Abstract
Objective
To identify challenges in mapping internal International Classification of Disease, 9th edition, Clinical Modification (ICD-9-CM) encoded legacy data to Systematic Nomenclature of Medicine (SNOMED), using SNOMED-prescribed compositional approaches where appropriate, and to explore the mapping coverage provided by the US National Library of Medicine (NLM)'s SNOMED clinical core subset.
Design
This study selected ICD-CM codes that occurred at least 100 times in the organization's problem list or diagnosis data in 2008. After eliminating codes whose exact mappings were already available in UMLS, the remainder were mapped manually with software assistance.
Results
Of the 2194 codes, 784 (35.7%) required manual mapping. 435 of these represented concept types documented in SNOMED as deprecated: these included the qualifying phrases such as ‘not elsewhere classified’. A third of the codes were composite, requiring multiple SNOMED code to map. Representing 45 composite concepts required introducing disjunction (‘or’) or set-difference (‘without’) operators, which are not currently defined in SNOMED. Only 47% of the concepts required for composition were present in the clinical core subset. Search of SNOMED for the correct concepts often required extensive application of knowledge of both English and medical synonymy.
Conclusion
Strategies to deal with legacy ICD data must address the issue of codes created by non-taxonomist users. The NLM core subset possibly needs augmentation with concepts from certain SNOMED hierarchies, notably qualifiers, body structures, substances/products and organisms. Concept-matching software needs to utilize query expansion strategies, but these may be effective in production settings only if a large but non-redundant SNOMED subset that minimizes the proportion of extensively pre-coordinated concepts is also available.
Keywords: Controlled vocabularies, ICD-9, SNOMED, vocabulary mapping
The World Health Organization's International Classification of Disease, 9th edition, Clinical Modification1 (ICD-9-CM) is widely used in the USA for billing and public health reporting. However, it has numerous terminological weaknesses, most of which are described in Cimino's classic paper on controlled vocabulary desiderata.2 These include numerous clinically obsolete terms (‘benign congestive heart failure’); semantically ambiguous ‘not elsewhere classified’/‘other’ terms3; excessive pre-coordination (eg, creating concepts by combining concepts with prepositional phrases such as ‘with’, ‘without’ or ‘without mention of’)4; inability to compose new concepts from existing ones and failure to support multi-hierarchy.2 ICD-10,5 ICD-9's successor, remedies only the obsolescence problem.
Importantly, ICD lacks sufficient granularity to capture nuances of the clinical encounter that impact therapy/prognosis6; this impacts subsequent data analysis for purposes such as outcomes research.7 8 By contrast, the Systematic Nomenclature of Medicine (SNOMED clinical terms; CT)9 has been shown in numerous studies10–15 to be significantly superior to any other single-source biomedical terminology for encounter encoding. SNOMED CT was originally developed by the College of American Pathologists and is now managed by the International Health Terminology Standards Development Organization (IHTSDO), an organization of organizations that includes the US National Library of Medicine (NLM).
Given, however, that most organizations have large volumes of existing (legacy) ICD-encoded data, several efforts focus on mapping between ICD and SNOMED CT, with the hope of eventually substituting the former with the latter. However, several challenges influence SNOMED CT's ultimate deployment; this paper explores some of these issues.
We explore issues in representing our legacy ICD-9 coded data with SNOMED building blocks, using a SNOMED CT-prescribed compositional approach where appropriate, in order to achieve interoperability with newer systems that may rely on SNOMED CT.
SNOMED CT is a ‘reference’ terminology, primarily intended to capture the semantics of the medical domain; it is unsuitable as an ‘interface terminology’,16 that is, one intended for end-user access. Being 40 times as large as ICD-9, its production use mandates electronic assistance. Neither commercial nor open-source software systems, however, have been evaluated for suitability in letting non-informatics-trained clinical users identify target SNOMED CT concepts accurately, selectively and rapidly. We analyze some software-assisted concept-match failures reported in the literature and identify currently unsolved problems that such software must overcome.
To reduce the impact of terminology size, one may create subsets. The NLM's SNOMED CT core problem list/diagnosis subset contains 5.2 K concepts, selected from the most frequent (95%) concepts encountered in clinical text from at least one of seven collaborating institutions. By design, the core does not include the following categories of concepts: qualifiers, procedures, substances, organisms, body structures. We describe the coverage this subset provided for our own mapping effort and identify areas that should possibly be covered in future releases of this subset.
Background
SNOMED CT-related bridging efforts: a brief history
In 2004, the President's Information Technology Advisory Committee recommended SNOMED CT's use instead of ICD. However, the American Health Information Management Association17 expressed their concerns that SNOMED CT's use was impossible without fully computerized health care. The Healthcare Information and Management Systems Society additionally pointed out18 that the conversion of ICD codes to SNOMED CT codes was not yet automated.
