Abstract
Colorectal cancer (CRC) screening rates are low despite confirmed benefits. The authors investigated the use of natural language processing (NLP) to identify previous colonoscopy screening in electronic records from a random sample of 200 patients at least 50 years old. The authors developed algorithms to recognize temporal expressions and ‘status indicators’, such as ‘patient refused’, or ‘test scheduled’. The new methods were added to the existing KnowledgeMap concept identifier system, and the resulting system was used to parse electronic medical records (EMR) to detect completed colonoscopies. Using as the ‘gold standard’ expert physicians' manual review of EMR notes, the system identified timing references with a recall of 0.91 and precision of 0.95, colonoscopy status indicators with a recall of 0.82 and precision of 0.95, and references to actually completed colonoscopies with recall of 0.93 and precision of 0.95. The system was superior to using colonoscopy billing codes alone. Health services researchers and clinicians may find NLP a useful adjunct to traditional methods to detect CRC screening status. Further investigations must validate extension of NLP approaches for other types of CRC screening applications.
Colorectal cancer (CRC) is the third most common cancer found in men and women in the USA, and is the second leading cause of cancer deaths.1 Screening for CRC is recommended for average-risk individuals aged 50 years and older.2 Current screening rates, however, are suboptimal; recent national studies report that only 40–60% of eligible patients receive proper screening.1 3
Whereas computerized decision support tools have the potential to improve CRC screening rates, the critical challenge is to identify quickly and accurately patients in need of screening. Current methods for determining CRC screening status (patient self-report, physician report, billing data, and manual chart abstraction) are time-consuming, expensive, and often inaccurate. Studies have shown that billing data underestimated CRC screening rates.4 Manual chart abstraction is expensive, and references to completed CRC screening tests are often located within the text of clinic notes, making them difficult to find. An Institute of Medicine report highlighted the need for automated data collection systems that could process natural language clinical texts to address challenges such as these.5
This study investigated the use of a natural language processing (NLP) system to detect the timing and receipt of colonoscopies, the most commonly recommended CRC screening test at many institutions, including the study institution. The authors developed new algorithms to detect the timing and status of colonoscopy references within Vanderbilt Medical Center electronic medical record (EMR) system documents. Vanderbilt's EMR comprises an integrated longitudinal system that receives data from more than 100 diverse sources such as laboratory and radiology reports, typed and dictated notes, interdisciplinary clinician-maintained problem lists, and inter- and intra-office messaging records.
Background
Algorithms employing NLP scan unstructured, ‘free-text’ documents, such as EMR notes, and apply syntactic and semantic rules to extract computer-understandable information, typically into a targeted, standardized terminology representation. Among many uses, researchers have successfully applied NLP to identify references to infections6–8 and cancers9–11 from radiology and pathology reports, to detect adverse events reported within clinical notes,12 13 and, more recently, to assess the quality of clinical care.14 15
Detecting mentions of CRC screening procedures and results within EMR involves unique NLP challenges. Ideally, the NLP system should pinpoint the timing of target events, even though clinicians often reference them using relative time expressions (‘five years ago’). While investigators have studied NLP extraction of temporal references from natural language texts for more than two decades,16–18 the topic remains an active subject of research, especially in the biomedical domain. In addition, many EMR documents contain ‘oblique’ references to CRC screening, such as discussions that clinicians have with patients regarding CRC screening testing, plans for the scheduling of CRC screening tests, reminders to physicians to examine future results, and records of patients' refusals to undergo CRC screening. The authors use the term ‘status indicators’ for descriptors that give information about such CRC screening epiphenomena. An important aspect of NLP recognition of status indicators is negation (‘no colonoscopy performed’). Previous researchers explored automated recognition of concept negation, and corresponding certainty modifiers, for a variety of clinical document types.19–22 Such algorithms use lists of regular expressions19 or syntactic parsing20 22 to identify ‘negating’ concepts. Fewer studies have evaluated broader recognition of other status indicators, such as future planned scheduling and past or current patient refusals. The medical language extraction and encoding system (MedLEE) developed by Friedman and colleagues,21 identifies some status modifiers (such as ‘need’ and ‘future’) and certainty metrics for many clinical concepts. Other related work has explored the recognition of broader definitions of concept certainty, hypothetical modifiers (‘if abnormal colonoscopy’), and detection of the experiencer (or subject) of a concept.23–25
