Abstract
We aim to build and evaluate an open-source natural language processing system for information extraction from electronic medical record clinical free-text. We describe and evaluate our system, the clinical Text Analysis and Knowledge Extraction System (cTAKES), released open-source at http://www.ohnlp.org. The cTAKES builds on existing open-source technologies—the Unstructured Information Management Architecture framework and OpenNLP natural language processing toolkit. Its components, specifically trained for the clinical domain, create rich linguistic and semantic annotations. Performance of individual components: sentence boundary detector accuracy=0.949; tokenizer accuracy=0.949; part-of-speech tagger accuracy=0.936; shallow parser F-score=0.924; named entity recognizer and system-level evaluation F-score=0.715 for exact and 0.824 for overlapping spans, and accuracy for concept mapping, negation, and status attributes for exact and overlapping spans of 0.957, 0.943, 0.859, and 0.580, 0.939, and 0.839, respectively. Overall performance is discussed against five applications. The cTAKES annotations are the foundation for methods and modules for higher-level semantic processing of clinical free-text.
Introduction
The electronic medical record (EMR) is a rich source of clinical information. It has been advocated that EMR adoption is a key to solving problems related to quality of care, clinical decision support, and reliable information flow among individuals and departments participating in patient care.1 The abundance of unstructured textual data in the EMR presents many challenges to realizing the potential of EMRs. For example, most EMRs record a narrative describing the history of the current illness for an episode of care. Clinical researchers leverage this information by employing a number of domain experts to manually curate such narratives. This process can be both error-prone and labor-intensive. Automating it is essential to developing an effective EMR infrastructure. Natural language processing (NLP) techniques have demonstrated successes within the clinical domain (for an overview see Meystre et al2). However, their widespread adoption rests on developing comprehensive clinical NLP solutions based on open standards and software.
Our goal is the development of a large-scale, comprehensive, modular, extensible, robust, open-source NLP system designed to process and extract semantically viable information to support the heterogeneous clinical research domain and to be sufficiently scalable and robust to meet the rigors of a clinical research production environment. This paper describes and evaluates our system—the clinical Text Analysis and Knowledge Extraction System (cTAKES).
Background
The clinical narrative has unique characteristics that differentiate it from scientific biomedical literature and the general domain, requiring a focused effort around methodologies within the clinical NLP field.2 Columbia University's proprietary Medical Language Extraction and Encoding System (MedLEE)3 was designed to process radiology reports, later extended to other domains,4 and tested for transferability to another institution.5 MedLEE discovers clinical concepts along with a set of modifiers. Health Information Text Extraction (HITEx)6 7 is an open-source clinical NLP system from Brigham and Women's Hospital and Harvard Medical School incorporated within the Informatics for Integrating Biology and the Bedside (i2b2) toolset.8 IBM's BioTeKS9 and MedKAT10 were developed as biomedical-domain NLP systems. SymText and MPLUS11 12 have been applied to extract the interpretations of lung scans13 to detect pneumonia14 and central venous catheters mentions.15 Other tools developed primarily for processing biomedical scholarly articles include the National Library of Medicine MetaMap,16 providing mappings to the Unified Medical Language System (UMLS) Metathesaurus concepts,17 18 those from the National Center for Text Mining (NaCTeM),19 JULIE lab,20 and U-compare,21 with some applications to the clinical domain.22 Within the Cancer Biomedical Informatics Grid23 initiative, the University of Pittsburgh's Cancer Tissue Information Extraction System24 25 aims at extracting information from surgical pathology reports using the National Cancer Institute Enterprise Vocabulary System26 and MetaMap.
In the general domain, a number of open-source NLP toolsets exist. The OpenNLP suite27 implements a maximum entropy (ME) machine learning (ML) classifier,28–30 with probabilities that maximize information entropy and that are derived from a dataset to include a diverse feature set.31 32 Buyko and colleagues33 adapted the OpenNLP to the biomedical scientific literature and demonstrated that the performance of OpenNLP's components on biotexts represented by GENIA34 and PennBioIE35 is comparable to that on the newswire text.
Our unique contribution is an NLP system specifically tailored to the clinical narrative that is large-scale, comprehensive, modular, extensible, robust, open-source and tested at component and system levels.
Design and system description
The cTAKES is a modular system of pipelined components combining rule-based and machine learning techniques aiming at information extraction from the clinical narrative. The gold standard datasets for the linguistic labels and clinical concepts are created on content that is a subset of clinical notes from the Mayo Clinic EMR. Standard evaluation metrics are used to measure the quality of the gold standards and cTAKES performance.
The cTAKES system
The cTAKES consists of components executed in sequence to process the clinical narrative with each component incrementally contributing to the cumulative annotation dataset (see figure 1 for a running example). This provides the foundation for future cTAKES modules for higher-level semantic processing of the clinical free-text.
Figure 1.