Several efforts have aimed to create maps between ICD-9 and SNOMED CT. An early joint American Health Information Management Association/College of American Pathologists effort19 recorded exact-match and broader-to-narrower mappings. (Exact-match mappings resulting from this effort are available from UMLS's MRSO table, and currently account for 11.6 K (66%) of ICD-9 concepts.) This initiative, which antedates SNOMED CT's current emphasis on description logic and compositionality,20 forms the basis of some commercial offerings, for example.21 Broader-to-narrower mappings may not, however, provide sufficient value for clinical decision support or most analytical purposes. Berg and Campbell.22 describe a joint WHO/IHTSDO plan to map SNOMED CT to ICD-10. The resulting map will provide semi-automated generation of ICD-10 codes from a SNOMED-CT-encoded clinical record.
Composing new concepts using SNOMED CT rules
The rules for composing concepts in the SNOMED CT user's guide, chapter 4, use pairs of concepts combined with an attribute. The rules are currently defined in narrative prose only, but may be implemented as a look-up table.23 The concept hierarchy to which the attribute applies is called the domain, and the permissible set of concepts that can represent the value of the attribute is called the range. For a given domain, only specific attributes and ranges for each attribute apply. A new concept's definition thus becomes a kind of miniature semantic net, in which an edge—an attribute—connects two nodes, a domain concept and a range concept. The guidelines have been developed (relatively recently) because of the realization that without carefully chosen attributes, the composite concept is too ambiguous to support electronic reasoning.
We explain with an actual example from Rosenbloom et al.24 The concept ‘Pony cart accident’, a type of vehicular accident and a term from the MEDCIN interface terminology25 26 used by the Department of Defense, could be composed by simply combining the SNOMED CT concepts ‘pony’, ‘cart’ and ‘accidental physical contact’. Without specific attributes, however, the concept's interpretation must be performed entirely through human knowledge: computationally, there is insufficient information to differentiate it, in a veterinary context, from a pony injured by an accidental collision with a cart. SNOMED CT methodology requires the following steps:
The new concept must relate to an accident, part of the event hierarchy (not a pony or a cart, which belong, respectively, to the organism and physical object hierarchies).
According to SNOMED CT, an event may be associated-with a physical-object. That is, domain=‘event’, attribute=‘associated-with’, and range=‘physical-object’.
We create a new concept, ‘pony cart (physical-object)’ under ‘303965009|Horse-drawn vehicle (physical object)’. Currently no SNOMED CT guidelines exist for fully defining physical objects, so we do not try to refine it further.
We next create ‘pony cart accident (event)’ under ‘242693000|Accident due to physical impact or mechanical violence (event)’.
We link ‘pony cart accident’ to ‘pony cart’ using the attribute ‘associated-with’.
This process is understandably time-consuming because one must compose intermediate concepts, which must be placed correctly within SNOMED CT's concept hierarchy; this requires interactive hierarchy exploration.
Materials and methods
Source data
Mapping from ICD-9 to SNOMED CT is manually intensive, so as part of a pilot institutional effort exploring an eventual SNOMED CT implementation, we decided to work with high-frequency ICD codes in our own data rather than try to map every unmapped ICD code. The second author gathered ICD diagnosis and/or procedure codes occurring at least 100 times in either problem lists or in diagnoses in Geisinger Health System (GHS) data for 2008 from the problem lists/diagnoses tables in GHS's EMR (EpicCare). The electronic medical record is deployed across a network of primary, secondary and tertiary-care centers across rural central Pennsylvania, so that the data represent a mix of care settings across diverse specialties; however, diseases such as HIV are underrepresented compared with data originating from urban settings.
This list also included 11 local codes without ICD-9 counterparts, such as ‘Patient not seen—left before examination’. Of the 2194 unique codes, 1410 were mapped automatically through simple table look-up by UMLS mappings. (These will not be discussed further: we assume UMLS's cross-mappings are accurate.) We attempted to map the remaining 784 concepts (35.7%) using a software-assisted manual approach. The NLM SNOMED CT core subset was obtained from the UMLS knowledge server (https://umlsks.nlm.nih.gov).
When a pre-coordinated ICD-9 concept needed mapping to multiple SNOMED CT concepts, we followed the SNOMED CT-prescribed compositional approach described above.
Software
While we will make our software freely available on request, the choice of software is not critical to the present work, which focuses mainly on mapping/coverage issues. Alternative content searchers, such as LexGrid,27 and alternative query expansion approaches, for example, based on NLM's lexical variant generator28 could have been employed.