System description
System overview
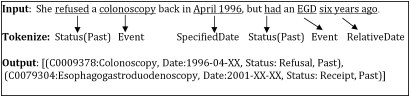
The authors applied their existing, locally developed KnowledgeMap concept identifier (KMCI) to detect concepts related to colonoscopy that were documented in clinical EMR notes. The KMCI, a general-purpose biomedical NLP system supporting concept identification and negation, has been in use at Vanderbilt and other sites over the past 9 years.26–28 In the current study, authors developed new algorithms to identify and interpret time descriptors (eg, ‘6/2003’ or ‘5 years ago’) and properly associate them with corresponding clinical events, such as colonoscopies; and assign values for certainty and status (eg, ‘never had colonoscopy’ or ‘discussed a colonoscopy’) for each identified colonoscopy concept occurrence. Figure 1 illustrates how the augmented KMCI applies date and status information to a recognized concept. The authors developed and refined the new NLP components on a training set including all text documents from 300 randomly selected patients. The core of the KMCI concept identification algorithm was not modified or tweaked for colonoscopy concepts. After refining the timing and status parameters on the training set, KMCI was applied to a test set composed of all text documents from 200 randomly selected patients whose colonoscopy statuses were unknown to the study at the time of selection, and whose records system developers had not previously reviewed or analyzed (details follow below). The study employed two board-certified internal medicine physicians to determine the ‘gold standard’ characterizations of colonoscopy timing and status for each test case, by means of a manual review of case-related EMR data.
Figure 1.
Example assignment of date and status events. The figure assumes a note date in 2007 for the relative date calculation. Only the ‘colonoscopy’ event would have been evaluated in this study.
The first step in the study's NLP ‘pipeline’ is KMCI concept identification, which analyzes an individual EMR document and outputs an XML file containing its highest-ranked ‘recognized’ Unified Medical Language System (UMLS) concepts—including their semantic types, part-of-speech information (derived from a library provided by Cogilex R&D, Inc), noun phrases, normalized word forms, and other information. The study integrated the new KMCI timing and status determination algorithms into a single Perl program. This program first parses the traditional (previous) KMCI output to identify temporal and status ‘tokens’. The program interprets the tokens and then links them to nearby recognized concepts (limited to colonoscopy concepts in the current study). The algorithms use a series of heuristic, linguistic, and semantic rules, as described in more detail below.
Identification of colonoscopy concepts
The authors used concept hierarchies derived from the UMLS metathesaurus to identify relevant concepts pertaining to colonoscopy.29 In addition, the authors manually queried a database of all words found in the clinical notes and messages that had been entered on the 300 training set patients to find other possible synonyms or common abbreviations/misspellings related to colonoscopy. The authors manually identified, with the help of the training set of EMR documents, 26 UMLS concepts (ie, concept unique identifiers) related to colonoscopy (see appendix 1, available online only, for a list of the concept unique identifiers used). Five new terms were added as local synonyms for existing UMLS concepts; they were ‘cscopy’, ‘C scope’, ‘C scopy’, ‘cscope’, and ‘colonscopy’. No changes were made to the core KMCI concept-identification algorithm for this study.
Identification of time descriptors
The authors developed and applied a general-purpose temporal extraction algorithm to identify and assign dates to colonoscopies, using previous work by other investigators as a guide.17 18 30 Time descriptors such as ‘last colonoscopy—5/4/04’ or ‘The patient remembers having a c-scope 5 years ago’ commonly appear in medical narratives. The study KMCI algorithm interprets temporal references in three steps: detection of time descriptors (eg, ‘2002’ and ‘2 years ago’); conversion of these descriptors into a standard representation of date and time; and linkage of time descriptors to the corresponding EMR CRC screening test concept.
The temporal algorithm identifies three categories of date information: fully and partly specified dates (‘3/5/03’ and ‘2002’, respectively); past and future relative date references (‘five years ago’, ‘next week’); and, time period references (‘this past year’, ‘3–5 years ago,’ or ‘last decade’). The study-related KMCI modifications used sets of regular expressions that grouped temporal phrases into ‘tokens’ when parsing the sentence.