Example sentence processed through cTAKES components ‘family history of obesity but no family history of coronary artery diseases.’ Fx, family history.
The cTAKES accepts either plain text or clinical document architecture-compliant36 XML documents. The current open-source release consists of the following components/annotators:
Sentence boundary detector
Tokenizer
Normalizer
Part-of-speech (POS) tagger
Shallow parser
Named entity recognition (NER) annotator, including status and negation annotators.
The sentence boundary detector extends OpenNLP's supervised ME sentence detector tool. It predicts whether a period, question mark, or exclamation mark is the end of a sentence.
The cTAKES tokenizer consists of two subcomponents. The first splits the sentence internal text stream on the space and punctuation. The second, the context-dependent tokenizer, merges tokens to create date, fraction, measurement, person title, range, roman numeral, and time tokens by applying rules (implemented as finite state machines) for each of these types.
The cTAKES normalizer is a wrapper around a component of the SPECIALIST Lexical Tools37 called “norm,” which provides a representation for each word in the input text that is normalized with respect to a number of lexical properties, including ‘alphabetic case, inflection, spelling variants, punctuation, genitive markers, stop words, diacritics, symbols, and ligatures.’38 Normalization makes it possible to map multiple mentions of the same word that do not have the same string representations in the input data. We did not separately evaluate the off-the-shelf normalizer, which is used to improve the recall defined in equation (2) of the NER annotator. Each word in the text is normalized, and both normalized and non-normalized forms are used by the dictionary look-up described below.
The cTAKES POS tagger and shallow parser are wrappers around OpenNLP's modules for these tasks. We provide new supervised ME models trained on clinical data.
The cTAKES NER component implements a terminology-agnostic dictionary look-up algorithm within a noun-phrase look-up window. Through the dictionary look-up, each named entity is mapped to a concept from the terminology. We use a dictionary that is a subset of UMLS,39 version 2008AB, to include SNOMED CT40 and RxNORM41 concepts guided by extensive consultations with clinical researchers and practitioners. Each term in the dictionary belongs to one of the following semantic types as defined in42: disorders/diseases with a separate group for signs/symptoms, procedures, anatomy, and drugs, the latter includes terms from the Orange Book43 that have an RxNORM code. This dictionary was enriched with synonyms from UMLS and a Mayo-maintained list of terms. On the basis of the output from the shallow parser, the algorithm finds all noun phrases, which become the look-up window. The dictionary is interrogated for permutations of variations of the head and modifiers within the noun phrases to account for non-lexical variations. The NER component does not resolve ambiguities that result from identifying multiple terms in the same text span.
The negation annotator implements the NegEx algorithm,44 which is a pattern-based approach for finding words and phrases indicating negation near named entity mentions. The status annotator uses a similar approach for finding relevant words and phrases that indicate the status of a named entity.
Each discovered named entity belongs to one of the dictionary semantic types and has attributes for (1) the text span associated with the named entity (‘span’ attribute), (2) the terminology/ontology code the named entity maps to (‘concept’ attribute), (3) whether the named entity is negated (‘negation’ attribute), and (4) the status associated with the named entity with a value of current, history of, family history of, possible (‘status’ attribute). These semantic types and their attributes were selected in consultation with clinical researchers and practitioners and supported by an analysis of clinical questions and retrieval requests where the most frequent UMLS types and groups were disorders, clinical drug, sign and symptom, and procedures. Any future event is considered hypothetical; hence, the status value will be set to ‘possible’. Allergies to a given medication are handled by setting the negation attribute of that medication to ‘is negated’. Non-patient experiences are flagged as ‘family history of’ if applicable.
The cTAKES is distributed with the best-performing modules and machine learning models for the results reported below. It is released open-source under an Apache License, Version 2.0, as part of the Open Health Natural Language Processing45 Consortium. The cTAKES runs on Apache Unstructured Information Management Architecture (UIMA)46 47 and Java 1.5. It has been tested for scalability in a cloud computing environment.48
Corpus
Absent shared community annotated resources for the clinical domain,49 we built our own gold standard datasets for named entity39 and linguistic annotations as described in the Gold Standard Corpus and Inter-annotator Agreement subsection. Both datasets are created on content derived from the Mayo Clinic EMR. In addition, we used the gold standard linguistic annotations of the Penn TreeBank (PTB)50 and GENIA34 corpora.
