Abstract
Objective
To determine whether a factorized version of the complement naïve Bayes (FCNB) classifier can reduce the time spent by experts reviewing journal articles for inclusion in systematic reviews of drug class efficacy for disease treatment.
Design
The proposed classifier was evaluated on a test collection built from 15 systematic drug class reviews used in previous work. The FCNB classifier was constructed to classify each article as containing high-quality, drug class-specific evidence or not. Weight engineering (WE) techniques were added to reduce underestimation for Medical Subject Headings (MeSH)-based and Publication Type (PubType)-based features. Cross-validation experiments were performed to evaluate the classifier's parameters and performance.
Measurements
Work saved over sampling (WSS) at no less than a 95% recall was used as the main measure of performance.
Results
The minimum workload reduction for a systematic review for one topic, achieved with a FCNB/WE classifier, was 8.5%; the maximum was 62.2% and the average over the 15 topics was 33.5%. This is 15.0% higher than the average workload reduction obtained using a voting perceptron-based automated citation classification system.
Conclusion
The FCNB/WE classifier is simple, easy to implement, and produces significantly better results in reducing the workload than previously achieved. The results support it being a useful algorithm for machine-learning-based automation of systematic reviews of drug class efficacy for disease treatment.
Keywords: PubMed, knowledge bases, review, systematic, evidence-based medicine
Introduction
This paper describes a computer system that assists people involved in building systematic reviews, which are among the basic tools of evidence-based medicine (EBM). According to the Centre for Evidence-Based Medicine, ‘Evidence-based medicine is the conscientious, explicit and judicious use of current best evidence in making decisions about the care of individual patients.1
Evidence-based medicine involves three distinct steps: (1) identifying evidence from the scientific literature that pertains to a clinical question; (2) evaluating this evidence; and (3) applying the evidence to the clinical problem.2
Since the body of the scientific literature is growing extremely fast (500 000 new abstracts are added to Medline every year), practicing EBM is so challenging and labor-intensive that tools are needed to support it. Advanced information technologies should be developed and implemented to support EBM by reducing the labor required while capturing high-quality evidence. The practice of EBM integrates individual clinical expertise with the best available external clinical evidence from systematic research. Systematic reviews are one of the main tools of EBM.
A systematic review is a highly structured process for reviewing literature on a specific topic or group of topics to distill a targeted subset of knowledge or data. Experts first review identified documents and complete a series of forms designed to screen out non-relevant documents. Then core data are extracted from the screened-in documents.
Commonly, the screening process consists of an initial screening phase, referred to as ‘broad screening’, and a final phase often referred to as ‘strict screening’. Usually, broad screening requires two reviewers who review each abstract. The goal is to retrieve 100% of relevant abstracts, while excluding the obvious non-relevant ones. Abstracts need the approval of only one of two reviewers to pass to strict screening, while abstracts are excluded only if both reviewers stipulate exclusion. Although retrieving as many relevant documents as possible is crucial, it is not the only goal of broad screening. The success of broad screening also requires minimizing the inclusion of non-relevant abstracts.
Articles that pass the broad screening phase are subjected to strict screening. In strict screening, reviewers read the article abstracts and the complete articles with the goal of achieving 100% precision.
Systematic reviews are costly, as they take considerable effort from domain experts. They are also error prone, extremely hard to manage, very difficult to keep up-to-date since new evidence continues to emerge, and take a long time to complete. Consequently, there is significant demand for tools that will enhance and facilitate systematic reviews. Machine-learning (ML) techniques could provide appropriate tools.3 In this paper we present novel methods for this task, achieving better results than in previous work.
Background
While there is a wealth of literature on automatic text classification,4–6 it has limited usefulness for classifying medical abstracts in systematic reviews, because it targets mainly topic identification, as in classifying newswire articles by topic. This task is relatively uncomplicated, since the vocabularies used in newswire articles differ significantly by topic. In systematic reviews, the situation is quite different. Initially, abstracts are preselected using standard information retrieval keyword-based queries on a bibliographic database, and the broad screening is performed on the result of these queries. The keywords with which the queries are built are therefore present in all the selected abstracts, regardless of their class. Therefore, the vocabularies in relevant and non-relevant abstracts exhibit fewer differences than in the newswire topic classification. Automatic text classification applied to systematic reviews is therefore more challenging than the standard topic classification task. For this reason, we limited the comparison for our work to previous attempts to use text classification for systematic reviews.
To our knowledge, Aphinyanaphongs et al3 was the first study to apply ML to systematic reviews. The work focused on improving performance over the clinical query filters first proposed by Haynes et al.7–9 The authors used the data derived from the ACP Journal Club as their corpus. After experimenting with a variety of ML techniques, they found that the support vector machine (SVM) achieved the best performance with those data. Their research showed that ML could be successfully applied to creating systematic reviews.