After token extraction, the system normalized dates into a standard ‘year–month–day’ format. Ambiguity was represented with placeholders: ‘2004–03–XX’ for ‘March 2004’ or ‘199X’ for the decade of the 1990s. For dates specified by a time range (‘2–4 years ago’), the algorithm approximated the date by selecting the average between the two dates defining the interval. In addition, the system does store the start and stop dates specifying the interval.
As described by Zhou and Hripcsak,30 interpretation of relative dates was necessary (eg, ‘five years ago’, ‘last Thursday’). Such descriptors often contain temporal prepositions (‘in’, ‘at’, or ‘on’), adverbs (‘ago’), or temporal phrases (‘in the past’). The authors developed a lexicon of these temporal phrases and their likely indication of past or future dates with respect to the other chart-based time references and concepts in the note. The actual date of the event was then calculated with date subtraction or addition using the note's date of service.
Assignment of temporal references to concepts
After KMCI normalized a temporal expression, it linked the expression to the best-matching target concept (defined by a set of concepts of interest). The algorithm considered each sentence as an independent entity; temporal references by presumption could only modify CRC screening ‘concepts of interest’ contained within the same sentence. The authors had previously determined, by manual review using the training set, that this presumption was rarely violated. Empirical analysis of the training set indicated that defining a ‘window’ of allowed words between a date and event led to worse performance, because other concepts and status phrases often occurred between the event and the date (eg, ‘In 2002, the patient experienced some rectal bleeding and later had a colonoscopy’). The current study used a simple, empirically derived approach that assigned each temporal reference to its nearest ‘event’, as measured by the number of tokens (eg, words or punctuation) between the date reference and the event. An ‘event’ was defined as any UMLS concept in the list of colonoscopy concepts, any concept with semantic types ‘therapeutic or preventive procedure’ or ‘diagnostic procedure’, and any concept containing words such as ‘surgery’ or ‘repair.’ Multiple events in a list joined by a coordinating conjunction (eg, ‘flex sig, mammogram, and colonoscopy in 2001’) received the same date reference. Similarly, the algorithm assigned a list of dates connected by a coordinating conjunction to the same event (eg, ‘colonoscopies in 1995 and 2005’). The algorithm treated intervening semicolons between a date and an event as boundaries that prevented assignment of the date to the event.
Detection of concept certainty and status
To determine accurately if a patient had undergone colonoscopy, the system had to establish concept certainty (eg, ‘never had a colonoscopy’) and one of six categories of status (see table 1). For example, the algorithm distinguished between ‘had a colonoscopy’, ‘declined colonoscopy’, and ‘scheduled a colonoscopy’.
Table 1.
Types of status indicators.
| Status type | Example phrases |
| Scheduling | ‘Referred for’, ‘ordered’ |
| Considering | ‘Would like to wait’ |
| Discussion | ‘Discussed’, ‘explained’, ‘recommended’ |
| In need of | ‘Due for’, ‘recommended’ |
| Receipt | ‘Had’, ‘underwent’ |
| Refusal | ‘Refused’, ‘declined’ |
To detect status indicators, the authors created a lexicon of base word forms for each status category, which included single words (eg, ‘schedule’, ‘arrange’) and short phrases (eg, ‘overdue for’, ‘set up to have’). The algorithm used each word's part of speech to create negated forms for document processing purposes. For verbs, the algorithm correspondingly created additional verb forms representing different conjugations, tenses, and voices (such as the addition of auxiliary verbs to create the passive form of the verb). Therefore, for the lexicon verb form ‘schedule’, the algorithm would automatically generate other verb forms, such as ‘scheduled’, ‘will be scheduled’, and ‘was not scheduled’. See appendix 2, available online only, for an example of the full list of status words and examples of related variant phrases.
Assignment of certainty and status to concepts
The algorithm uses the part of speech and verb type to assist in assigning the status to an event. For example, transitive verbs modify the event following them; passive verbs modify the events before them. However, if a status indicator appeared as the first or last phrase in a sentence, it could be applied to the concept that came after or before it, respectively, contrary to expected behavior. For example, in the sentence ‘Colonoscopy pt refused’, the algorithm expected the transitive verb ‘refused’ to modify a concept following the verb, but, lacking one, instead applied the ‘declined’ status to ‘colonoscopy’ because the sentence was a cryptic rewording of ‘patient refused colonoscopy’. To assign a status to an event, the algorithm required that the status indicator occur within four words of the event.
