Abstract
Objective
To develop an automated system to extract medications and related information from discharge summaries as part of the 2009 i2b2 natural language processing (NLP) challenge. This task required accurate recognition of medication name, dosage, mode, frequency, duration, and reason for drug administration.
Design
We developed an integrated system using several existing NLP components developed at Vanderbilt University Medical Center, which included MedEx (to extract medication information), SecTag (a section identification system for clinical notes), a sentence splitter, and a spell checker for drug names. Our goal was to achieve good performance with minimal to no specific training for this document corpus; thus, evaluating the portability of those NLP tools beyond their home institution. The integrated system was developed using 17 notes that were annotated by the organizers and evaluated using 251 notes that were annotated by participating teams.
Measurements
The i2b2 challenge used standard measures, including precision, recall, and F-measure, to evaluate the performance of participating systems. There were two ways to determine whether an extracted textual finding is correct or not: exact matching or inexact matching. The overall performance for all six types of medication-related findings across 251 annotated notes was considered as the primary metric in the challenge.
Results
Our system achieved an overall F-measure of 0.821 for exact matching (0.839 precision; 0.803 recall) and 0.822 for inexact matching (0.866 precision; 0.782 recall). The system ranked second out of 20 participating teams on overall performance at extracting medications and related information.
Conclusions
The results show that the existing MedEx system, together with other NLP components, can extract medication information in clinical text from institutions other than the site of algorithm development with reasonable performance.
Introduction
Medication information is an important type of clinical data in electronic medical record (EMR) systems. Obtaining an accurate medication profile of a patient is a common and critical task for clinical research (eg, to investigate drug toxicity and efficacy) and clinical operations (eg, medication reconciliation, the process for creating a complete and accurate list of a patient's medications at each transition point of care). As large amounts of medication data are stored as free-text in clinical notes, there is a need to develop automated methods to extract structured medication information from clinical narratives.
A number of studies have focused on extracting medication information from clinical notes. Some studies have focused on extracting and encoding drug names.1–3 A recent study by Gold et al4 reported a regular expression-based approach for extracting drug names and signature information, such as dose, route, and frequency. Jagannathan et al assessed four commercial natural language processing (NLP) systems for their ability to extract medication information (including drug names, strength, route, and frequency) and they reported a high F-measure of 93.2% on capturing drug names, but lower F-measures of 85.3%, 80.3%, and 48.3% on retrieving strength, route, and frequency, respectively.5 At the Vanderbilt University Medical Center (VUMC), we have developed a medication extraction system called MedEx.6 The system achieved F-measures over 90% on extracting drug names, strength, route, and frequency information in discharge summaries and clinic visit notes from VUMC's EMR.
In this paper, we describe how we extended MedEx and integrated it with other existing NLP tools for the 2009 Informatics for Integrating Biology and the Bedside (i2b2) NLP challenge,7 which sought to extract medication-related information from discharge summaries.
Methods
System overview
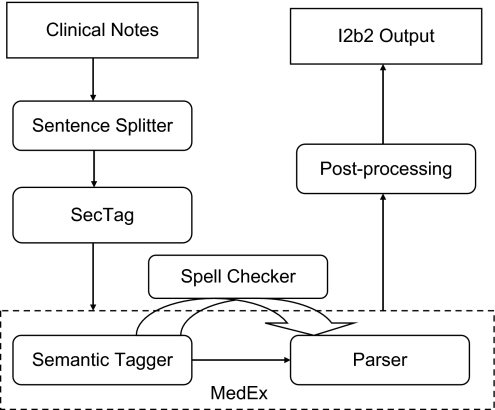
We built an integrated system that leveraged several existing NLP components developed at VUMC (Figure 1). The system consists of the following components: (1) a sentence boundary detection program, which was developed at VUMC and modified for the challenge; (2) an existing section identification program (SecTag); (3) an existing medication extraction system (MedEx); (4) a newly developed spell checker for drug names based on Aspell8; and (5) a newly developed post-processing program that maps MedEx outputs into the i2b2 output format.
