Abstract
Objective
While essential for patient care, information related to medication is often written as free text in clinical records and, therefore, difficult to use in computerized systems. This paper describes an approach to automatically extract medication information from clinical records, which was developed to participate in the i2b2 2009 challenge, as well as different strategies to improve the extraction.
Design
Our approach relies on a semantic lexicon and extraction rules as a two-phase strategy: first, drug names are recognized and, then, the context of these names is explored to extract drug-related information (mode, dosage, etc) according to rules capturing the document structure and the syntax of each kind of information. Different configurations are tested to improve this baseline system along several dimensions, particularly drug name recognition—this step being a determining factor to extract drug-related information. Changes were tested at the level of the lexicons and of the extraction rules.
Results
The initial system participating in i2b2 achieved good results (global F-measure of 77%). Further testing of different configurations substantially improved the system (global F-measure of 81%), performing well for all types of information (eg, 84% for drug names and 88% for modes), except for durations and reasons, which remain problematic.
Conclusion
This study demonstrates that a simple rule-based system can achieve good performance on the medication extraction task. We also showed that controlled modifications (lexicon filtering and rule refinement) were the improvements that best raised the performance.
Introduction
Patient records typically contain a vast amount of information; much of it written in free text. This makes it difficult for clinicians to rapidly access the whole content of a record. Medical data in textual form is also difficult to use in health information systems. Yet it can be useful in many contexts. Main.php-related information is one type of data physicians need to access. We took the opportunity of the 2009 i2b2 (Informatics for Integrating Biology and the Bedside) challenge on medication extractioni to develop a medication extraction system.
Natural language processing (NLP) methods1–17 have been used to extract a variety of information from clinical texts. Prior to the i2b2 challenge, few studies had investigated the extraction of medication-related information. We classify them in two categories according to the variety of information they aim to extract. From a technical point of view, most approaches are rule-based and make extensive use of lexicons.
One approach is to extract one specific piece of information, such as drug names11 12 or dosage.13 Other studies try to detect a wider scope of information. First, a model to represent medication information is defined to be filled with the extracted information (drug names associated with dosage, route, etc). Evans et al14 designed the first system and defined the following drug model: a drug name associated with dose level, route, frequency, and necessity. The system is rule-based and relies on lexicons and NLP processes (stemming, part-of speech (POS) tagging, etc.). More recently, Gold et al15 used the same drug model to build their lexicon and rule-based system Merki. Jagannathan et al16 combined several commercial engines to extract medications. Xu et al17 defined a more fine-grained representation model and developed a system based on lexicons, rules, and a semantic grammar.
Our work lies in the framework of the i2b2 2009 challenge on medication information extraction,18 which specifies the following medication model: drug names associated with dosage, mode of administration, frequency, duration, reason for prescription, and whether the medication was found in a list or in a narrative passage. This paper describes our system, and gives insight on which types of tactics are the most beneficial to medication extraction by detailing various strategies to improve the system after the challenge.
Material and methods
Corpus description
The corpora used to develop and test our system were provided within the context of the challenge and were composed of narrative patient records. The development corpus consists of 696 records (17 were annotated for training purposes). The test corpus includes 547 records; 251 of them annotated to be used as gold standard.
System overview
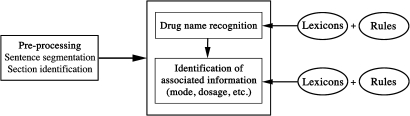
Our system performs medication extraction based on lexicons and rules (see figure 1 for an overview of the system).
Figure 1.
Overview of the system.
Lexicon compilation
Our system relies on rules that use semantic classes of words expressing the pieces of information to extract (drug names, modes, dosages, etc). Words in these classes are stored in a lexicon; since drug names and reasons are much more numerous, we list them in separate lexicons for convenience.