Determination of colonoscopy completion (receipt)
After processing all notes with the modified version of KMCI, the study evaluated ‘completed colonoscopies’ by comparing the algorithm's output with the gold standard physician review categorization. The study definition for KMCI-based colonoscopy completion determination was that all UMLS-derived colonoscopy concepts were associated with a past date or ‘today’, and that each had a status of either ‘receipt’ or ‘NULL’. Negated concepts were removed from consideration with respect to receipt.
Evaluation
The authors conducted a preliminary evaluation of the modified KMCI system. The primary study outcome measures were recall and precision for the algorithm-assigned determination of the dates of completed colonoscopies. The ability to recognize dates of colonoscopies is critical to providing real-time CRC screening-related decision support, and for enabling clinical research on screening compliance.
The evaluation, approved by Vanderbilt's institutional review board, randomly selected 200 patients who were aged 50 years or over who had also had more than one primary care clinic visit in the previous year. Authors then used KCMI to identify colonoscopy concepts within all clinical EMR notes from the 200 patients (NB, not all patients had such references in their EMR records). Two physician-reviewers examined all of the sentences containing KMCI-identified references to colonoscopy. The reviewers did not examine any sentences without KMCI-identified colonoscopy concepts. Reviewers scored the algorithm timing and status outputs using a spreadsheet that showed each original sentence in its entirety, highlighted the algorithm-identified date and status words, and indicated the algorithm's interpretation of the date and status strings. Discrepancies between reviewers' determinations were resolved by consensus decision. Reviewers scored each algorithm-identified timing and status reference in each sentence as being true positive (TP, colonoscopy status and timing correctly coded by KMCI), false positive (FP, wrong status or timing descriptor or improperly indicated by KMCI as applying to the patient), true negative (TN, colonoscopy status correctly coded by the algorithm as not done or not known), or false negative (FN, colonoscopy status incorrectly coded by the algorithm as not done or not known when information in the sentence indicated to reviewers that the procedure was done or a timing or status indicator had not been picked up correctly by the algorithm). Recall (sensitivity) was calculated as TP/(TP+FN). Precision (positive predictive value) was calculated as TP/(TP+FP). F measure was calculated as the harmonic mean of recall and precision.
Physician reviewers scored each algorithm-identified temporal tag and status tag independently, so that recall and precision metrics could be calculated separately for each component of the algorithm. Then, reviewers determined from the original sentence whether each sentence indicated that the patient had received a colonoscopy (or not) on a given date (the gold standard). Therefore, for the sentence ‘colonoscopy was rescheduled from originally scheduled date, 3/04’, reviewers would consider a temporal tag a true positive if the algorithm correctly associated ‘colonoscopy’ with ‘2004–03–XX’ even if the status algorithm failed to identify ‘scheduling’ as a modifier for ‘colonoscopy’. Conversely, reviewers could mark the algorithm-assigned status as correct, even if the date was incorrectly interpreted. The reviewers also recorded date and status references omitted by the algorithm (false negatives).
Status report and preliminary results
Of the 200 patients in the test cohort, patients were 62% female with a median age of 64 years and 78% were white, 16% black, and 6% other racial groups. Patients had been followed in the primary care clinics for a median of 5 years. There was an average of 149 notes per patient (inclusive of all available electronic documentation such as admission notes, clinic visits, and clinical messaging) comprising 29 770 total notes. The test set of notes contained 1 112 952 sentences, from which there were 1208 colonoscopy references identified by KMCI concept query; all of these were judged to be true references to colonoscopy-related concepts by the physician reviewers. These 1208 colonoscopy references came from 793 unique notes written by 311 different providers. Colonoscopy references were found in 147 of the 200 (74%) patients in the study sample. Physician reviewers identified 538 (45%) temporal references and 518 (43%) status modifiers associated with the 1208 identified references to colonoscopy concepts. There were 367 references to 156 unique completed colonoscopies in the test set. Correspondingly, there were 841 references to colonoscopy concepts not related to completed colonoscopies (eg, references to recommendations for future screening or discussions about colonoscopies not performed).