Cohen et al10 also applied ML to systematic reviews, focusing on reducing the workload of reviewers during broad screening by eliminating as many non-relevant documents as possible while including no less than 95% of the relevant documents. They introduce a new measure, work saved over sampling (WSS). They define work saved as ‘the percentage of papers that meet the original search criteria that the reviewers do not have to read (because they have been screened out by the classifier)’. They then specify that the work saved must be greater than the work saved by random sampling; thus work saved over sampling is proposed as an evaluation measure. When training data are only available for a single topic, Cohen et al10 11 represent state of the art in automating systematic reviews. The classifier performance scores they obtained with the voting perceptron (VP) algorithm,10 the algorithm that they used for the classification task, are used as a baseline for our current research.
In applications, besides the accuracy-based performance, it is often interesting to look at the interpretability of the classifier. A classifier is interpretable (understandable) if the user of this classifier is able to interpret its meaning, and reason about the explanation of the decision made by the classifier for a given instance. The perceptron, which is a special case of a neural network, consists of a set of equations of hyperplanes, with words as variables. Decisions of Bayesian classifiers are presented in terms of frequency of occurrence of words within a given abstract. It is well known, for example,12 that neural networks have low interpretability and require complex transformations to extract interpretable knowledge from them. Following,13 we believe that classifications by Bayesian classifiers are more understandable by the medical end users; therefore they have an interpretability advantage over the perceptron classifiers.
Recently, Cohen and collaborators have published two papers in which SVM with an n-gram-based representation is used to classify systematic reviews.11 14 Direct comparison is difficult, as the evaluation measure is different (area under the curve in Cohen11 and Cohen et al14), and that work uses a different training protocol based on cross-topic training. Therefore the most recent work with which we can directly compare the proposed approach is the study of Cohen et al.10 Furthermore, it is generally recognized in the literature that SVM classifiers give an excellent performance in text classification. Recent experimental research,15–18 26 however, indicates that advanced, heuristic modifications of the classical naïve Bayes classifier, for example, multinomial naïve Bayes (MNB) and complement naïve Bayes (CNB), yield a performance equaling that of SVM, while being many times faster.
Our motivation in undertaking the present study was to demonstrate that a simple, more easily interpretable classifier can be used in the systematic reviews application and yield results comparable to other more complex and less interpretable methods. The recent progress in Bayesian classifiers, in particular the MNB classifier and the CNB classifier, resulted in classification tools with topic classification performance comparable to SVM and neural networks. We wanted to investigate whether these results extend to the challenging and practically important context of systematic review classification.
Methods
Our task was to build a classifier that, once trained, will classify previously unseen abstracts as either relevant or non-relevant to the topic of the systematic review, with (1) very high recall and (2) sufficient precision in removing the non-relevant articles that, although human effort will still be required, there will be significant labor savings.
We built and tested the automated classification system for systematic reviews in five phases: (A) building the text collection by extracting source data; (B) preparing the text collection for ML and applying feature engineering, such as removing noisy features or modifying weights; (C) text classification; (D) classifier performance evaluation; (E) tuning the classifier to specific systematic review performance requirements. Below we discuss each of the five phases.
Text collection
To make our research results comparable with previously obtained results, as well as with future investigation results, we used static and publicly available data. In particular, we built a text collection that is as close as possible to that built by Cohen et al.10 That collection contained 15 evidence reports produced by the Oregon Evidence Based Practice Centre (EPC), evaluating the efficacy of medications in several drug classes (opioids, skeletal muscle relaxants, estrogen replacement). These reports were mapped into a public domain collection of Medline records from 1994 to 2003—that is, the abstracts of papers evaluated and triaged by the EPC were fetched from that collection. Because Cohen et al10 have published the intermediate results of their data extraction (as a Drug Review Journal Citation Records file19), we did not need to repeat all the extraction process steps they used to obtain the Drug Review Journal Citation Records file (see online appendix A for details). We used this file as input for our data extraction process.
Table 1 gives information about the 15 drug groups for which systematic reviews were built. These are the same data as used by Cohen et al.10 We list in this table the number of abstracts in each group, as well as the percentage of the relevant abstracts among all the abstracts. We can observe that, in general, the data are imbalanced: many more abstracts are judged non-relevant by the EPC than are found relevant. This pattern makes it important to use a classifier that will not become overwhelmed by the prevailing non-relevant abstracts. When dealing with imbalanced data, the existing data mining terminology used in this paper calls the class that contains most of the abstracts (here, the non-relevant class) the majority class, and the other class, containing the relevant abstracts, the minority class.
Table 1.
Datasets' description (source Cohen et al1)
| Drug class review | No of abstracts | % judged relevant (the Included class) |
| ACEInhibitors | 2544 | 1.6 |
| ADHD | 851 | 2.4 |
| Antihistamines | 310 | 5.2 |
| AtypicalAntipsychotics | 1120 | 13.0 |
| BetaBlockers | 2072 | 2.0 |
| CalciumChannelBlockers | 1218 | 8.2 |
| Estrogens | 368 | 21.7 |
| NSAIDs | 393 | 10.4 |
| Opioids | 1915 | 0.8 |
| OralHypoglycemics | 503 | 27.0 |
| ProtonPumpInhibitors | 1333 | 3.8 |
| SkeletalMuscleRelaxants | 1643 | 0.5 |
| Statins | 3465 | 2.5 |
| Triptans | 671 | 3.6 |
| UrinaryIncontinence | 327 | 12.2 |
Our text repository consists of a subset of Medline abstracts in the XML format, defined by the Text Retrieval Conference (TREC). To be consistent with Cohen et al,10 we extracted from the text repository a collection of 15 drug review topics based on the Drug Class Review names, and then extracted data from the text repository, including PMID, title, abstract, Publication Type (PubType), Medical Subject Headings (MeSH) tagsi, and the class label. Our data-extraction schema selected only articles with PMIDs that are included in the Drug Review Journal Citation Records dataset.