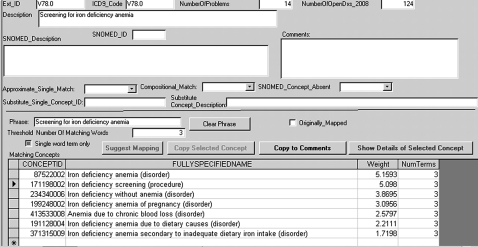
The first author developed a SNOMED CT content searcher/browser using Microsoft structured query language server 2008 to host the Jan 2009 SNOMED CT content, with a stored-procedure visual basic.net library and a browsing front-end created with Microsoft Access 2003. The user specifies search terms either by pasting the ICD-9 term into a text box or entering keywords manually. Concepts matching one or more words in the search phrase are returned using the ‘term frequency*inverse document frequency’29 approach for relevance-ranking of search results, see figure 1. The user must select from the concepts returned, because there will typically be multiple candidate matches.
Figure 1.
The concept-matching interface. The ‘Phrase’ textbox is populated from the contents of the International Classification of Disease, 9th edition (ICD-9) ‘Description’ field. When the user clicks ‘Suggest Mapping’, matching Systematic Nomenclature of Medicine (SNOMED) concepts are presented in descending order of weight (computed relevance), and the number of lemmatized words in a term associated with the concept that are the same as lemmatized words in the search phrase are also shown. The search can be tuned by specifying the number of words that must match (by default, this is N/2+1, where N is the number of non-stop words in the search phrase). In the screen-shot, there is no exact phrase match for the ICD-9 concept, but the concept ‘171198002|Iron deficiency screening’ is semantically equivalent.
We improved search speed by indexing SNOMED CT fully specified names and synonymous descriptions. We split each first phrase into individual words. We eliminated stop words (common words such as articles, pronouns, prepositions and conjunctions, with minimal search value) using the PubMed stop-word set.30 A keyword index was built from the remaining words after lemmatization,31 that is, conversion to root forms (lemmas) by eliminating variations in tense and person, as discussed below. Search terms were lemmatized before consulting the keyword index.
To improve both computational and human search efficiency, we implemented a technique described in Manning et al32 of allowing the user to specify optionally that terms for returned concepts must contain at least N words that are present in the search phrase. This minimized the number of non-relevant concepts returned when the phrase contained a common word such as ‘neoplasm’. We also allow the user to limit optionally the search to concepts with terms comprising single words; this was necessary to support search of ‘qualifiers’: concepts such as ‘secondary’, ‘idiopathic’, ‘iatrogenic’, ‘chronic’, etc, which are very commonly parts of other concepts and serve to qualify the condition described.
Query expansion approach
Medical terms make abundant use of word variants (eg, singular vs plural), and so we employed the following approaches to increase search sensitivity.
Lemmatization
We used morph, part of Wordnet,33 combined with part-of-speech (POS) information from the Moby POS list,34 available through Project Gutenberg.35 Morph combines table look-up (an exceptions list of approximately 4800 words) with word-transformation rules. The lemmas of homographs (words with different meanings but identical spellings) may change according to the part of speech; for example, the lemma of ‘leaves’ can be ‘leaf’ or ‘leave’, depending on whether ‘leaves’ is a plural noun or a verb. In such cases we use both lemmas. Our approach is similar to that of Denny et al36 for KnowledgeMap.
Synonyms
We produced a single-word synonym table by identifying all sets of single-word terms in SNOMED CT mapping to the same concept. This was augmented with a manually created list of Greco-Latin/common-word and other equivalences (eg, hepatic/liver, gastric/stomach, neoplasm/tumor); this information was missing in SNOMED CT. Note that while NLM's SPECIALIST lexicon,37 which the lexical variant generator uses, handles certain Greco-Latin forms (eg, stromata is normalized to stroma), it does not link adjectival and noun forms of anatomical structures (eg, pontine is not related to pons).
Efficiency of the mapping process
The above mapping process has a manual component—the user's selection of a concept from several candidate matches. In some cases, very few candidates (eg, two to three) were returned, allowing the top-ranked concept to be picked immediately within a couple of seconds. In other cases, however, none of the returned concepts matched either partly or completely to the phrase of interest, and we had to search SNOMED CT content by manually providing alternative search terms based on our own knowledge of medicine or English—often, repeatedly so after multiple match failures. A few phrases took 15–20 minutes of searching to map and compose, so that search efficiency varied by a factor of at least 300.
Results
Of the 784 concepts that needed manual mapping, we could map all except one (local) concept, which we were unable to locate in SNOMED CT: ‘Medication Use Agreement’—a document signed by an ambulatory patient who is prescribed narcotics, agreeing to stringent conditions for medication use. The closest approximation was ‘288834001|Agreeing on care plan (procedure)’ combined with ‘360204007|Opiate (product)’ by the attribute ‘using-substance’, but this does not capture the concept's nuances.