Temporal extraction
Physician review of the temporal descriptors related to colonoscopies identified 488 true positive temporal references, 32 false negatives and 25 false positives (table 2). The recall and precision were thus 0.91 and 0.95, respectively, for identifying and assigning the timing descriptors to the correct colonoscopy-related event.
Table 2.
Recall and precision of temporal extraction algorithm
| Gold standard | True positives | False positives | False negatives | Recall | Precision | F measure | |
| Past dates | 373 | 349 | 10 | 14 | 0.93 | 0.97 | 0.95 |
| Specified | 297 | 285 | 3 | 7 | 0.96 | 0.99 | 0.97 |
| Relative | 78 | 64 | 7 | 7 | 0.82 | 0.90 | 0.86 |
| Future dates | 123 | 101 | 8 | 14 | 0.82 | 0.93 | 0.87 |
| Specified | 30 | 19 | 2 | 9 | 0.63 | 0.90 | 0.75 |
| Relative | 93 | 82 | 6 | 5 | 0.88 | 0.93 | 0.91 |
| Recurring dates | 13 | 9 | 0 | 4 | 0.69 | 1.00 | 0.82 |
| Present dates | 29 | 29 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| Totals | 538 | 488 | 25* | 32 | 0.91 | 0.95 | 0.93 |
True positives represent dates correctly identified and assigned to individual colonoscopy concepts. Specified dates are any explicit date formats (eg, ‘3/06’, ‘early 1990s’). Relative dates include any reference that requires a calculation (eg, ‘5 years ago’, ‘last Monday’).
Seven false positives were identified from sentences without a colonoscopy date (n=670).
Authors analyzed the reasons for the ‘missed’ temporal classifications. The majority of false negatives (75%) occurred in sentences containing multiple time references linked to the same colonoscopy concept. In most cases, the algorithm correctly identified the date reference but did not assign it to the event because of one or more of the following: distance of descriptor from the concept, presence of boundary words or characters that disrupted concept recognition or assignment, and date formatting issues. Ten false positives resulted from the misinterpretation of relative date strings (eg, the algorithm interpreted ‘after February 15’ as indicating a previous event from the previous February instead of its intended meaning for a future scheduling date). Eight errors resulted from unanticipated time reference formats within clinic notes (eg, the use of periods instead of commas or slashes in dates), resulted in date miscalculations less than 1 year in length (eg, ‘1.2005’ was interpreted as ‘2005–XX–XX’ instead of ‘2005–01–XX’).
Identification of status descriptors
With respect to the gold standard physician determinations, the status identification algorithm generated 424 true positives, 94 false negatives and 23 false positives. Of the 518 physician-identified status modifiers, 202 (39%) were associated with dated colonoscopies. The recall and precision of the algorithm to detect status indicators were 0.82 and 0.95, respectively. The most common physician-identified status assignments were colonoscopy receipt (n=202), indications of need (n=130), and scheduling references (n=76). Only 35 physician-identified references to colonoscopy-related events were negations. The most common causes of unrecognized status indicators (eg, false negatives) were the absence of certain status-indicating words or phrases from the status lexicon (which had been held constant during the evaluation) and colonoscopy concepts located too far (beyond the defined window of four words) from the status indicator within a sentence. The use of multiple status modifiers together often caused the system to assign incorrect status to colonoscopy events (resulting in false positives, eg, ‘recommend he undergo’ was classified as ‘receipt’ instead of ‘recommend’).
Identification of colonoscopy completion (receipt)
By combining both the temporal extraction (date) and status algorithms, the system correctly assigned the ‘colonoscopy completed’ status more accurately than using either negation detection or status detection alone (F measures of 0.94 vs 0.48 or 0.55, as per table 3). The ability to detect unique completed colonoscopy events from multiple references to such events benefitted from KMCI inclusion of both timing and status descriptors when determining completion status and colonoscopy timing. Of the 156 unique colonoscopies, 147 (92%) were identified by NLP algorithms using status and date detection. A separately performed query of the billing system to determine from billing codes when colonoscopies had been performed on test set patients identified 106 (67%) colonoscopy events. One colonoscopy was detected by billing codes and not by the NLP algorithms (due to a ‘scanned’ non-parseable procedure report for that patient). Therefore, KMCI using EMR notes alone detected 147 of 157 completed colonoscopy events that could be determined by either EMR review or billing record review.