We built a drug lexicon from various sources: the Food and Drug Administrationii and RxListiii websites, and the Unified Medical Language System (UMLS) Metathesaurus, which we queried for terms with the following semantic types: clinical drug, pharmacologic substance, antibiotic, vitamin, and hormone. We acquired 183 941 drug names. Additionally, we compiled a list of 102 therapeutic classes from the WHO websiteiv.
We gathered a lexicon of reasons by querying the UMLS for terms with the Sign and Symptom semantic type and flagged as MetaMap NLP View (which identifies terms useful for NLP). We also manually added examples from the development corpus. This resulted in a list of 10 625 entries.
A smaller lexicon for other semantic classes was derived from the list of biomedical abbreviations compiled by Bermanv and was extended with examples from the development corpus. It contains 161 entries, each paired with its semantic class (see table 1).
Table 1.
Abbreviations and expressions for dosage, mode, frequency and duration (excerpt)
| Entry | Attribute |
| tablet | dosage |
| mg | dosage |
| po | mode |
| topical | mode |
| daily | frequency |
| tid | frequency |
| month | duration |
| wk | duration |
po, orally; tid, three times a day; wk, week
Rule-based medication extraction: ‘the i2b2 challenge system’
We present the system as it was run for the official evaluations of the challenge.
Sentence segmentation and section identification
As preprocessing steps, we segment the texts into sentences and tag the different sections (eg, medications on admission, discharge medications, etc.) based on the identification of potential section titles using rules (our corpus study revealed that these titles exhibit little variation in their form).
Drug name recognition
Each sentence is scanned to find drug names matching an entry in the drug lexicon exactly. We expand drug recognition with rules to detect drug strengths (eg, 5%), dose forms (eg, oral solution), and release modes (eg, extended release) immediately following the name. We also recognize two drug names as one when they refer to a single drug and the second name follows the first one parenthetically (eg, tylenol (acetaminophen)).
As per the challenge's guidelines, we then exclude drugs that cause allergies or that are part of a diet by looking for keywords (eg, diet, allergies) occurring in the same sentence as the drug name.
Additionally, we mark the medication as belonging to a ‘list’ if it occurs in a section where medication lists are found (eg, medications on admission, discharge medications). Elsewhere, it belongs to a ‘narrative’ passage.
Identification of associated information
In order to detect related information, we hypothesized that medication-related information is most often found in the portion of text following a drug name (eg, Lasix 50 mg qdvi). Thus, we split each sentence according to the identified drug names so that each subpart of a sentence begins with a drug name.
We then look for each type of associated information inside these subparts—relying on a set of extraction rules (see examples in table 2) and the previously built lexicons.
Table 2.
Example extraction rules
| Rule | Examples |
| Mode (mode)? | iv, po |
| (0–9)(−0–9)* dose | 20 mg, 2–3 tabs |
| q.(0–9) freq | q.6 h |
| Freq (per | a) freq | twice a week, once per day |
| (for | ×) (0–9) dur | for 3 weeks, ×5 days |
Zero or more items.
dur, duration; freq, frequency; iv, intravenous; po, orally; tabs, tablets.
Additional rules were designed to process multiple information elements associated to a given drug. They search for a drug followed by two modes, doses, and/or frequencies (metformin 1000 mg po qam and 500 mg po qhsvii). We also look for several entries of the reason lexicon contained in a portion (eg, simethicone prnviii for abdominal gas and bloating).
To deal with straightforward cases of anaphora, we look for phrases potentially related to medications (this was increased/decreased/discontinued) in the portion of text following a drug name.
Testing methods to improve the original system
After the challenge, we had access to the annotated test corpus, which we used with the official evaluation software to assess progress while modifying our system. We used this corpus without looking at its contents to avoid overfitting the system to this corpus; therefore, keeping the results meaningful. Our goal in this phase was both to improve our system and to study in a systematic way which methods can bring the largest improvements to this information extraction task.