Our computed mappings are supplied in the accompanying data supplement available online only at http://jamia.bmj.com.
Of the manually mapped codes, 435 (55.5%) had one or more of the following embedded phrases: ‘NEC/Not elsewhere classified’ (36); ‘unspecified’ (213); ‘without mention of’ (91); ‘other specified’ (65); ‘other’ without ‘unspecified’ (236). (Some phrases are double counted.) There were 302 ‘NOS/not otherwise specified’ concepts in the original dataset, but all except one of these had SNOMED CT/UMLS-provided mappings. These mappings essentially substituted the ambiguous concept with the non-ambiguous super-concept thus: 789.00, ‘ABDOMINAL PAIN, SITE NOS’ had been mapped to 207205003|‘Abdominal pain (situation)’. We decided to take a similar approach, by substituting the closest semantically related super-concept, eg, with the ‘NEC’ or the ‘without mention of’ qualifier removed. (Strictly speaking, this approach is not entirely valid for NEC concepts, because of the semantic drift problem. However, given that SNOMED's curators actively discourage the creation of NEC concepts, this is the only way to preserve information partly for legacy NEC data; in production scenarios, a Boolean flag needs to be associated with NEC codes to indicate partial information loss.)
Two hundred and sixty-four (33.7%) of the manually mapped codes were composite when represented in SNOMED CT; most could be represented using a pair of SNOMED CT concepts, but 30 of these required three concepts and three ICD concepts required four. Mapping 45 of these concepts required introducing a disjunction (‘or’) operator. Many of these concepts are the equivalent of what SNOMED CT terms ‘navigational concepts’: concepts that exist merely to provide intermediate nodes in a concept hierarchy, often linking independent concepts below them. An example of a doubly navigational concept is 843.9, ‘Sprain and strain of unspecified site of hip and thigh’. Strains and sprains may co-occur, but they must be differentiated clinically if occurring singly because they affect different tissues (muscle–tendon junctions and ligaments, respectively), and require different long-term management approaches. Note also that ICD-9 tends to use ‘and’ imprecisely for what would be ‘or’ in Boolean logic—‘hip and thigh’ in the above example means an injury of either the hip or the thigh. The SNOMED CT technical implementation guide states that ‘A navigation concept plays no part in the semantic definitions of any other concept’, and ideally should not be used for clinical documentation either because of the possibility of ambiguity; in the above case, the precise site and nature of injury would need recording.
Ten composite concepts required introducing a set difference operator (a form of qualified negation), for example, 959.4, ‘Injury, other and unspecified, hand, except finger’ 600.90, ‘Unspecified hyperplasia of prostate without urinary obstruction and other lower urinary tract symptoms’ and 347.00, ‘Narcolepsy without cataplexy’. The first concept excludes another concept ‘injury of finger’, while the latter two concepts model combinations of main findings with significant negative findings.
Coverage provided by the NLM's core SNOMED CT subset
Among the SNOMED CT concepts used to create the composites, there were 442 unique SNOMED CT concepts. Of these, only 148 concepts were in the core subset. Table 1 summarizes the distribution of the missing concepts by concept hierarchy.
Table 1.
Concepts necessary to create new composite concepts, but which were not in the July 2009 release of the NLM SNOMED clinical core subset.
| Category | Count |
| Disorder | 93 |
| Qualifier*** | 45 |
| Body structure*** | 34 |
| Finding | 33 |
| Procedure*** | 31 |
| Situation | 12 |
| Morphologic abnormality | 11 |
| Substance/product *** | 11 |
| Regime/therapy | 6 |
| Observable entity | 5 |
| Organism*** | 4 |
| Attribute* | 3 |
| Navigational concept | 2 |
| Event | 2 |
| Person | 2 |
| Physical object* | 1 |
These are grouped by category (Systematic Nomenclature of Medicine (SNOMED) concept hierarchy). Asterisks indicate hierarchies that are absent in the core subset; a triple asterisk indicates hierarchies that are absent as a consequence of core's original design goals.
NLM, US National Library of Medicine.
To quantify coverage, we must subtract from 442 the counts of those hierarchies deliberately omitted from core: qualifiers, procedures, body structures, organisms, and substances. The total of these is 125. Therefore, the true coverage is 148/(442−125)=148/317=46.7%.
On inspection, we found that concepts in the ‘findings’ and ‘disorders’ were represented in core with finer granularity than we needed: among the necessary coarse-grain concepts were ‘Primigravida’, ‘gangrene’, ‘ulcer’ and ‘drug intolerance’.