Table 3.
Different NLP approaches to determining from EMR records whether a colonoscopy was actually performed (completed)
| Concept-match only* | Concept-identification with: | ||||
| Negation† | Status‡ | Date§ | Date & Status | ||
| Manually verified no of completed colonoscopies from result set identified by method (TP) | 367 | 367 | 358 | 349 | 340 |
| No of ‘completed colonoscopies’ inferred from method (TP+FP) | 1208 | 1174 | 940 | 396 | 359 |
| Recall | 1.00 | 1.00 | 0.98 | 0.95 | 0.93 |
| Precision | 0.30 | 0.31 | 0.38 | 0.88 | 0.95 |
| F measure | 0.47 | 0.48 | 0.55 | 0.91 | 0.94 |
Test sample contained 1208 sentences extracted verbatim from electronic medical records (EMR). Each of the 1208 sentences contained at least one reference to a project-specified colonoscopy-related concept. Manual review determined that of the 1208 sentences, only 367 referred to colonoscopies actually performed. Note that status and date methods were able to eliminate references to future not-yet-completed events, discussions of a patient's need for a colonoscopy, and other remarks not pertaining to actual, completed colonoscopies.
‘Concept-match only’ simply identifies a ‘colonoscopy event’ based on straightforward concept name matching (with synonymy), independent of surrounding text describing corresponding colonoscopy status, state of negation, or reference date(s).
‘Negation’ refers to ‘concept-match only’, as above, augmented by a negation tagger that removed any negated concept from consideration.
‘Status’ refers to application of a status algorithm to remove non-receipt statuses (eg, scheduled, performed).
‘Date’ refers to use of the date detection algorithm to include only present-day and past events.
FP, false positive; NLP, natural language processing; TP, true positive.
Discussion
Findings
The authors developed a method to identify the timing and status of colonoscopy events, as a component of CRC screening. The system detected text references to completed colonoscopies with a precision of 0.95. This study found that the use of timing and status detection algorithms significantly improved the accuracy of completed colonoscopy detection from a baseline concept query that employed only negation detection. Furthermore, the study's enhanced NLP approach identified 29% more completed colonoscopies than a query of the institution's billing records, primarily because the EMR text records included mentions of patients' colonoscopies performed at other institutions. In a real-world clinical setting, detecting differences between text EMR references to completed colonoscopies compared with billing records for such events might enable reminders to obtain patient consent for CRC testing. Using a diverse set of dictated and typed clinical notes and problem lists as data sources, the KMCI temporal extraction algorithm correctly assigned dates to references to planned or completed colonoscopies, with an overall recall and precision of 0.91 and 0.95, respectively.
Most of the temporal descriptors in the clinical notes referred to past events (72%). These often contained an exact or partial date (eg, ‘last March’) reference. In contrast, most future date references were relative periods of time (eg, ‘five years from now’). As might be expected, the algorithm more accurately interpreted exact dates than relative dates. Most temporal errors occurred due to incorrectly determining whether a reference such as ‘colonoscopy in March’ referred to a past or a future event.
This study is one of the first to evaluate algorithm-identified status indicators related to clinical procedural concept descriptors within EMR documents. The study suggests that several categories of status indicators beyond simple negation are useful to determine the completion status of clinical procedures better. In the current study, an NLP approach incorporating status indicators identified 147 of 157 possible completed colonoscopies out of a total of 1208 colonoscopy references. Only 35 references to colonoscopy events were negated, whereas 206 colonoscopies were ‘discussed’ or ‘scheduled’. The MedLEE in an analogous manner identifies certainty statuses for some clinical events, but not with the granularity for procedural events in the present study.21 The status algorithm in this study performed with good precision (0.95), although its recall must improve through future research. Progress is possible, as most of the study's status assignment errors resulted from terms not in the system lexicon and the algorithm's use of a fixed window for detection that was at times insensitive to the distance of the target concept from the status indicator. Despite these limitations, the use of status indicators did improve the classification of colonoscopies as completed or not.