To measure the quality of the in-house developed annotations, we report inter-annotator agreement (IAA) as the positive specific agreement (PSA)51 and κ52 53 defined as:
| (1) |
where P(a) is the relative observed agreement among the annotators and P(e) is the hypothetical probability of chance agreement. P(e) is computed from a contingency matrix representing agreements and disagreements. P(e) is the sum of the proportion of annotator A1 assigned tags to the positive row over all assignments and the proportion of A2 assigned tags to the positive column over all assignments, which is added to the proportion of annotator A1 assigned tags to the negative row and the proportion of annotator A2 assigned tags to the negative column.53 P(e) for the shallow parse task follows.52
Evaluation metrics
We used standard metrics:
| (2) |
| (3) |
| (4) |
| (5) |
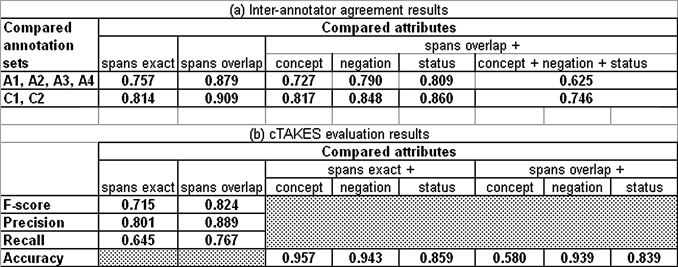
Because the NER module allows multiple annotations per span (exact and overlapping), we also computed precision, recall, and F-score for the partial matches, which we report as ‘spans overlap’ in table 1B. This measure has been referred to as soft F1-score in.54
Table 1.
(A) Inter-annotator agreement (IAA) as positive specific agreement on all annotations in compared sets (A1, A2, A3, A4 are the annotators; C1 is the gold standard created by A1 and A2; C2 is the gold standard created by A3 and A4). The overall agreement between C1 and C2 when the match criteria require that the spans overlap and the concept, negation, and status are the same is 0.746. (B) F-score and accuracy for system output compared to disorder mention gold standard. Results for attributes should be read ‘given that the spans are exact, the accuracy for attribute X is Y’ or ‘given that the spans overlap, the accuracy for attribute X is Y.’
|
The two OpenNLP parameters for number of iterations (the number of times the training procedure should iterate when finding the model's parameters) and a cut-off (the number of times a feature must have been seen in order to be considered in the model) are selected by a 10-fold cross validation. The results for the cTAKES ML components are reported on the basis of a 10-fold cross-evaluation by averaging the results from each fold. The tests for statistical significance use a t test for paired-two sample means with no-difference null hypothesis.
Status and report
Gold standard corpus and inter-annotator agreement
Two types of gold standard annotations were created. The linguistic annotations were used to train and evaluate cTAKES machine learning models. The named entity gold standard annotations were used to evaluate the NER component.
Linguistic gold standard
In addition to the widely tested PTB and GENIA corpora, our linguistic gold standard included annotations generated on a clinical corpus sampled from the Mayo Clinic EMR. The Mayo-derived linguistically annotated corpus (Mayo) was developed in-house and consisted of 273 clinical notes (100 650 tokens; 7299 sentences; 61 consult; 1 discharge summary; 4 educational visit; 4 general medical examination; 48 limited exam; 19 multi-system evaluation; 43 miscellaneous; 1 preoperative medical evaluation; 3 report; 3 specialty evaluation; 5 dismissal summary; 73 subsequent visit; 5 therapy; 3 test-oriented miscellaneous).
For the POS tags and shallow parses, we extended the PTB annotation guidelines55 56 to the clinical domain, with specifics for the annotations of numbering, roman numerals, drug names, abbreviations, drug-related attributes, and symbols. Guidelines for annotating the shallow parses (or chunks) included specifics for the annotations of medications, key clinical phrases, negation, and shared modifiers. Examples are in tables A1 and A2 of the online data supplement at http://jamia.bmj.com.
Three linguistic experts performed the annotation task on the Mayo corpus. The corpus was split into 10 sets of roughly the same size. One set was additionally split into three subsets: (1) for developing the guidelines (150 sentences), (2) for training (400 sentences), and (3) for measuring IAA (200 sentences). The IAA is reported on the jointly annotated third subset consisting of 2416 tokens and 1535 chunks (about 2.5% of the entire corpus).
Named entity gold standard
The clinical named entity corpus consists of 160 notes (47 975 tokens) randomly selected from the Mayo Clinic EMR. Altogether, 1466 gold standard Disorder UMLS Semantic group named entity annotations were discovered in it (Ogren et al39; for IAA summary see table 1A). The most important datum that best represents the overall consistency of the gold standard on the entire annotation task is the agreement between C1 and C2 (0.746) for the match criteria that require the spans to overlap and the concept code, negation, and status values to match. These Disorder gold standard named entity annotations are used to evaluate the NER component.
Inter-annotator agreement
The three linguistic annotators (LA) are represented as LA1, LA2, and LA3. The POS tags IAA is excellent, with the same result of 0.993 for positive specific agreement and κ because P(e) is extremely small due to the large number of possible values (details in table A3 of the online data supplement at http://jamia.bmj.com). The main source of disagreement is associated with RP (particle) and RB (adverb) part-of-speech tags (eg, ‘She needs to lie down,’ where the bolded word was assigned an RB tag by LA1 and RP tag by LA2). Another source of disagreement is the VBG (verb, gerund) and NN (noun, singular or mass) pairing (eg, ‘Her husband is disturbed by her sleeping’).
