.Chapman WW. NegEx version 2: a simple algorithm for identifying pertinent negatives in textual medical records. http://www.dbmi.pitt.edu/chapman/NegEx.html (accessed 28 Jan 2009).
- 16.International Health Terminology Standards Development Organisation SNOMED Clinical Terms® Transforming Expressions to Normal Forms. 2007. January 31 http://www.ihtsdo.org (accessed Sep 2008).
- 17.Lawley M, Vickers D, Hansen D. Converting Ad Hoc terminologies to SNOMED CT extensions. Proceedings of the health informatics conference; 2008, Melbourne, Australia; 2008:133 [Google Scholar]
- 18.Fong KM, Bowman R, Fielding D, et al. Queensland integrated lung cancer outcomes project (QILCOP): initial accrual and preliminary data from the first 30 months. The Thoracic Society of Australia and New Zealand Annual Scientific Meeting; 4–9 April 2003, Adelaide, Australia; 2003 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.