This study combined several NLP features to identify the timing of completed colonoscopies. In this regard, it was similar to other NLP research on tasks such as the identification of adverse events from discharge summaries,12 the recognition of important findings from radiology reports,31 or recent Informatics for Integrating Biology and the Bedside (i2b2) ‘NLP challenges’ regarding the detection of smoking status,32 obesity and related comorbidities,33 or medication descriptors.34 These applications used a variety of methods combining NLP techniques with rule-based or machine learning approaches, with the best systems achieving F measures greater than 0.90. The NLP approach in the current study used linguistic and heuristic rules that may require minimal or even no training when applied to other datasets. With respect to the clinical goal of the quasi-automated determination of when and what types of CRC screening a given patient should next undergo, the current algorithm must be extended to recognize other forms of CRC screening (eg, barium enemas, flexible sigmoidoscopies) and to determine their timing, statuses, and relevant results (eg, adenomatous polyps of a given size), which alter recommendations for follow-up.
Although the authors focused on colonoscopy concepts, the methods the study employed to identify timing and status references are likely to generalize to other clinical procedural events. Future development should extend the current methods of date and status extraction beyond colonoscopies and related CRC screening procedures. As observed by others, many ‘events’ (including procedures, diagnoses, and other findings) are often mentioned with timing in medical narratives.16–18 30 To assign dates to events, the algorithm relies on UMLS semantic type to define an event. This early analysis suggests that the semantic types in the UMLS may need manual curation to define which could be appropriately linked to date references—many procedures and surgeries were not identified by semantic types ‘therapeutic or preventive procedure’ or ‘diagnostic procedure’ and were captured in this study by simple string matches. Furthermore, the classes of status indicators relevant for a given task are likely to depend on the concept type. For example, medication status indicators would include ‘start’, ‘increase’, and ‘discontinue’.
Limitations
The current study has several limitations. The algorithms were only tested for colonoscopies, and may not perform as well with other concepts. Use of the current system for CRC screening decision support will require extension to include all CRC screening tests such as flexible sigmoidoscopy, barium enema, and fecal occult blood testing. The authors did not compare the colonoscopy concept extraction algorithm with manual chart review to find free-text references to colonoscopies that may have been missed by KMCI. In addition, the temporal extraction algorithm assumed the date references provided in the narratives were correct; however, earlier research has shown this is not always the case.35 Another potential source of error in electronic notes created with a ‘cut-and-paste’ feature are relative references such as ‘five years ago,’ which could be propagated over multiple years of notes. In addition, the algorithm chose the midpoint of a range (eg, selecting ‘4 years’ from the text ‘3–5 years’) when calculating the date. This method could be problematic for large ranges or particular time-sensitive clinical applications. In addition, patients' and providers' estimations of ranges can be inaccurate.35 The current system also could not resolve date co-references, such as assigning a correct antecedent date for temporal phrases such as ‘at that time’. Finally, the current system is based on the output from KMCI, which, like all algorithms, has its own biases and limits on applicability.27 Nevertheless, extending KMCI to detect such concepts as described in the current study only required a list of concepts and their location, the semantic types of each concept, and part-of-speech information about the other words around the concept. The approach to status and date detection used in the current study might thus be integrated into other frameworks, such as the unstructured informatics management architecture,36 to allow broader use within other NLP and information retrieval systems.
Conclusions
Using NLP algorithms to detect the timing and status of procedural concepts within EMR records identified patients who had received past colonoscopies with recall and precision of greater than 90%. The NLP approach detected more references to completed colonoscopy tests than a billing records query alone. The study results suggest that a robust system to identify CRC screening testing should incorporate NLP methods.
Supplementary Material
Footnotes
Funding: This study was funded by National Cancer Institute grant R21 CA116573 and National Library of Medicine grant R01 LM007995.