The omission of qualifiers is justifiable: core is intended to be deployed in production along with qualifiers (of which there were 45 in our data). However, the omission of other categories from core is more disputable; these comprise essential building blocks for many common concepts, and we believe that lists of these should be available to the end-user, even if only in abbreviated form.
Issues in searching SNOMED CT for appropriate matches
Despite query expansion, we found that search of SNOMED CT still required knowledge of both medical and English synonymy, and took time because several search patterns had to be composed manually. Medical knowledge is needed to ascertain that the phrases ‘surgical’ (as in ‘surgical dressing’) and ‘primary’ (as in 89.81, ‘Primary hypercoagulable state’) refer to the respective SNOMED CT concepts ‘Postoperative state’ and ‘idiopathic’; within SNOMED CT they are defined only as synonyms of ‘applying to the surgical specialty’ and ‘principal’. Similarly, mapping the local code ‘Patient left—not seen’ to a composite term that is the intersection of ‘162650008|Patient not examined (situation)’ and ‘398090008|Patient unavailable (finding)’ requires search by English-language synonyms.
Despite its very large clinical coverage of the clinical domain, SNOMED CT lacks certain general concepts. Therefore, ‘monoarthritis’ (unqualified) is missing, while ‘Monoarthritis, unspecified’, a deprecated concept, is present. Certain concepts are hierarchically ambiguous: 384709000|‘Sprain’ is classified (possibly incorrectly) as a morphologic abnormality, which descends from body structure, while sprains related to specific body structures, such as 44465007|‘Sprain of Ankle’ are disorders, which descend from clinical finding, a separate hierarchy.
Discussion
This study has the limitations that the set of high-frequency concepts are necessarily unique to our organization and the manual mappings may have errors. The theme of the paper is not the codes or the mapping accuracy per se, but the difficulties that institutions are likely to face when dealing with legacy ICD-9 data. Many ICD-9 codes must necessarily be converted to SNOMED CT equivalents manually, and the harder it is to map these codes into SNOMED CT (especially when requiring complex compositional mapping), the more difficult it will be to work with the converted codes.
More than half of the high-frequency concepts in our data that required manual mapping were concepts with aspects flagged by SNOMED CT as deprecated. Most clinicians, however, are not trained in controlled terminology principles, and will select from concepts presented to them; we believe that our institution is not anomalous in this regard. Any strategy for dealing with legacy ICD-9 data must provide a migration path for such concepts.
Our experiences are relevant to ongoing or future work aimed at making SNOMED CT the basis of interface terminologies. The level of coding necessary for the documentation of clinical findings and symptoms is necessarily far more detailed than for billable diagnoses or procedure codes alone; without high-quality software assistance, the process may be unacceptably burdensome. We discuss related issues below.
Coverage by subsets: how many concepts are enough?
The SNOMED CT framework is not intended to be an interface terminology, and its curators advocate using subsets: the NLM's core subset is valuable. The 95% coverage threshold used to select the concepts in the core implies that approximately 5% of the time, the clinician/coder would have to search the full SNOMED CT (or a much larger subset) semimanually.
In many areas of software-assisted information capture, however, 95% coverage is now considered unacceptable; the best optical character recognition software achieves accuracy of 99.5% at the word level for printed Latin-character text,38 while Microsoft claims a dictation accuracy for Vista speech recognition, if the software has been trained for a specific speaker, that may exceed 99%.39 More important, for both optical character recognition and speech-to-text, both error detection and manual error correction are relatively fast. Searching for concepts not present in a subset, by contrast, takes much more time, because the time required to probe the larger terminology dominates the coding process. However, the core subset is a work in progress, and may grow with time.
The process of encoding is especially onerous if a concept encountered in text does not exist in the terminology; it takes considerable effort to ascertain that this is indeed the case, because one rarely has a priori knowledge that the terminology lacks a given concept. We have cited our own worst-case performance of 15–20 minutes for a concept match (although the screen-shot of figure 1 took approximately only 3 seconds). Most busy clinicians would abandon the search much sooner; we provide some examples from the literature shortly to show that even trained informaticians may mistakenly denote certain concepts as missing in SNOMED CT when they are not.
Composing new concepts
Our experience indicates that users in time-constrained situations may not be able to compose concepts accurately. This process, which also means defining concepts accurately and placing them correctly within the SNOMED CT hierarchy, is very difficult to automate; at best, software can only assist content browsing. In production situations, coders/clinicians might instead have to record the concepts that they could not map, so that individuals intimately familiar with the larger terminology (and responsible for local terminology maintenance) can create them off-line.