Figure 1.
Components of the integrated medication extraction system.
Sentence splitter
We used a sentence as a basic unit for extracting medication information. Thus, it was necessary to determine sentence boundaries accurately. One challenge in the i2b2 documents was ambiguous newline characters, which could be introduced by the writer during document creation or as a text-formatting technique by the system, which automatically added newline characters after a line exceeds its length limit. A user-entered newline usually indicates the end of a sentence, but a system-introduced newline character typically does not. In this dataset, we found that the insertion of system-introduced newline characters varied greatly. Dictated notes tended to insert newline characters after 50–75 characters, but typed notes would include inserted newlines anywhere from about 30–75 characters based on whether text had been (apparently) auto-inserted from another system. We developed a rule-based sentence splitter to determine whether a newline character is the end of a sentence or not. It takes into account abbreviations containing periods, such as ‘b.i.d’, which will not be treated as the end of sentence markers.
Section identification
We used SecTag,9 a program developed using VUMC EMR data, to identify section headers in the i2b2 data set without modifying SecTag code or retraining its probabilities. We used outputs of SecTag for two tasks: (1) excluding false-positive medications by removing drug mentions occurring in the ‘allergy’, ‘lab’, or ‘study’ sections, and (2) determining ‘list or narrative’—if a drug is mentioned in a ‘medication‘ section, it was marked as ‘list’ otherwise it was marked as ‘narrative’. In addition, we developed a customized section tagger for the i2b2 clinical notes based on regular expressions to identify limited sections such as ‘allergy’. We submitted results from SecTag and the customized section tagger as different runs.
Extended MedEx
The MedEx6 creates structured medication-related outputs specified by 11 different semantic types (table A1 of the online supplementary material at http://jamia.bmj.com). It consists of two main components: (1) a semantic tagger that labels words or phrases with a semantic category and (2) a chart parser10 that uses a context-free grammar to parse textual sentences into structured forms based on pre-defined semantic patterns. Drug names in MedEx are derived from the RxNorm11 and the Unified Medical Language System (UMLS).12
Despite a similarity between the MedEx output and the i2b2 challenge goals, there were several important changes required to adapt the MedEx output to the i2b2 challenge. First, the output format of MedEx is different from the format that the i2b2 challenge required, because MedEx uses a different representation model of medication information. For example, ‘fluocinonide 0.5% cream’ is marked as one drug name in i2b2 annotation, but it will be marked as three parts in MedEx: ‘fluocinonide—DrugName’, ‘0.5%—Strength’, and ‘cream—Form’. Second, MedEx does not report the offsets of drug-related findings. Third, some types of information required by i2b2 challenge, such as ‘list/narrative’ and ‘reason’, are not captured by MedEx.
To minimize the changes to MedEx, we developed a post-processing program to map MedEx outputs into i2b2′s formats, as well as to extract additional information such as reasons. Major extensions to MedEx included: (1) a function to locate the offsets of extracted drug findings; and (2) adding additional lexicons such as a list of biological substances and drug classes extracted from UMLS to the MedEx lexicon. We did not modify the core MedEx code for this challenge.
Spell checker
We implemented a spell checker for drug names based on the Aspell algorithm.8 We created a lexicon for Aspell based on all single words found in the UMLS Metathesaurus and all words in the drug lexicon file. We automatically ‘corrected’ any word identified as misspelled by Aspell if the most likely single-word correction was a medication name. The spell checker was placed after the semantic tagger in MedEx.