The shallow parses IAA is very strong (κ > 0.8) (details in tables A4 and A5 of the online data supplement at http://jamia.bmj.com). The phrasal category with the lowest agreement is PRT (particle phrase) (IAA range is 0.500–0.727). For example, in the sentence: ‘She is up for short periods’, LA1 annotated ‘up’ as a PRT, while LA2 as an adverbial phrase (ADVP). This ambiguity affects the annotations of the ADVP category, which has the second lowest agreement (IAA range is 0.841–0.899). The disagreements for the adjectival phrase (ADJP) category were mainly in spans that included a modifier or a post-modification (eg, ‘Bowel movements otherwise regular’—one annotator chose the span for ‘otherwise regular’ while the other selected ‘regular’ as the ADJP).
The IAA results for the named entity corpus are summarized in table 1A with details described by Ogren et al.39
Sentence boundary detector
Experiments of various corpus combinations were conducted (table 2A). We experimented with iteration values ranging from 100–350 and cut-off values from 1–7. The values that resulted in the maximal accuracies in table 2A are 350 iterations and a cut-off of 6 for GENIA, 350 iterations and a cut-off of 4 for PTB, 100 iterations and a cut-off of 1 for Mayo, and 100 and 4 for the combined data.
Table 2.
Accuracy results from the OpenNLP ME classifier. Rows represent training data; columns are test data. 10-fold cross validation with 80/20 data split for each fold
| GENIA | PTB | Mayo | |
| (a) Accuracy for the openNLP sentence boundary detector | |||
| GENIA | 0.986 | 0.646 | 0.821 |
| PTB | 0.967 | 0.944 | 0.940 |
| Mayo | 0.959 | 0.652 | 0.947 |
| GENIA+PTB+Mayo | 0.986 | 0.942 | 0.949 |
| (b) Accuracy results for the openNLP POS TAGGER | |||
| GENIA | 0.986 | 0.764 | 0.804 |
| PTB | 0.851 | 0.969 | 0.878 |
| Mayo | 0.812 | 0.844 | 0.940 |
| GENIA+PTB+Mayo | 0.984 | 0.969 | 0.936 |
ME, maximum entropy; POS, part-of-speech.
There are several major sources of errors: (1) sentences starting with titles, especially ‘Dr’ and ‘Mr’; (2) sentences starting with abbreviations (eg, ‘US’) and month names; (3) numbers and numbered lists; and (4) initials of person names. The first case constitutes roughly 73% of all the false negatives in the PTB corpus and nearly 30% in the Mayo corpus, but it is not present in GENIA because it consists of biomedical articles. The second, third, and fourth cases are frequent in the Mayo corpus because the clinical reports contain more shorthands that are not consistently followed by periods, which creates a challenge for the machine learning models. The clinical data also includes more unconventional abbreviations and short sentences (1–3 words), which offers an explanation for the low cut-off value.
Our results are similar to those reported by Buyko et al.33
Tokenizer
The rule-based tokenizer achieves an accuracy of 0.938 without the context-dependent tokenizer (CDT) and 0.949 with it. The remaining errors are generally related to punctuation (the tokens ‘140–150/60–85’ and ‘S/P’ are tokenized into three tokens) or due to lack of relevant rules (the tokens ‘p.o.’ and ‘a.m.’ are tokenized into four tokens). A baseline space-delimited tokenizer achieves an accuracy of 0.716.
Part-of-speech tagger
We determined an optimal cut-off of 4 with an iteration cycle of 100. Using these values, different combinations of training and testing corpora were used to build and evaluate the models (table 2B). All results were obtained by using gold standard tokens.
The accuracy of 0.986 on GENIA is comparable to the score of 0.989 on the same corpus reported by Buyko et al.33 The best results for each training corpus were obtained when the model was tested on the same corpus. One could consider this result the ceiling of the tagger performance: 0.969 for PTB, 0.986 for GENIA, and 0.940 for Mayo dataset. This supports results previously reported in the literature33 57 58 and the explanation that each corpus has its specific sublanguage. The differences between the accuracy on PTB+GENIA+Mayo and ceiling accuracy are statistically significant (p<0.01). A conclusion could be drawn that the POS model built using data from PTB, GENIA, and Mayo dataset could be successfully ported across these three domains. We conclude that the optimal parameters for a POS tagger using the OpenNLP technology are a corpus of combined data, a frequency cut-off of 4, and iterations of 100. The per-tag results are high for the adjectival tags (JJ) (accuracy=0.887), noun tags (NN) (accuracy=0.938), and verb tags (VB) (accuracy=0.925), which are prerequisites for a high-performance shallow parser. Error analysis shows that the most frequent failure sources are incorrect NN (5%) or VBN (2%) tags for JJ, incorrect JJ tags (2.5%) for NN and incorrect NN (4.6%), and NNP (1%) tags for VB (details in table A6 of the online data supplement at http://jamia.bmj.com).