The latter individuals' productivity will be enhanced by having rapid access to concepts that are common building blocks. We have identified certain categories of concepts that might need to be incorporated eventually into the NLM core. We have also mentioned our need to introduce disjunction and set difference operators. Many description logics, including the description-logic subset of the web ontology language (OWL-DL),40 as well as SNOMED CT, do not support these operators because they introduce significant additional computational complexity in classification tasks. However, there are efforts to support such operations in a variety of software, including the Protégé OWL plug-in.41 Set difference, a very restricted form of negation, is implemented efficiently in structured query language,42 because it operates on an initial subset of the data; thus, patients with narcolepsy but without cataplexy are much fewer than all patients without cataplexy.
Limitations of existing software-assisted approaches
We have reason to believe that the issue of worst-case search performance is not due to limitations of our own software per se, but due to insufficient synonymy information in existing clinical vocabularies. Rosenbloom et al,24 using terminology server software developed by the Mayo Clinic Biomedical Informatics group, reported difficulties in mapping the concepts ‘tremor observed’ and ‘auscultated’, and consequently asserted that the concepts of detection of a finding by observation or auscultation do not exist in SNOMED CT. While it is true that the words ‘observed’ and ‘auscultated’ are not present as single-word terms in SNOMED CT, the respective equivalent terms 32750006|‘Inspection (procedure)’ and 37931006|‘Auscultation (procedure)’ do exist; the first has the synonymous term ‘Visual observation’, and both concepts are immediate children of 315306007|‘Examination by method (procedure)’.
To be honest, we spent approximately 10 minutes on this exercise by exploring SNOMED CT using alternative terms and synonyms in order to locate these concepts. If a team of experienced informaticians could make occasional mistakes using well-regarded software, typical clinicians or coders working under time constraints, and who have much less familiarity with SNOMED CT content (or with medical synonyms, if they are coders), are likely to make many more mistakes. The use case for SNOMED CT in encoding clinical encounters anticipates many more concepts to be encoded per clinical document than in ICD-9 usage scenarios: busy clinicians/coders may abandon the search for a concept if they cannot find it readily, resulting in incomplete documentation.
Query-expansion software strategies will need to be augmented considerably. This approach by itself is double-edged, trading increased sensitivity for lower specificity and presenting many more partial matches to the user, and prolonging concept selection time. We believe that concept search will also be facilitated by streamlining the content of the larger terminology, for access in cases in which matches to a ‘core’ subset are not found. We are probably not the first to advocate the creation of a much larger, interface-oriented subset for this purpose. While each new release of SNOMED CT has consistently had better content quality than the previous version, elimination of redundancy, which can interfere with end-user usability, should possibly be made a high priority if SNOMED CT's deployment is intended to become widespread and eventually mandated.
One possible approach is the use of natural language processing (NLP) techniques that suggest concepts after scanning the clinical narrative. Because the window of text scanned at a time by NLP approaches is limited (typically a sentence), concepts that are extensively pre-coordinated, such as those involving negation—for example, 28082003|‘Chronic duodenal ulcer without hemorrhage AND without perforation but with obstruction (disorder)’ are hard to recognize. Recognition failure can occur for two reasons. First, the negation may not even be mentioned in the narrative because of the circumstance—for example, an ambulatory, non-distressed duodenal ulcer patient is unlikely to have ulcer perforation. Second, even if the negated concept is mentioned, the mention may occur in a different sentence: for example, an operative note may indicate that the patient had a history of duodenal ulcer and later that, on endoscopy, no bleeding was observed.
Consequently, rather than attempt uncertain algorithmic contortions to try to match extensively pre-coordinated concepts that include negation, it may be simpler to eliminate such concepts from a working lexicon.
Conclusions
Giannangelo and Fenton43 surveyed 408 clinical-software vendors about SNOMED CT use. Of the 72 responders, only 14 supported SNOMED CT, and in only five were customers using it. If SNOMED CT use becomes mandated, its user base will grow by approximately two orders of magnitude, and issues such as the one discussed here will become prominent.
Given that ICD-9-to-SNOMED CT mapping necessarily involves a large manual (curatorial) component, focusing on ICD-9 codes that occur with relatively high frequency can optimize the use of scarce human expertise; codes that occur very rarely or not at all should have a much lower priority in mapping efforts.
The ease of composing concepts accurately will be facilitated greatly by IHTSDO's ongoing efforts with the SNOMED CT machine-readable concept model,44 concurrent with an expansion of the number of existing relationships (as seen with the SNOMED CT observables hierarchy).45 However, the challenges of devising concept-encoding software (whether using NLP approaches or otherwise) that is sufficiently responsive to the needs of non-informatician clinicians and clinical coders remain formidable.