Post-processing
The post-processing program converts MedEx's outputs into i2b2′s outputs using a set of heuristic rules. For example, medication names were determined by combining semantic types of DrugName, Strength, and Form in MedEx. One of the rules for medication names is ‘If DrugName followed by Strength then Medication Name = DrugName + Strength’. More details of the post-processing rules can be found in the online appendix (available at http://jamia.bmj.com). Negated medications were identified in a similar way as the NegEx algorithm.13
Evaluations and results
Evaluations
The data sets, the annotation guideline,14 and the evaluation metrics for the 2009 i2b2 challenge are described in detail in.7 Standard precision, recall, and F-measure are calculated vertically or horizontally at the patient and system levels as described in Uzuner et al.7
Results
The best results of our system on the ground truth containing 251 notes are shown in table 1. The system achieved an F-measure of 0.821 (exact) and 0.822 (inexact) at the system level and 0.810 (exact) versus 0.807 (inexact) at the patient level. This was the second best score among the 20 participating teams. Medication names, dosage, mode, and frequencies achieved higher F-measures (most above 0.85), but reason and duration were very poor with F-measures below 0.4. Our system had an F-measure of 0.588 on ‘narrative’ and an F-measure of 0.855 on ‘list’ status, which were ranked as the third place in list/narrative determination.
Table 1.
Evaluation results of Vanderbilt's system for 2009 i2b2 challenge
| Exact | Inexact | |||||||
| F-measure | Pre | Rec | F-measure | Pre | Rec | |||
| Horizontal | System-level | System | 0.821 | 0.839 | 0.803 | 0.822 | 0.866 | 0.782 |
| Horizontal | Patient-level | System | 0.810 | 0.840 | 0.792 | 0.807 | 0.863 | 0.770 |
| Vertical | System-level | Dosage | 0.855 | 0.895 | 0.818 | 0.880 | 0.930 | 0.835 |
| Vertical | Patient-level | Dosage | 0.830 | 0.878 | 0.802 | 0.857 | 0.915 | 0.823 |
| Vertical | Ssystem-level | Frequency | 0.868 | 0.879 | 0.858 | 0.859 | 0.902 | 0.820 |
| Vertical | Patient-level | Frequency | 0.860 | 0.881 | 0.852 | 0.855 | 0.900 | 0.834 |
| Vertical | Ssystem-level | Mode | 0.887 | 0.918 | 0.858 | 0.882 | 0.926 | 0.841 |
| Vertical | Patient-level | Mode | 0.842 | 0.883 | 0.820 | 0.839 | 0.888 | 0.811 |
| Vertical | System-level | Medication | 0.856 | 0.842 | 0.871 | 0.893 | 0.895 | 0.891 |
| Vertical | Patient-level | Medication | 0.855 | 0.849 | 0.870 | 0.884 | 0.892 | 0.886 |
| Vertical | System-level | Reason | 0.360 | 0.459 | 0.296 | 0.367 | 0.517 | 0.285 |
| Vertical | Patient-level | Reason | 0.344 | 0.455 | 0.319 | 0.360 | 0.522 | 0.335 |
| Vertical | System-level | Duration | 0.361 | 0.364 | 0.358 | 0.405 | 0.458 | 0.364 |
| Vertical | Patient-level | Duration | 0.369 | 0.405 | 0.395 | 0.423 | 0.491 | 0.451 |
‘Exact’ and ‘inexact’ matching are two different ways to determine whether an extracted textual finding is correct or not.
Standard precision, Recall and F-measure were reported for each individual type such as medication names, dosage, and frequency (termed the ‘vertical’ analysis), as well as for all outputs regardless of types (termed the ‘horizontal’ analysis).
In addition, those measurements were also calculated at two different levels: patient and system levels.
The patient level calculated precision, recall, and F-measure for each note and reported the averages across all notes, while the system level calculated them based on all entries from all notes.
Discussions
In this study, we developed an integrated medication extraction system based on the existing MedEx system and other NLP components such as SecTag. We successfully applied this system to the 2009 i2b2 NLP challenge and achieved an F-measure of 0.821 on 251 annotated discharge summaries from Partners Healthcare, ranking second in the overall i2b2 challenge.