Shallow parser
The OpenNLP ME model is trained on IOB-formatted data, where B indicates the beginning of the phrase, I the elements within the phrase, and O the elements outside of the phrase. We determined an optimal cut-off of 4 with an iteration cycle of 100, which were used for training the models represented in table 3. The accuracy and F-score were computed using the methodology and evaluation script from the Conference on Computational Natural Language Learning (CoNLL) 2000 shared task. All results were obtained by using gold standard POS tags.
Table 3.
Accuracy and F-score results for the OpenNLP shallow parser. Rows represent training data; columns are test data. 10-fold cross validation with 80/20 data split for each fold
| Accuracy and F-score (from CoNLL script) | PTB | GENIA | Mayo |
| PTB | 0.969/0.954 | 0.917/0.884 | 0.879/0.846 |
| GENIA | 0.888/0.826 | 0.956/0.935 | 0.839/0.768 |
| Mayo | 0.880/0.834 | 0.901/0.858 | 0.945/0.912 |
| PTB+ GENIA+Mayo | 0.969/0.953 | 0.953/0.932 | 0.952/0.924 |
The F-score of 0.93462 on GENIA is comparable to the score of 0.9360 on the same corpus reported by Buyko et al33—a state-of-the art figure compared to the performance figures from CoNLL 2000. The best performance on the Mayo dataset is achieved through training on the combined corpus, with an overall F-score of 0.924. However, domain-specific data is critical to achieving best performance. As expected, corpus size is critical to explain the result from training Mayo/testing Mayo experiment. There is significant difference between the results for training PTB/testing PTB and training PTB+GENIA+Mayo/testing PTB, and training Mayo/testing Mayo and training PTB+GENIA+Mayo/testing Mayo (p<0.0001). Comparing the results from training GENIA/testing GENIA and training PTB+GENIA+Mayo/testing GENIA yields a p value of 0.04, which suggests that GENIA has distinct patterns from those in PTB and Mayo.
The categories essential for deep parsing (NP, PP, VP) and NER (NP and PP) have high recognition rates (0.908, 0.954 and 0.956; details in table A7 of the online data supplement at http://jamia.bmj.com).
Error analysis shows that about 7% of the NP errors, 2.6% of the PP errors, and 2.3% of the VP errors are due to incorrect B, I, and O assignments.
Named entity recognition (NER)
We reported preliminary results in the paper by Kipper-Schuler et al59 and table 1B summarizes our most recent evaluation. These results represent an overall cTAKES system evaluation, as the NER module produces the annotations of greatest interest to applications that use cTAKES and because it relies on the output of all the other components. The NER performance is reported in terms of F-score, while the assignment of attribute values for each discovered named entity in terms of accuracy because each attribute is assigned a label. The best results achieve an F-score of 0.715 for exact matches and 0.824 for overlapping matches.
Consistent with our preliminary results,59 mapping to the UMLS concept unique identifier (CUI) accuracy is high for exact span matches. However, it drops when span matching is relaxed to require only that they overlap. This follows from the nature of dictionary-based approaches in which the span identified in the text is used to look up the concept. If the span identified is different than what is in the gold standard, it is no surprise that the phrases map to different terms in the dictionary. For the gold standard annotation of ‘degenerative joint disease’ with CUI C0029408, the algorithm produces the right-boundary match ‘joint disease’ with CUI C0022408 as well as the exact match ‘degenerative joint disease’ with CUI C0029408. As the boundary match is a broader term than the manual annotation, it is considered a CUI non-match. Negation detection achieves an accuracy of around 0.94 for both exact and overlapping spans. Status annotation has an accuracy of 0.859, which is comparable to the IAA of 0.860. We categorize the error sources as algorithmic errors, dictionary problems, and conceptual problems.59
Discussion
Remaining challenges
Although fast, our permutational dictionary look-up NER approach requires maintaining a lexically variant-rich dictionary and fails at recognizing complex levels of synonymy. In our paper, Li et al,60 we investigated an alternative ML approach through conditional random fields (CRFs)61 62 and support vector machines63 that showed that CRFs with multiple features outperform a single feature of dictionary look-up (of note, the ML NER module is not part of the current cTAKES release). Alternatively, a linguistic approach could be considered in which variants of the NP candidate are generated, mapped to a dictionary, and results ranked to produce the final mapping.16 These approaches are a trade-off between rich dictionary inclusion and computational cost. Although we report positive specific agreement values (identical to F-scores51) of 0.398 and 0.457 for exact and overlapping spans with the default MetaMap parameters in our paper, 39 these results should not be considered as directly comparable to our cTAKES NER component as we did not attempt to optimize MetaMap parameters. We plan to conduct a detailed comparative study of a variety of approaches to eventually combine their strengths.