Supplementary Material
Footnotes
Funding: This work was supported in part by Centers for Disease Control and Prevention grant no U10 OH008225, and by institutional funding from Geisinger Health System.
Competing interests: None.
Ethics approval: This study was conducted with expedited approval from Geisinger Health System institutional review board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.World Health Organization International Classification of Diseases. 9th edn Geneva: Switzerland, 1977 [Google Scholar]
- 2.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403 [PMC free article] [PubMed] [Google Scholar]
- 3.International Health Terminology Standards Development Organization SNOMED Clinical Terms (SNOMED CT) Technical Implementation Guide. Copenhagen, Denmark: IHTSDO, 2009 [Google Scholar]
- 4.Bodenreider O, Smith B, Burgun A. The ontology-epistemology divide: a case study in medical terminology. Proceedings of the Third International Conference on Formal Ontology in Information Systems, 2004. Amsterdam: 2004:185–95 http://lhncbc.nlm.nih.gov/lhc/docs/published/2004/pub2004064.pdf (accessed Jun 2010). [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization International Classification of Diseases, 10th edn Geneva, Switzerland, 1992 [Google Scholar]
- 6.Brouch K. AHIMA project offers insights into SNOMED, ICD-9-CM mapping process. J AHIMA 2003;74:52–5 [PubMed] [Google Scholar]
- 7.Nanovic L, Kaplan B. Reliability of Medicare claim forms for outcome studies in kidney transplant recipients: epidemiology in clinical outcome trials. Clin J Am Soc Nephrol 2009;4:1156–8 [DOI] [PubMed] [Google Scholar]
- 8.Stein H, Nadkarni P, Erdos J, et al. Exploring the degree of concordance of coded and textual data in answering clinical queries from a clinical data repository. J Am Med Inform Assoc 2000;7:42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Health Terminology Standards Development Organization. SNOMED Clinical Terms (SNOMED CT). 2009. http://www.snomed.org (accessed 2 Jan 2009). [Google Scholar]
- 10.Chiang M, Casper D, Cimino J, et al. Representation of ophthalmology concepts by electronic systems: adequacy of controlled medical terminologies. Ophthalmology 2005;112:175–83 [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Flaitz C, Johnson T. Comparison of accuracy captured by different controlled languages in oral pathology diagnoses. AMIA Annu Symp Proceeding; Austin, Texas, 30 November–1 December 2005:918. [PMC free article] [PubMed] [Google Scholar]
- 12.Warren J, Collins J, Sorrentino C, et al. Just-in-time coding of the problem list in a clinical environment. Proc AMIA Symp; Washington DC, 7–11 November 1998:280–4 [PMC free article] [PubMed] [Google Scholar]
- 13.Vardy D, Gill R, Israeli A. Coding medical information: classification versus nomenclature and implications to the Israeli medical system. J Med Systems 1998. 22:203–10 [DOI] [PubMed] [Google Scholar]
- 14.Chute C, Cohn S, Campbell K, et al. The content coverage of clinical classifications. For the Computer-Based Patient Record Institute's Work Group on Codes and Structures. J Am Med Inform Assoc 1996;3:224–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell J, Payne T. A comparison of four schemes for codification of problem lists. Proc Annu Symp Comput Appl Med Care; Washington DC, 4–7 November 1994:201–5 [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbloom ST, Miller RA, Johnson KB, et al. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc 2006;13:277–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.American Health Information Management Association AHIMA's Open Letter to the President's Information Technology Advisory Committee, 2004. http://www.ahima.org/dc/AHIMACommentstoPITAC.asp (accessed Jun 2010). [Google Scholar]
- 18.Gibbons P. Comments on the PITAC Draft Recommendation on ICD-10/SNOMED (4/13/04). 2004. http://www.himss.org/content/files/PITAC_ICD10_SNOMED_gibbons05182004.pdf (accessed Jun 2010).
- 19.Imel M. A closer look: the SNOMED clinical terms to ICD-9-CM mapping. J AHIMA 2002;73:66–9 [PubMed] [Google Scholar]
- 20.Spackman KA, Campbell KE. Compositional Concept Representation Using SNOMED: Towards Further Convergence of Clinical Terminologies. Proc AMIA Fall Symposium; 7–11 November 1998 [PMC free article] [PubMed] [Google Scholar]
- 21.Intelligent Medical Objects IMO Announces Enhanced ICD-9 Encoded Problem List Vocabulary with Mapping to SNOMED® CT 2004 [cited 8/17/09]. http://www.e-imo.com/press/10440.aspx (accessed Jun 2010).