Preprocessing the data and weight engineering (WE)
We used the bag-of-words (BOW) representation to code each text collection. Each article in a text collection is represented as a vector in an N-dimensional space where N is the total number of terms (features) extracted from the text collection. Terms are single words, not multiword phrases. Terms extracted from MeSH and PubType represent the content of the relevant tags, which could include multiword phrases.
The whole text collection should be represented as a (N+1)xJ matrix where J is the number of articles in the text collection. Each line j of this matrix is an N-dimensional BOW vector for the article j, plus the class label for the article j.
If a feature i does not occur in the article j, then the relevant matrix element is zero: aij=0; otherwise aij is assigned a positive value. The way to calculate this value depends on the method used for feature representation. The value aij is 0 or 1 for the binary method. For the frequency method, the value is the number of occurrences of the feature i in the document j. There are other methods to compute aij such as tf.idf (term frequency–inverse document frequency method) feature representation.21
To be consistent with Cohen et al,10 we applied some modifications to the classical BOW approach. Technically, these modifications address the way we built the BOW terms. Because of the nature of the Medline information, our collection includes both flat texts, such as the abstracts, and structured texts, such as the MeSH tags.
We grouped the extracted Medline fields into three categories. Each category was ‘tokenized’ and preprocessed in a special way. The first category consisted of titles and abstracts. The second was composed of the MeSH tags. The last one included PubTypes. We processed each category according to our understanding of how preprocessing was performed in Cohen et al,10 because we wanted to apply the learning algorithm to the same data as Cohen et al.10 The steps we applied to build the final list of terms (features for ML) from titles and abstracts are presented in appendix B (available as an online data supplement).
It should be noted that the use of a frequency-based representation versus a binary representation requires some thought, as it was not immediately obvious which feature representation method would work better for systematic review preparation data. On the one hand, we can assume that the frequency-based representation or more sophisticated frequency-related methods such as tf.idf would work better than the binary representation for flat text fields, such as abstracts. On the other hand, since MeSH-based and PubType-based features do not occur in an article more than once, the binary representation method might be more suitable for this type of data. As abstracts are narrative text and MeSH tags and PubType are discrete values, the frequency-based representation works better for abstracts, and the binary representation is more suitable for MeSH-based and PubType-based features. We have also experimented with the commonly used tf.idf, and we have found that the performance of this representation is equal to that of a representation using ‘raw’ (unnormalized) frequencies. This is generally consistent with the findings of Cohen et al.10 Since a representation using frequencies alone, without weighting features by inverse document frequencies, is simpler and more efficient to compute, we have decided to represent the abstracts by word frequencies, and the Pub-type and MeSH features as binary. Cohen et al10 tried both the binary and the frequency feature weighting methods. In their experiments, the binary method achieved better performance for the VP classifier they used. In the experimental comparisons between FCNB and FCNB/WE in the Results section, we therefore use Cohen's binary representation for the VP classifier and the representation described above for our methods.
In ML, all features are usually treated as equal by the learning algorithms. In some tasks, however, it makes sense to give some features weights bigger than other features, if this is likely to improve the performance of a given learning algorithm. Such tuning of weights is known as ‘weight engineering’. Weight engineering modifies the existing data by weighting some of the attributes more than others.
Some MeSH-based and PubType-based features included in our data are highly informative for correct classification decisions (table 9 in Cohen et al10), but the frequency weighting method can assign less weight to them than to abstract-based features that occur in an article more than once. This ‘unfair’ weighting could account for the poor performance of the frequency-based representation scheme in the experiments of Cohen et al.10
Therefore, to increase the importance of PubType and MeSH features, we used a simple WE scheme: each PubType-based and MeSH-based feature weight was modified by multiplying it by a fixed weight multiplier (WM). We tried multiple WM values starting with the value 2 by increments of 1. Finally, we assigned each drug review group the WM value that resulted in the best WSS performance. The procedure is fully automatic; only the final selection was performed manually, but this could also be easily automated. We called this value the best WM value for the current drug review group. If more than one value yielded the best performance, we selected the smallest value. We observed that, while different datasets could have different best WM values, those WM values were all between 2 and 21. Although in our experiments each WM value was computed on the training set, it could as well be determined from a separate hold-out set, so that it would be independent of the training data.