One of the most frequent error sources in the NER component is the selection of one unique meaning to an named entity, which potentially maps to several concepts.64 We investigated supervised ML Word Sense Disambiguation and concluded that it is a combination of features unique for each ambiguity that generates the best results65 suggesting potential scalability issues. The cTAKES currently does not resolve ambiguities. Another cTAKES limitation lies in coordination structure interpretations—for example, the phrase ‘bladder and bowel habits’ should be parsed into two concepts (‘bladder habits’ and ‘bowel habits’), which cTAKES currently processes incorrectly as ‘bladder and bowel habits’ and ‘bowel habits’.
The cTAKES named entity attributes are similar to MedLEE's. Unlike MedLEE, the current cTAKES release does not include a module for asserting the relation between a disease/disorder, sign/symptom or procedure, and an anatomical site (MedLEE's BodyLoc modifier). We view this as a post-NER relation assertion task, which we will address in the future in the broader context of UMLS relation discovery from the clinical narrative. Another future challenge is expanding the values of the status named entity attribute with levels of granularity to express uncertainty and relatedness to patient to reflect non-patient experiences other than ‘family history of’.
Global evaluation and applications
We have conducted global evaluations of cTAKES for two large-scale phenotype-extraction studies: (1) ascertaining cardiovascular risk factors for a case-control study of peripheral arterial disease using the EMR within the eMERGE66 and (2) treatment classification for a pharmacogenomics breast cancer treatment study within the PGRN.67 Agreement results are in the low 90s when compared to an expert-abstracted gold standard, which we describe in separately submitted, under-review manuscripts. We present a cTAKES application and extension to the discovery of disease progression from free-text neuroradiology notes. 68 We are conducting a global system evaluation of cTAKES output against a manually abstracted gold standard for patient cohort identifications for 25 clinical research studies, which will be described in another paper.
We extended cTAKES to participate in the first i2b2 NLP challenge for the task of identifying the document-level patient's smoking status69 and have extended it further to patient-level summarization.70 Some of the limitations are a finer-grained certainty detection and temporal resolution. An independent evaluation of cTAKES constituted our entry in the second i2b2 NLP challenge71 highlighting cTAKES portability to data from other institutions. For a summary of the performance of the systems that participated in the first and second i2b2 challenges see Uzuner et al72 and Uzuner et al.73
Current and future developments
We are actively working on cTAKES modules for coreference,74 temporal relation discovery,75 and certainty assertion, to be implemented as downstream cTAKES components following NER. These components are expected to contribute to a refined set of named entity attributes. The cTAKES is being integrated within the Ontologies Development and Information Extraction tool.76 We recently ported the HITEx sectionizer into cTAKES, which highlights cTAKES's modularity. In collaboration with University of Colorado investigators, we are extending cTAKES for semantic processing. The cTAKES will be further enhanced for data normalization as part of a Strategic Health IT Advanced Research Project (SHARP) focusing on secondary use of the EMR. By making cTAKES available open-source, we present the community with the opportunity to collaboratively develop the next generation clinical NLP systems for large-scale intelligent information extraction from the clinical narrative.
Supplementary Material
Acknowledgments
We would like to acknowledge our annotators, Barbara Abbott, Debra Albrecht, Pauline Funk, and Donna Ihrke, for their excellent work; Dingcheng Li's work towards the linguistic annotations; Serguei Pakhomov for conceptualization; Patrick Duffy for brilliant programming; and Jim Buntrock for being an integral part of this project. The work was partially supported by a 2007 IBM UIMA grant. We would like to thank our alpha testers, Dr Leonard D'Avolio, VA Boston, and Dr Cheryl Clark, MITRE Corporation, who endured the pains of the initial code. Our appreciation goes to the three anonymous JAMIA reviewers whose detailed comments greatly improved the manuscript.