- 22.Berg L, Campbell J. Mapping SNOMED CT to ICD-10 – a joint task of IHTSDO and WHO–FIC. 2008. http://www.tc215wg3.nhs.uk/pages/docs/isotc215wg3_n362.pdf (accessed 8 Apr 2009).
- 23.Nadkarni P, Marenco L. Implementing description-logic rules for SNOMED-CT attributes through a table-driven approach. J Am Med Inform Assoc 2010;17:182–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenbloom ST, Brown SH, Froehling D, et al. Using SNOMED CT to represent two interface terminologies. J Am Med Inform Assoc 2009;16:81–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goltra PS. MEDCIN: A New Nomenclature for Clinical Medicine. Ann Arbor, MI: Springer-Verlag, 1997 [Google Scholar]
- 26.Medicomp Corporation MEDCIN terminology. 2009. http://www.medicomp.com (accessed 8 Jan 2009).
- 27.Pathak J, Solbrig HR, Buntrock JD, et al. LexGrid: a framework for representing, storing, and querying biomedical terminologies from simple to sublime. J Am Med Informat Assoc 2009:305–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Divita G, Browne AC, Rindflesch TC. Evaluating lexical variant generation to improve information retrieval. Proceedings/AMIA Annual Symposium; 7–11 November 1998:775–9 [PMC free article] [PubMed] [Google Scholar]
- 29.Baeza-Yates R, Ribeiro-Neto B. Modern information retrieval. Harlow, UK: Addison-Wesley Longman, 1999 [Google Scholar]
- 30.National Center for Biotechnology Information. The PubMed stop-word list. 2009 [cited 7/8/09]. http://www.ncbi.nlm.nih.gov/entrez/query/static/help/pmhelp.html#Stopwords <http://www.ncbi.nlm.nih.gov/entrez/query/static/help/pmhelp.html> (accessed Jun 2010).
- 31.Jurafsky D, Martin JH. Speech and language processing. 2nd edn Englewood Cliffs, NJ: Prentice-Hall, 2008 [Google Scholar]
- 32.Manning C, Raghavan P, Schuetze H. Introduction to Information Retrieval. Cambridge, UK: Cambridge University Press, 2008 [Google Scholar]
- 33.Miller GA, Fellbaum C, Tengi R, et al. Wordnet: a lexical database for the English Language. 2002. http://www.cogsci.princeton.edu/∼wn/ (accessed Jun 2010).
- 34.Ward G. The MOBY part of speech List. Project Gutenberg 2009. http://www.gutenberg.org/etext/3203(accessed 7 Jan 2009). [Google Scholar]
- 35.Project Gutenberg Project Gutenberg Home Page. 2009. http://www.gutenberg.org (accessed 7 Jan 2009).
- 36.Denny J, Smithers JD, Miller RA, et al. Understanding¡¨ Medical School Curriculum Content Using KnowledgeMap. J Am Med Inform Assoc 2003;10:351–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCray A. The nature of lexical knowledge. Methods Inf Med 1998;37:353–60 [PubMed] [Google Scholar]
- 38.von Ahn L, Maurer B, McMillan C, et al. reCAPTCHA: High transcription accuracy. 2009. http://recaptcha.net/digitizing.html (accessed 8 Jan 2009).
- 39.21talks.net. Windows Vista: Speech recognition accuracy reaches 95–99.x%. 2007 [cited 8/2/09]. http://21talks.net/voip/windows-vista-speech-recognition (accessed Jun 2010).
- 40.World Wide Web Consortium OWL Web Ontology Language: Overview. 2004. http://www.w3.org/TR/owl-features/ (accessed 9 Jan 2009).
- 41.Protege Users' group Protege OWL announces addition of negation and disjunction. 2008. https://mailman.stanford.edu/pipermail/protege-owl/2008-December/008991.html (accessed 8 Feb 2009).
- 42.Melton J, Simon AR, Gray J. SQL 1999: understanding relational language components. San Mateo, CA: Morgan Kaufman, 2001 [Google Scholar]
- 43.Giannangelo K, Fenton SH. SNOMED CT survey: an assessment of implementation in EMR/EHR applications. Perspect Health Inf Manag 2008;5:7. [PMC free article] [PubMed] [Google Scholar]
- 44.International Health Terminology Standards Development Organization SNOMED CT machine-readable concept model. 2010. https://thecap.basecamphq.com/projects/388747/file/35466849/xres_MachineReadableConceptModel_Core-Full_INT_20091001.zip (accessed 3 Jan 2009). (this URL requires IHTSDO membership to access; membership, however is freely granted).
- 45.Spackman K. Snomed CT Observables Concept Model 2010. https://thecap.basecamphq.com/P29878266 (accessed 3 Jan 2009). (this URL is available only to IHTSDO members: membership, however, is free).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.