Text classification
We used a novel, modified version of the CNB classifier16 as a classification algorithm, which we called factorized CNB (FCNB). We used CNB as a basis for our classification process because the CNB classifiers implement state-of-the-art modifications of the MNB17 classifier for a classification task with highly imbalanced data. Because the pre-selected systematic review data usually contain a large majority of non-relevant abstracts (sometimes more than 99%), we needed to use classifiers that take this problem into account. The CNB classifier modifies the standard MNB classifier by applying asymmetric word count prior probabilities that reflect the skewed class distribution.16 The CNB algorithm is also fast and easy to implement. Although many other classifiers could have been used, Bayesian methods are simple and highly efficient. Besides using VP in one study,10 Cohen et al report in another study11 good results with the use of SVMs. We find, however, that the available implementations of SVM are two orders of magnitude slower (in run time) than Bayesian methods.15 Moreover, SVM parameters require understanding of the SVM algorithm—for example, the choice of the kernel or the setting of the slack parameters coefficient—whereas the parameters of Bayesian classifiers are based on the properties of the data—for example, the importance of individual features or classes.
Evaluation of classification performance in systematic reviews
A systematic review ML classification system has two main objectives: (1) to minimize the number of relevant documents excluded by the classifier; (2) to reduce the reviewers' workload by excluding the maximum number of irrelevant documents. While the first objective could be formalized by assigning a required recall threshold, the second one requires a special measure for workload savings. The use of precision as the rate of correctly classified relevant articles could indicate the efficiency of reducing workload, but it does not take into account the achieved recall and the number of excluded non-relevant documents. Cohen et al10 introduced a new measure, WSS, that is designed to quantify the reduction in workload in systematic review preparation when using a classifier. That is, the classification system can be said to save work only if the work saved is greater than the work saved by simple random sampling.
WSS is defined as:
| (1) |
where TN (true negatives) is the number of negative (non-relevant) abstracts correctly classified, FN (false negatives) is the number of positive (relevant) abstracts incorrectly classified as negatives (non-relevant), N is the total number of abstracts in the test set, and R is the recall. This could be equivalently expressed as:
| (2) |
where TP (true positives) is the number of positive (relevant) documents correctly classified. As Cohen et al10 used recall and WSS as their main measure, we did also.
In more detail, Cohen et al10 used the special modification of the WSS measure called WSS@95%. Where WSS@95% is a WSS interpolation for recall at 0.95. We have similarly approximated WSS@95% with the highest obtained WSS score with recall no less than 0.95. (We call it the Best WSS.)
Tuning the Bayesian classifier to specific systematic review performance requirements: the FCNB/WE algorithm
When running the original CNB,16 we were obtaining recall for the relevant class that was significantly lower than the required 95% level. Such results are not acceptable for a user building a real systematic review. We have therefore modified the classifier by adding a heuristic weight factorization technique to the CNB algorithm. A factor was added to the classification rule for choosing the label lFCNB of a document d, as follows:
| (3) |
where p(θc) is the class prior estimate, fi is the frequency count of feature i in document d, is the number of times feature i occurred in documents of classes other than c, and is the total number of feature occurrences in classes other than c, αi is a smoothing parameter (the common practice is to set αi=1); α denotes the sum of the αi (to ensure that the probability distribution sums up to 1). The p(θc) term preceding the ‘minus’ operator represents the prior probability of a given class, and is obtained from the probability distribution between the two classes in the training set. Two main features of the algorithm are implemented in this formula and need to be explained.
Firstly, as discussed above, the evidence of belonging to the minority class c is weak, as a frequency-based estimation of a probability of belonging to the minority class is very inexact. To focus the classifier on the minority class c, instead of measuring the evidence of an abstract belonging to c, we measure the better supported evidence of belonging to any class other than the class c. We then use the probability of the opposite event, which is the event of not belonging to a class other than c. That event is logically equivalent to belonging to c. This is implemented here with the expression following the ‘minus’ operator in the square bracket. It is important to observe that the minus in front of this expression means that we evaluate here the probability of the event complementary to belonging to a class other than c—that is, we evaluate the probability of belonging to c. Moreover, we further focus on the minority class by decreasing the weight of the evidence of not belonging to this class by multiplying the log expression by a factor Fc.
Secondly, to improve the recall on the class of relevant abstracts, we give the evidence of belonging to this class an additional weight with respect to the non-relevant class. This increases the weight for the relevant class, while keeping the weight of the non-relevant class.
Therefore Fc=1 will be used when we compute the above formula for the non-relevant class, and Fc<1 when c represents the relevant (minority) class. We refer to this algorithm as factorized CNB, or FCNB. To use FCNB in systematic review preparation, we had to select a factor value that could result in the best performance for a given drug review topic. We also had to evaluate how this factor would accommodate new data (eg, future articles and updates of the drug evidence review). Factor values were determined using 10-fold cross-validation on a separate hold-out set. Details are presented in appendix C (available as an online data supplement).