Footnotes
Funding: The work was partially supported by a 2007 IBM UIMA grant.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Hornberger J. Electronic health records: a guide for clinicians and administrators. Book and media review. JAMA 2009;(301):110 [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. IMIA Year book of Medical Informatics 2008;47(Suppl 1):128–44 [PubMed] [Google Scholar]
- 3.Friedman C. Towards a comprehensive medical language processing system: methods and issues. Proc AMIA Annu Fall Symp 1997:595–9 [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000:270–4 [PMC free article] [PubMed] [Google Scholar]
- 5.Hripcsak G, Kuperman G, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med 1998;37:1–7 [PubMed] [Google Scholar]
- 6.Health Information Text Extraction (HITEx) https://www.i2b2.org/software/projects/hitex/hitex_manual.html
- 7.Zeng Q, Goryachev S, Weiss S, et al. Extracting principal diagnosis, comorbidity, and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.HiTEXT. https://www.i2b2.org/
- 9.Mack R, Mukherjea S, Soffer A, et al. Text analytics for life science using the unstructured information management architecture. IBM Syst J 2004;43:490–515 [Google Scholar]
- 10.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a cancer disease knowledge model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 11.Haug P, Koehler S, Lau L, et al. Experience with a mixed semantic/syntactic parser. Ann Symp Comp Appl Med Care 1995:284–8 [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen L, Haug P, Fiszman M. MPLUS: a probabilistic medical language understanding system. Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain - Volume 3. 2002:29–36 Philadelphia, PA [Google Scholar]
- 13.Fiszman M, Haug P, Frederick P. Automatic extraction of PIOPED interpretations from ventilation/perfusion lung scan reports. Proc AMIA symp 1998:860–4 [PMC free article] [PubMed] [Google Scholar]
- 14.Fiszman M, Chapman W, Aronsky D, et al. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 2000:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trick W, Chapman W, Wisniewski M, et al. Electronic interpretation of chest radiograph reports to detect central venous catheters. Infect Control Hosp Epidemiol 2003;24:950–4 [DOI] [PubMed] [Google Scholar]
- 16.Aronson A, Bodenreider O, Chang H, et al. The NLM indexing initiative. Proc AMIA symp 2000:17–21 [PMC free article] [PubMed] [Google Scholar]
- 17.Unified Medical Language System (UMLS). http://www.nlm.nih.gov/research/umls/
- 18.UIMA MetaMap wrapper. http://sourceforge.net/projects/metamap-uima/
- 19.National Center for Text Mining (NaCTeM). http://www.nactem.ac.uk/index.php
- 20.JULIE Lab. http://www.julielab.de
- 21.U-compare. http://u-compare.org/index.html
- 22.Meystre S, Haug P. Evaluation of medical problem extraction from electronic clinical documents using MetaMap transfers (MMTx), in Connecting Medical Informatics and Bio-Informatics. In. Proceedings of MIE2005-The XIXth International Congress of the European Federation for Medical Informatics. IOS Press, 2005:823–8 [PubMed] [Google Scholar]
- 23.Cancer biomedical informatics grid (caBIG). https://cabig.nci.nih.gov/
- 24.caTIES. https://cabig.nci.nih.gov/tools/caties
- 25.Crowley R, Castine M, Mitchell K, et al. caTIES - a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc 2010;17:253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCI Enterprise Vocabulary System (EVS). http://ncicb.nci.nih.gov/NCICB/infrastructure/cacore_overview/vocabulary
- 27.OpenNLP: http://opennlp.sourceforge.net/index.html
- 28.Berger A, Della Pietra S, Della Pietra V. A maximum entropy approach to natural language processing. Computational Linguistics 1996;22:39–71 [Google Scholar]
- 29.Rosenfeld R. A maximum entropy approach to adaptive statistical language modeling. Computer, Speech and Language 1996;10:187–228 [Google Scholar]
- 30.Manning C, Schutze H. Foundations of statistical natural language processing. Cambridge, Mass: MIT Press, 1999 [Google Scholar]
- 31.Sha R, Pereira F. Shallow parsing with conditional random fields. NLT-NAACL 2003:134–41 [Google Scholar]
- 32.Ratnaparkhi A. Maximum entropy models for natural language ambiguity resolution. PhD thesis, University of Pennsylvania, 1998 [Google Scholar]
- 33.Buyko E, Wermter J, Poprat M, et al. Automatically adapting an NLP core engine to the biology domain. ICBM 2006:65–68 [Google Scholar]
- 34.GENIA. 2009. http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/home/wiki.cgi?page=GENIA+Project
- 35.PennBioIE. http://www.ldc.upenn.edu/Catalog/CatalogEntry.jsp?catalogId=LDC2008T20
- 36.Clinical Document Architecture (CDA) http://hl7book.net/index.php?title=CDA
- 37.LVG. http://www.SPECIALIST.nlm.nih.gov
- 38.LVG user guide. http://www.lexsrv3.nlm.nih.gov/SPECIALIST/Projects/lvg/current/docs/userDoc/index.html
- 39.Ogren P, Savova G, Chute C. Constructing evaluation corpora for automated clinical named entity recognition. Language Resources and Evaluation Conference, Proc LREC 2008:3143–50. http://www.lrec-conf.org/proceedings/lrec2008/ Marrakesh, Morocco.
- 40.Snomed CT. http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html
- 41.RxNORM. http://www.nlm.nih.gov/research/umls/rxnorm/
- 42.Bodenreider O, McCray A. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Electronic Orange Book. http://www.fda.gov/drugs/informationondrugs/ucm129689.htm
- 44.Chapman W, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 45.Open Health Natural Language Processing consortium (OHNLP) http://www.ohnlp.org
- 46.UIMA. http://incubator.apache.org/uima/
- 47.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng 2004;10(3:4):327–48 [Google Scholar]
- 48.Baldwin D, Carrell D. Rapidly deployable, highly scalable natural language processing using cloud computing and an open source NLP pipeline. https://www.cabig-kc.nci.nih.gov/Vocab/uploaded_files/7/73/Rapid_Scalable_NLP_And_Cloud_Computing.doc (accessed 2010).