Though the i2b2 challenge required extraction of six types of drug-related findings, medication name is the most important since all other finding types are dependent on it. Our system did quite well on recognizing drug names (with F-measure 0.852 for exact matching and 0.893 for inexact matching). However, MedEx's performance was lower than previously reported performance on recognizing drug names,2 6 where F-measures were over 0.9. The main reason could be related to how drug names are defined in the annotation guideline. As we described above, MedEx recognizes drug names differently than required by the i2b2 challenge. The post-processing program combined individual name components recognized by MedEx into i2b2 drug name phrases, but it made occasional errors in which MedEx had correctly identified medication information according to its native output format. Failure to properly combine medicine name tokens was the primary reason inexact matching results (F-measure of 0.893) were higher than those from exact matching (0.856). We evaluated 100 randomly selected drug name errors to determine causes of false positive and false negative drug name assertions. Drug name false positives mainly resulted from incomplete drug names matching (54%); for example, the system identified ‘aspirin’ and ‘xalatan’ as drug names instead of ‘enteric-coated aspirin’ and ‘xalatan eye’, respectively, as per the challenge requirements. Another major type of false positives (35%) was caused by wrong drug names in our lexicon file, such as ‘ecg’, ‘salt’, ‘igg’. We also noticed that some diet names were falsely recognized as drug names, which could be easily removed based on identifying the ‘diet’ section using SecTag. Drug name false negatives were mainly due to missing lexicon entries (48%), such as abbreviated or uncorrected misspelled drug terms (eg, ‘czi’, ‘amio’, ‘zolof’), and incomplete drug name matching (36%) as described in the false positives above. Some errors were caused by incorrectly typed clinical text, such as ‘Norvasc.Consider’, ‘integ/hep’, where missing spaces caused errors in tokenization, and our spelling correction algorithm only consider single word replacements. The i2b2 challenge also asked to keep negative drug names if they indicated historical profile of patients; our system did not differentiate them from regular negated drug names. In the future, the drug lexicon derived from UMLS and RxNorm would be improved by removing unlikely drug names and adding new entries such as drug name abbreviations.
False positive errors in five types of medication modifiers can be summarized into three categories: (1) incorrectly recognized terms—for example, we labeled ‘tube’ in ‘chest tube discontinued’ as a ‘mode’ modifier; (2) incorrectly linked modifiers to corresponding medications; and (3) incompletely recognized terms—for example, we identified a duration phrase of ‘for 7 day’ but it should be ‘for 7 day course’. Errors of the latter type resulted in both false positives and false negatives per guidelines. Most false negatives were due to missing lexicon entries. For example, we do not have the term ‘pre-meal’ in our lexicon file as a frequency term. Such terms have since been added to the MedEx lexicon.
As MedEx was developed to extract prescription type of drug mentions in clinical text, it was not surprising that the extended system had good performance on extracting drug signature information, including dosage, mode, and frequency. However, contextual level information, including duration and reason, was more difficult to extract (F-measure <0.4). We found that these types of information were often loosely attached with medication mentions. Often, they were not contained within the same sentence. Furthermore, determining the boundary for duration and reason phrases was difficult. Based on the challenge guidelines, duration was a ‘noun phrase, prepositional phrases, or clauses’, and reason phrases were ‘the most informative adjective phrase or the longest base noun phrase’. Even human annotators had very low performance on determining duration and reason phrases (F-measures of 0.60–0.73 based on the statistics released by i2b2) compared to the final ground truth. Currently, our system only used simple rules to identify duration/reason phrases and link duration/reason to medications. More sophisticated methods, which use syntactic information of sentences or knowledge bases of drug-indication relations, may improve the performance of the system.
Sentence boundary detection is an important first step for our system. However, it is not straightforward to define sentence boundaries in clinical text as many sentences only contain short phrases. For specific tasks, such as medication extraction, an error in sentence detection may not affect the final results. An in-depth analysis of sentence detection is out of the scope of this study. But we did look into errors caused by the rule-based sentences splitter. We randomly picked up 10 notes and manually reviewed sentences generated by the sentence splitter. Among 1206 generated sentences, we found 10 instances (about 0.83%) that would cause the wrong interpretation of medication information. For example, the sentence ‘Number of doses required (approximate): 6’ was broken into two sentences: ‘Number of doses required (approximate):’ and ‘6’, which caused the duration information ‘6’ to be missed in our outputs.