Our method, which we call FCNB/WE, is therefore a modification of FCNB that applies the FCNB algorithm to data that were previously modified by performing WE on the features (a method that boosts MeSH and PubType features). As WE modifies (increases) the frequency values of the MeSH and PubType features, it affects frequency-related variables in equation (3), namely fi, and for MeSH and PubType features. We have implemented FCNB/WE on the basis of the Weka open-source software.22
To be able to compare our classifier performance with previously obtained results, we followed the 5×2 cross-validation scheme used in the previous work.10
In 5×2 cross-validation, the dataset is randomly split in half. One half (the training dataset) is used to train the classifier, and then the classifier is evaluated using the other half as a test dataset. Then the roles of the two datasets are exchanged, with the second half used as a training set and the first half used as a test set. The results are accumulated from both halves of the split. The scoring process is repeated 5 times; each half of the data is used twice—first for training and then for testing—which produces 5×2=10 sets of scoring results. The 5×2 cross-validation approach produces more realistic estimates of the actual performance than the 10-fold cross-validation method, another common approach, which often overestimates performance.23
We used stratified folds for cross-validation—that is, the classes for each fold are represented in approximately the same proportion as in the full dataset. We applied the FCNB classification algorithm with various factor values. We used 22 values for the factor, from 0.78 to 0.99 in increments of 0.01.
Results
We performed the experiments in two batches, to evaluate first the effects of focusing the classifier by means of using the factor in CNB, and then to evaluate the combined effect of a factorized classifier with WE: first, FCNB without WE, and then FCNB/WE. For FCNB, we completed two sets of experiments with the FCNB classifier: factor validation and 5×2 cross-validation. The results of the factor validation experiments are presented in appendix C.
Cohen et al10 published two sets of experimental results. The first set was obtained without any stemming or use of a stop word list to the representation of the data; the second set of results used the data after removal of the 300 words from the stop word list and after applying the Porter stemmer.24
Table 2 contains the main detailed results of our experiments in comparison with the results of Cohen et al10 obtained using stemming and a stop word list, while table D1 in appendix D (online supplement) compares our results with those of Cohen et al10 obtained without stemming or use of a stop word list. As usual, we refer to the approach presented in Cohen et al10 as the VP approach. The three columns to the right of the drug topic review column present the results of the 5×2 cross-validation experiments. All the results are expressed using the WSS measure, unless indicated otherwise. The first column (FCNB) includes average WSS scores calculated as the average of all FCNB 5×2 cross-validation WSS scores for the current drug review. (There are five splits with two training/testing pairs each. Therefore, 10 WSS scores were used to calculate each average WSS.) To compare our FCNB results with the results of Cohen et al10 using the VP approach, the second column (‘best VP for recall > 0.95’) shows the results in Cohen et al10 with the best WSS for recall over 0.95. The seventh column shows that the summed difference between FCNB WSS and VP WSS for all the drug reviews is +57.8%. The average difference per review is 3.9%.
Table 2.
Work saved over sampling results, in percentages, for 5×2 cross-validation experiments with a factorized complement naïve Bayes (FCNB) classifier and with a FCNB plus weight engineering (FCNB/WE) classifier
| Drug review topics | FCNB | Best VP for recall >0.95 | FCNB/WE | FCNB/WE – VP | FCNB/WE – FCNB | FCNB –VP |
| ADHD | 49.4 | 68.4 | 62.2 | –6.2 | +12.8±12.2 | –19.0 |
| UrinaryIncontinence | 29.4 | 19.0 | 29.6 | +10.6 | +0.2±9.7 | +10.4 |
| Opioids | 45.9 | 15.4 | 55.4 | +40.0 | +7.5±5.5 | +30.5 |
| ACEInhibitors | 29.9 | 59.2 | 52.3 | –6.9 | +22.4±7.9 | –29.3 |
| Estrogens | 22.0 | 12.8 | 37.5 | +24.7 | +15.5±6.6 | +9.2 |
| SkeletalMuscleRelaxants | 12.5 | 0.0 | 26.5 | +26.5 | +14.0±11.0 | +12.5 |
| BetaBlockers | 27.0 | 22.0 | 36.7 | +14.7 | +9.7±7.7 | +5.0 |
| OralHypoglycemics | 6.1 | 3.4 | 8.5 | +5.1 | +2.4±1.9 | +2.7 |
| Statins | 22.1 | 20.3 | 31.5 | +11.2 | +9.4±5.9 | +1.8 |
| ProtonPumpInhibitors | 5.2 | 17.8 | 22.9 | +5.1 | +17.7±9.6 | –12.6 |
| Antihistamines | 5.6 | 0.0 | 14.9 | +14.9 | +9.3±4.5 | +5.6 |
| CalciumChannelBlockers | 17.3 | 13.9 | 23.4 | +9.5 | +6.1±6.5 | +3.4 |
| NSAIDS | 36.7 | 9.0 | 52.8 | +43.8 | +16.1±8.9 | +27.7 |
| Triptans | 14.1 | 0.9 | 27.4 | +26.5 | +13.3±15.3 | +13.2 |
| AtypicalAntipsychotics | 11.7 | 15.0 | 20.6 | +5.6 | +8.9±6.0 | –3.3 |
| Sum | 334.9 | 277.1 | 502.2 | +225.1 | +165.3 | +57.8 |
| Average | 22.3 | 33.5 | +15.0 | +11.2 | +3.9 |
VP denotes the Voting Perceptron classifier used in Cohen et al.1 The fifth column reports t-test CIs at the 95% level. Data areis subject to stemming and removal of words from a stoplist.