- 49.Cohen K, Fox L, Ogren P, et al. Empirical data on corpus design and usage in biomedical natural language processing. Proc AMIA Symp 2005:156–60 [PMC free article] [PubMed] [Google Scholar]
- 50.Marcus M, Santorini B, Marcinkiewicz M. Building a large annotated corpus of English: the Penn treebank. Computational Linguistics 1994;19:313–30 [Google Scholar]
- 51.Hripcsak G, Rothschild A. Agreement, the F-Measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poesio M, Vieira R. A corpus-based investigation of definite description use. Computational Linguistics 1998;24:183–216 [Google Scholar]
- 53.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20:37–46 [Google Scholar]
- 54.Settles B. ABNER: an open source tool for automatically tagging genes, proteins, and other entity names in text. Bioinformatics 2005;21:3191–2 [DOI] [PubMed] [Google Scholar]
- 55.Santorini B. Part-of-speech tagging guidelines for the Penn Treebank Project, Technical report number MS-CIS-90–47. Department of Computer and Information Science. University of Pennsylvania, 1990 [Google Scholar]
- 56.Bies A, Ferguson M, Katz K, et al. Bracketing guidelines for Treebank II Style Penn Treebank project. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.31.9795 Accessed 24 July 2010.
- 57.Coden A, Pakhomov S, Ando R, et al. Domain-specific language models and lexicons for tagging. J Biomed Inform 2005;38:422–30 [DOI] [PubMed] [Google Scholar]
- 58.Liu K, Chapman W, Hwa R, et al. Heuristic sample selection to minimize reference standard training set for a part-of-speech tagger. J Amer Med Inform Assoc 2007;14:641–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kipper-Schuler K, Kaggal V, Masanz J, et al. System evaluation on a named entity corpus from clinical notes. Language Resources and Evaluation Conference, LREC 2008:3001–3007. http://www.lrec-conf.org/proceedings/lrec2008/ Marrakesh, Morocco.
- 60.Li D, Schuler K, Savova G. A comparison between CRFs and SVMs in Disorder Named Entity Recognition in Clinic Texts. Intelligent Data Analysis in Biomedicine and Pharmacology (IDAMAP), 2008. A colloquium in conjunction with the American Medical Informatics Association Annual Symposium 2008 in Washington, DC, USA: 57–62. [Google Scholar]
- 61.Lafferty A, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. In 8th international conference on machine learning (ICML-2001). 2001:282–9
- 62.McCallum A, Li W. Early results for name entity recognition with conditional random fields, feature induction and web-enhanced Lexicons. ACM Trans Comput Log 2003:188–191 [Google Scholar]
- 63.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;10:273–97 [Google Scholar]
- 64.Schuemie M, Kors J, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol 2005;12:554–65 [DOI] [PubMed] [Google Scholar]
- 65.Savova G, Coden A, Sominsky I, et al. Word sense disambiguation across two domains: biomedical literature and clinical notes. J Biomed Inform 2008;41:1088–100 [DOI] [PubMed] [Google Scholar]
- 66.eMERGE. https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page
- 67.PGRN. http://www.nigms.nih.gov/Initiatives/PGRN
- 68.Cheng L, Zheng J, Savova G, et al. Discerning tumor status from unstructured MRI reports—completeness of information in existing reports and utility of natural language processing. J Dig Imag Soc Imag Inform Med 2009;23(2):119–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savova G, Ogren P, Duffy P, et al. Mayo clinic system for patient smoking status classification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sohn S, Savova G. Mayo clinic smoking status classification system. AMIA Annu Symp Proc 2009. San Francisco, CA. 619–23 [PMC free article] [PubMed]
- 71.Savova G, Clark C, Zheng J, et al. The Mayo/MITRE system for discovery of obesity and its comorbidities. In. The Second i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Washington, DC, 2008
- 72.Uzuner Ö, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savova G, Chapman W, Zheng J, et al. Annotation schema for anaphoric relations in the clinical domain. AMIA Annu Symp Proc;2009. San Francisco, CA. [Google Scholar]
- 75.Savova G, Bethard S, Styler W, et al. Towards temporal relation discovery from the clinical narrative. AMIA Annu Symp Proc;2009. San Francisco, CA. 568–72 [PMC free article] [PubMed] [Google Scholar]
- 76.ODIE Ontology development and information extraction (ODIE) toolset. http://www.bioontology.org/ODIE
- 77.Health insurance portability and accountability act (HIPAA). http://www.hhs.gov/ocr/privacy/index.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.