In this study, we used two methods to identify sections in test data set: a customized section tagger developed specifically for this task and SecTag, a general-purpose section tagger originally developed for VUMC's history and physical (H&P) documents. The customized section tagger produced a slightly better F-measure (0.821 vs 0.819) than the unmodified SecTag at the system level; however, SecTag was slightly better at the patient level (0.812 vs 0.810). Of note, the SecTag system, which uses a combination of rule and probabilistic methods to identify sections and their start and end boundaries, was run using a VUMC H&P training set. Although this specific application represents a very limited evaluation, it does suggest that the SecTag system and vocabulary may be portable across different institutions and note types. More details of the effects of section taggers on the final performance can be found in table A3 of the online supplementary material at http://jamia.bmj.com.
To evaluate the effectiveness of the spell checker program, we investigated the list of possibly misspelled drug names that were ‘corrected’ by the spell checker. Among 79 cases from 251 notes, 63 of them were truly misspelled drug names, which indicated a precision of 80% for the spell checker. Some common misspelled drug names included names for ibuprofen (‘ibuprfen’), augmentin (‘qugmentin’), and insulin (‘inuslin’). Our results showed that the spell checker improved the F-measure very little—about 0.1%, that is, 0.821 versus 0.820 for exact matching and 0.822 versus 0.821 for inexact matching. However, the potential impact of spell correction on individual drugs could be significant, especially for those that are often misspelled.
The integrated system ranked second overall and it was the best rule-based system in the challenge. We think the high performance is mainly from the existing MedEx system, which uses generalizable semantic patterns for extracting medication findings. In addition, the post-processing program based on the annotation guideline customizes the outputs of MedEx and further improves its performance. Other existing tools such as SecTag and Spell Checker also contribute to the system.
Conclusion
We adapted an existing medication extraction system for the i2b2 medication extraction challenge, while minimizing changes to the core NLP systems used in this study. Our results showed that the MedEx system, when combined with other NLP components such as SecTag, can be used to extract medication information from clinical notes at a different institution with reasonable performance.
Supplementary Material
Acknowledgments
We would like to thank the i2b2 challenge team for the development of training and test corpora.
Footnotes
Funding: This study was partially supported by grants from the US NIH: NHGRI U01 HG004603, NLM R01-LM007995-05, and NCI R01CA141307-01. The project described was supported in part by the i2b2 initiative, Award Number U54LM008748 from the National Library of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Chhieng D, Day T, Gordon G, et al. Use of natural language programming to extract medication from unstructured electronic medical records. AMIA 2007:908. [PubMed] [Google Scholar]
- 2.Levin MA, Krol M, Doshi AM, et al. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA 2007:438–42 [PMC free article] [PubMed] [Google Scholar]
- 3.Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. Pac Symp Biocomput 2005:308–18 [DOI] [PubMed] [Google Scholar]
- 4.Gold S, Elhadad N, Zhu X, et al. Extracting structured medication event information from discharge summaries. AMIA 2008:237–41 [PMC free article] [PubMed] [Google Scholar]
- 5.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009;78:284–91 [DOI] [PubMed] [Google Scholar]
- 6.Xu H, Stenner S, Doan S, et al. MedEx—a medication information exaction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner Ö SI, Cadag E. Extracting medication information from clinical text. In current issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson K. GNU Aspell. 2003. http://www.aspell.net
- 9.Denny JC, Spickard A, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc 2009;16:806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay M. Algorithm schemata and data structures in syntactic processing. text processing: text analysis and generation, text typology and attribution 1982:327–58
- 11.RxNorm. http://www.nlm.nih.gov/research/umls/rxnorm/
- 12.UMLS. http://www.nlm.nih.gov/research/umls/
- 13.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 14.Uzuner P, Solti I, Xia F. I2b2 Medication challenge annotation guidelines, i2b2 challenge google groups. http://www.groups.google.com/group/i2b2-medication-extraction-nlp-challenge [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.