To test the effects of factorization and WE, we applied the 5×2 cross-validation FCNB experiments using the WE techniques (see appendix C for details). Table 2 presents the following results:
FCNB/WE experimental results: in the third column (FCNB/ WE);
differences between FCNB/WE results and VP results: in the fourth column (difference between columns 3 and 2);
differences between FCNB results achieved with and without applying the WE techniques: in the fifth column (‘FCNB/WE – FCNB’ is the difference between columns 3 and 1. We also report 95% CIs for this comparison).
The lowest workload reduction for a topic achieved with FCNB/WE is 8.5% (compared with 5.2% with FCNB alone), the maximum is 62.2% (49.4% with FCNB), and the average per topic over the 15 topics is 33.5% (22.3% with FCNB). This makes it 11.2% higher than the average reduction workload per topic obtained with FCNB, and 15.0% higher than the results previously obtained with the VP algorithm used by Cohen et al.10
The summed difference between FCNB/WE WSS and VP WSS for all drug reviews is +225.1%. FCNB/WE showed the best performance relative to the results achieved by Cohen et al on the NSAIDS drug review topic (the WSS percentage difference is +43.8%). FCNB/WE's worst WSS score was obtained on ACEInhibitors (the WSS percentage difference is –6.9%).
The results in appendix D, table D1, are consistent with the above results. When the VP classifier works on data that are neither stemmed nor subjected to stop word removal, the differences in WSS performance between FCNB and VP in table D1 correspond to the respective differences in table 2.
Discussion
Our results show that FCNB and FCNB/WE can be recommended as algorithms for ML-based automation of systematic reviews for the drug class efficacy for disease treatment. In this section, we briefly summarize the main results, indicating that, overall, FCNB and FCNB/WE achieve a better WSS performance than the VP algorithm,10 discuss what makes FCNB, and particularly FCNB/WE, achieve better WSS scores than the VP approach for the majority of drug review topics and why the VP method possibly outperforms the FCNB/WE method for two specific drug groups, and present future work entailed by our research.
Summary of the main results
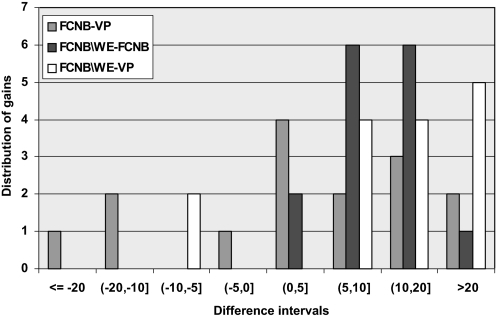
Figure 1 compares results obtained in the present work with those obtained for the VP approach.10 It summarizes the distribution of the gains achieved by FCNB over VP and FCNB/WE over FCNB. Using FCNB, we achieved an average workload reduction of 22.3% per drug review study. Using FCNB/WE, we achieved an average workload reduction of 33.5%—that is, 15.0% greater than the average workload reduction achieved in previous work. It also shows that WE really pays off: the differences in performance between FCNB/WE and FCNB are all in favor of FCNB/WE.
Figure 1.
Histogram summarizing the work saved over sampling results for factorized complement naïve Bayes (FCNB)/weight engineering (WE), FCNB and voting perceptron (VP). The x-axis shows the discretized differences between the methods (FCNB – VP, FCNB/WE – FCNB, FCNB/WE – VP) , and the y-axis shows for how many topics (drug reviews) the given difference in performance occurs. Looking at the white bars, we observe that most of the topics are to the right of 0 on the x-axis, visualizing the advantage of FCNB over VP. Looking at the light grey bars, we observe that, except for two reviews, they are to the right of 0, meaning that FCNB/WE performs better than VP on 13 out of 15 drug groups. Looking at the dark grey bars, we observe that all the bars are in the right half of the interval, visualizing the clear advantage of weight management when using FCNB.
Figure 1 shows that, in the majority of drug review groups (13 out of 15), the performance of FCNB/WE is better than the performance of the VP method (we address below the case of the two datasets, ADHD and ACEInhibitors, when this is not the case). This result is of practical importance, because it demonstrates that a simple and very efficient classifier performs the systematic reviews classification task better than the main published alternative, while being more efficient. The approach presented here could therefore be considered as a possible technique for automating the systematic review process. We want to emphasize that systematic reviews often start with datasets much larger than those considered by Cohen et al,10 with the broad screening phase including typically tens of thousands of abstracts. The matter of the efficiency of the classifier used to automate this phase is therefore of great practical importance.
Comparative analysis of performance
The work presented in Cohen et al10 makes some observations about the possible reasons for wide variations in the VP classification performance for the 15 drug reviews. The authors did not find any significant correlation between performance and sample size or fraction of positive samples. They also discussed the issue of the number of significant features selected with the χ2 test for the VP experiments.25 While 30 or fewer significant features are not enough to adequately model the triage process, the highest scoring topics did not necessarily have the highest number of significant features, and the correlation between the number of significant features and WSS was not statistically significant. The fact remains, however, that the VP method was used in conjunction with an intensive feature selection.