Competing interests: None
Ethics approval: This study was conducted with the approval of the Vanderbilt University.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Swan J, Breen N, Coates RJ, et al. Progress in cancer screening practices in the United States: results from the 2000 National Health Interview Survey. Cancer 2003;97:1528–40 [DOI] [PubMed] [Google Scholar]
- 2.Winawer S, Fletcher R, Rex D, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale—update based on new evidence. Gastroenterology 2003;124:544–60 [DOI] [PubMed] [Google Scholar]
- 3.From the Centers for Disease Control and Prevention Trends in screening for colorectal cancer—United States, 1997 and 1999. JAMA 2001;285:1570–1 [PubMed] [Google Scholar]
- 4.Freeman JL, Klabunde CN, Schussler N, et al. Measuring breast, colorectal, and prostate cancer screening with medicare claims data. Med Care 2002;40(8 Suppl):IV-36–42 [DOI] [PubMed] [Google Scholar]
- 5.Board on Health Care Services and Institute of Medicine Key capabilities of an electronic health record system. Washington DC, USA: Institute of Medicine, 2003 [Google Scholar]
- 6.Chapman WW, Fizman M, Chapman BE, et al. A comparison of classification algorithms to automatically identify chest X-ray reports that support pneumonia. J Biomed Inform 2001;34:4–14 [DOI] [PubMed] [Google Scholar]
- 7.Jain NL, Knirsch CA, Friedman C, et al. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp 1996:542–6 [PMC free article] [PubMed] [Google Scholar]
- 8.Elkin PL, Froehling D, Wahner-Roedler D, et al. NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Annu Symp Proc 2008:172–6 [PMC free article] [PubMed] [Google Scholar]
- 9.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 1997:829–33 [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, Anderson K, Grann VR, et al. Facilitating cancer research using natural language processing of pathology reports. Medinfo 2004;11:565–72 [PubMed] [Google Scholar]
- 11.Carrell D, Miglioretti D, Smith-Bindman R. Coding free text radiology reports using the Cancer Text Information Extraction System (caTIES). AMIA Annu Symp Proc 2007:889. [PubMed] [Google Scholar]
- 12.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005;12:448–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichley RM, Henderson KE, Currie AM, et al. Natural language processing to identify venous thromboembolic events. AMIA Annu Symp Proc 2007:1089. [PubMed] [Google Scholar]
- 14.Chen ES, Stetson PD, Lussier YA, et al. Detection of practice pattern trends through Natural Language Processing of clinical narratives and biomedical literature. AMIA Annu Symp Proc 2007:120–4 [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SH, Speroff T, Fielstein EM, et al. eQuality: electronic quality assessment from narrative clinical reports. Mayo Clin Proc 2006;81:1472–81 [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Parsons S, Hripcsak G. The evaluation of a temporal reasoning system in processing clinical discharge summaries. J Am Med Inform Assoc 2008;15:99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sager N, Lyman M, Bucknall C, et al. Natural language processing and the representation of clinical data. J Am Med Inform Assoc 1994;1:142–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschman L, Story G. Representation implicit and explicit time relations in narrative. Proc of the 7th International Joint Conferences on Artificial Intelligence. Vancouver, CA, 1981:289–95 [Google Scholar]
- 19.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 20.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001;8:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elkin PL, Brown SH, Bauer BA, et al. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak 2005;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savova GK, Ogren PV, Duffy PH, et al. Mayo Clinic NLP System for Patient Smoking Status Identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clegg AB, Shepherd AJ. Benchmarking natural-language parsers for biological applications using dependency graphs. BMC Bioinformatics 2007;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denny JC, Miller RA, Waitman LR, et al. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inf 2009;78(Suppl 1):S34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003;10:351–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denny JC, Bastarache L, Sastre EA, et al. Tracking medical students' clinical experiences using natural language processing. J Biomed Inform 2009;42:781–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Library of Medicine UMLS source vocabularies, 2010. http://www.nlm.nih.gov/research/umls/metab4.html (accessed Jun 2007).
- 30.Zhou L, Hripcsak G. Temporal reasoning with medical data—a review with emphasis on medical natural language processing. J Biomed Inform 2007;40:183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiszman M, Chapman WW, Aronsky D, et al. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 2000;7:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Third i2b2 Shared-Task and Workshop: Medication extraction challenge, 2009https://www.i2b2.org/NLP/Medication/ (accessed Feb 2010).
- 35.Hripcsak G, Elhadad N, Chen YH, et al. Using empiric semantic correlation to interpret temporal assertions in clinical texts. J Am Med Inform Assoc 2009;16:220–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgartner WA, Jr, Cohen KB, Hunter L. An open-source framework for large-scale, flexible evaluation of biomedical text mining systems. J Biomed Discov Collab 2008;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.