As FCNB does not normally require feature selection for classification, we did not have problems such as a performance decrease for small feature sets. This is in line with the general properties of Bayesian approaches, which do not need feature selection—non-relevant features result in small probability values and are eliminated by the argmax operator at the core of the naïve Bayes approach. This may be the main reason why the FCNB scores are higher than the results produced by the VP for the small drug review groups, such as Antihistamines.
As previously noted above, regarding the binary representation–frequency representation dilemma, while Cohen et al10 used a binary scheme, we used a frequency-based representation scheme. Both approaches have some advantages and disadvantages that depend on the nature of the data. We believe that the use of word frequencies for abstract-based features may be the second reason for our better results. We provided the classifier with more discriminative information for making better decision boundaries. When we did not use WE with FCNB, there was a risk of underestimating the weights for the PubType-based and MeSH-based features. (Unlike the abstract-based features, PubType-based and MeSH-based features cannot have a frequency greater than 1. This kind of ‘weight discrimination’ could ‘confuse’ the ML system.) The results of our experiments show that the possible negative impact of underweighting of MeSH-based and PubType-based features could be overcome by applying WE. The fifth column in table 2 shows the effects of WE combined with FCNB. The performance of FCNB/WE is consistently better than that of FCNB, although in three drug groups, UrinaryIncontinence, CalciumChannelBlockers and Triptans, the differences in favor of FCNB/WE are not large enough to be statistically significant.
Regarding the ACEInhibitors and ADHD data, for which FCNB/WE performs less well than VP, we found that the performance obtained with FCNB/WE on these groups was still quite acceptable. The FCNB/WE WSS scores for these data are much better than the average FCNB/WE WSS score obtained over 15 topics: the ADHD score was 62.2% and the ACEInhibitors score was 52.3%, whereas the average WSS over 15 topics was 33.5%. We believe that these two review groups are easy for the VP, as the performance on both is among the best of all 15 reviews. In general, owing to its geometry (a linear hyperplane that best separates the two classes), the VP approach is particularly suitable for data that are linearly separable—that is, data on which a linear classifier will do well in separating the two classes. We believe, on the basis of a recent paper,11 that the ADHD and ACEInhibitors drug groups are linearly separable. In that paper, Cohen reports that a linear classifier (SVMLight with the default, linear kernel) achieves some of its best performances among all the 15 groups on these two groups. The linear separability of these two groups would therefore explain why the VP method, which is designed to work well with such data, outperforms FCNB/WE on them.
We now briefly discuss the statistical significance of the improvement of our results over those in Cohen et al.10 As the experimental results for individual folds of the cross-validation are not published in that paper, we could not run statistical tests, such as a t test, to confirm that the differences between our results and their results are significant.
Future work
In future work, we plan to integrate FCNB/WE with other state-of-the-art ML approaches and techniques to create a fully automated document classification system in which FCNB/WE could be a part of a meta-algorithm that includes an ensemble of classifiers. In addition to the data representation which we used in the current research, in case the preferred name for a concept changes over time, the integrated system could include feature engineering based on using the Unified Medical Language System (UMLS).26 The UMLS is a knowledge source containing a metathesaurus, a semantic network, and the specialist lexicon for biomedical domain. The UMLS is likely to contain both the older and newer terms for a concept in a group of synonyms.
Another way to improve the FCNB-based text categorization technique for systematic reviews could be to find a robust algorithm to select factor values that match the current data. It could be done by using data characteristics, such as the imbalance rate for the available training data. An alternative approach could be to score each article according to the confidence the classifier has in its classification of this article. In other words, each article and its score will determine a cut-off point, such that only articles with a higher score will be included. We could provide the users with results ranked by the scores and leave them the option of choosing the level of confidence they need.
Weight engineering is not the only possible way to tune ML classifiers to both types of features, namely binary features (MeSH tags and PubType) and frequency features (abstracts and titles). For example, we could try to solve the weight-underestimation problem and improve performance by applying a meta-algorithm approach that combines FCNB with other algorithms designed for binary features. This could also be a useful topic for future research.
Finally, in this paper we have focused on comparing our efforts with what is currently the state of the art in automating the systematic review process. This included a performance evaluation using a cross-validation protocol, as was done by Cohen et al.1 In addition, we used the WSS measure developed by Cohen et al,1 which measures work saved over and above work saved due to random sampling. This kind of evaluation does not necessarily reflect the labeling effort needed in a fielded system embedded in a deployed systematic reviews system. We have addressed these issues and we discuss our results elsewhere.27
Conclusion
Our research has provided more evidence that automated document classification has strong potential for aiding the labor-intensive literature review process for systematic reviews and other similar studies.
We have demonstrated that CNB, which is designed specifically to address classification tasks with skewed class distribution, can be applied to the process of preparing systematic reviews. We have shown how to modify CNB to emphasize the high recall on the minority class, which is a requirement in classification of systematic reviews. The result, which we have called FCNB, is able to meet the restrictive requirement level of 95% recall that must be achieved. At the same time, we found that FCNB leads to better results in reducing the workload of systematic review preparation than the results previously achieved with the VP method. Moreover, FCNB can achieve even better performance results when machine-performed WE is applied. FCNB provides better interpretability than the VP approach,1 and is far more efficient than the SVM classifier also used for classifying medical abstracts for systematic reviews.14
Supplementary Material
Acknowledgments
This work was funded by the Precarn/Ontario Centres of Excellence Partnership, and by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Funding: Precarn/Ontario Centres of Excellence Partnership and the Natural Sciences and Engineering Research Council of Canada.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
MeSH (Medical Subject Headings) is a medical thesaurus,20 a hierarchical structure of descriptors (tags) representing the US National Library of Medicine's controlled vocabulary used for medical information indexing and retrieval.
References
- 1.Sackett DL, Rosenberg WM, Gray JA, et al. Evidence based medicine: what it is and what it isn't. BMJ 1996;312:71–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigby M. Evidence-based medicine in a nutshell. Arch Dermatol 1998;123:1609–18 [DOI] [PubMed] [Google Scholar]
- 3.Aphinyanaphongs Y, Aliferis CF. Text categorization models for retrieval of high quality articles in internal medicine. J Am Med Inform Assoc 2005;12:207–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis DD, Hayes PJ. Guest editorial—special issue on text categorization. ACM Trans Inf Syst 1994;12:231 [Google Scholar]
- 5.Sahami M, Craven M, Joachims T, et al., eds. Learning for text categorization. AAAI/ICML Workshop, WS-98-05. Menlo Park, CA, USA: AAAI Press, 1998 [Google Scholar]
- 6.Sebastiani F. Machine learning in automated text categorization. ACM Computing Surveys 2002;34:1–47 [Google Scholar]
- 7.Haynes RB, Wilczynski N, McKibbon KA, et al. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Inform Assoc 1994;1:447–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilczynski NL, Haynes RB. Developing optimal search strategies for detecting clinically sound causation studies in MEDLINE. AMIA Annu Symp Proc 2003;719–23 [PMC free article] [PubMed] [Google Scholar]
- 9.Wong SS, Wilczynski NL, Haynes RB, et al. Developing optimal search strategies for detecting sound clinical prediction studies in MEDLINE. AMIA Annu Symp Proc 2003;728–32 [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen AM, Hersh WR, Peterson K, et al. Reducing workload in systematic review preparation using automated citation\classification. J Am Med Inform Assoc 2006;13:206–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen A. Optimizing feature representation for automated systematic review work prioritization. AMIA Annu Symp Proc 2008:121–5 [PMC free article] [PubMed] [Google Scholar]
- 12.Craven M, Shavlik J. Extracting Comprehensible Concept representations from Trained Neural Networks. In: Proceedings of IJCAI 95 Workshop on Machine Learning and Comprehensibility. Montreal: 1995:61–75 [Google Scholar]
- 13.Blanco R, Inza I, Merino M, et al. Feature selection in Bayesian classifiers for the prognosis of survival of cirrhotic patients treated with TIPS. J Biomed Inform 2005;38:376–88 [DOI] [PubMed] [Google Scholar]
- 14.Cohen A, Ambert K, McDonagh M. Cross-topic learning for work prioritization in systematic review creation and update. J Am Med Inform Assoc 2009;16:690–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su J, Zhang H, Ling C, et al. Discriminative parameter learning for bayesian networks. The 25th International Conference on Machine Learning, 2008, 1016–23
- 16.Rennie JD, Shih L, Teevan J, et al. Tackling the poor assumptions of Naїve Bayes text classifiers. In: Proc Int Conf on Machine Learning; 2003:616–23 [Google Scholar]
- 17.Mccallum Andrew, Nigam Kamal. A comparison of event models for Naїve Bayes text classification, 1998 [Google Scholar]
- 18.Steidl S, Schuller B, Batliner A, et al. The Hinterland of Emotions: Facing the Open-Microphone Challenge. In: Proceedings of the International Conference on Affective Computing and Intelligent Interaction; Amsterdam: 2009:690–7 [Google Scholar]
- 19.Cohen AM, Drug review journal citation records file. http://medir.ohsu.edu/∼cohenaa
- 20.United States National Library of Medicine Medical subject headings. http://www.nlm.nih.gov/mesh/overview.html
- 21.Salton G, McGill MJ. Introduction to modern information retrieval. New York: McGraw-Hill, 1983, ISBN 007054484 [Google Scholar]
- 22.WEKA. http://www.cs.waikato.ac.nz/ml/weka/
- 23.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput 1998;10:1895–924 [DOI] [PubMed] [Google Scholar]
- 24.Martin Porter The Porter Stemming Algorithm. http://tartarus.org/∼martin/PorterStemmer/ [Google Scholar]
- 25.Yang Y, Pedersen J. A comparative study on feature selection in text categorization. The 14th International Conference on Machine Learning. 1997:412–20
- 26.National Library of Medicine Unified medical language system fact sheet. http://www.nlm.nih.gov/pubs/factsheets/umls.html
- 27.Kouznetsov A, Matwin S, Inkpen D, et al. Classifying Biomedical Abstracts Using Committees of Classifiers and Collective Ranking Techniques. Canadian Artificial Intelligence Conference, 2009
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.