Drug name recognition conditions the subsequent processing and, therefore, deserves special attention. Our first strategy was to test methods to improve this task. In a second phase, we attempted to refine the extraction of associated information.
Improvement of drug name recognition
We first filtered our existing lists by removing drug names belonging to a general English word list (89 402 inflected forms). We acquired new lists of drug names (which we filtered in the same way): a list used by the Merki software15 (17 363 entries), a list compiled from the drug interface terminology RxTerms,ix and one from the DailyMed websitex (approximately 7300 entries). We collected from the development corpus all relevant contexts preceding nouns, such as medication, meds, and regimen, in order to fill our lists with generic types of drugs (eg, hypertensive medications, bowel regimen).
To overcome missing drug names from our lexicon, we tried identifying unknown drug names based on affixes (eg, -azepam in lorazepam): we collected affixes used in the INN from WHO (14 prefixes and 165 suffixes) and an affix list created by applying general syllabic patterns to a drug lexicon (209 prefixes and 2133 suffixes). We considered as a potential drug name any word composed of a prefix/suffix from these lists and absent from the general-language word list.
To identify typing errors, we created a list of 167 common misspellings in drug names from a drug information website.xi They involve character deletion (warfin for warfarin), substitution (zanax for xanax), or insertion (flowmax for flomax).
Finally, we extended the rules detecting dose forms and release modes as part of drug names by adding new forms (eg, elixir, er) based on the development corpus. We created new rules to detect series of drug names separated by hyphens or slashes (eg, ASA/Coumadin/Plavix), and to detect negation (eg, do not take Lasix) based upon regular expressions triggered by words such as no, avoid, etc. The principle is similar to that of the NegEx algorithm,19 but while NegEx focuses on diseases and findings we customized trigger words for drug names.
Improvement of associated information extraction
We extended our list of abbreviations and expressions used in drug-related information by adding dosage and frequency expressions based on examples from the development corpus (eg, tbsp, nightly) and mode expressions using the drug route of administration attributes in RxNorm.
We extended rules for frequencies, modes, and dosages focusing on multiple information (eg, multiple dosages for one drug) and changes in dosage and frequency. This was done by looking at the development corpus for potential cases we did not take into account. Also, we handled negation for reasons.
Configurations to test improvements
We performed automatic evaluations on the test corpus to see whether the above changes were indeed improvements and which were the most beneficial, integrating one change at a time. This resulted in running several test configurations, each consisting of the baseline system with the addition of one specific modification:
A first test with filtered drug lexicons: run #1
-
Drug lexicon extension:
adding generic expressions of drugs: run #2
adding new lists: run #3
-
Unknown drug name detection:
relying on affixes: run #4
detecting misspellings: run #5
Improvement of rules recognizing drug names: run #6
Extension of the list of abbreviations: run #7
Improvement of rules detecting associated information: run #8
Results
Results were evaluated against the gold standard (the 251 annotated texts of the test corpus) in two ways: a global evaluation (horizontal level) to assess the extraction of all information together (a drug name and its associated information), and a field evaluation (vertical level) to assess the extraction of each item individually (medications, modes, dosages, etc).
Our original system ranked 8th out of 20 participants at the i2b2 challenge with a 77% F-measure (see table 3). Among the top ten systems, the three highest F-measures were 86%, 82%, and 81%, while the lowest was 76%. We achieved F-measures over 80% for all items except for durations and reasons, which obtained very low results as in all systems participating in the challenge.
Table 3.
Results obtained at the i2b2 challenge (original system)
| Precision (%) | Recall (%) | F-measure (%) | |
| Horizontal level | 82.7 | 72.5 | 77.3 |
| Medications | 80.2 | 79.3 | 79.8 |
| Dosages | 89.2 | 73.2 | 80.4 |
| Modes | 88.5 | 79.2 | 83.6 |
| Frequencies | 89.3 | 77.0 | 82.7 |
| Durations | 65.7 | 28.2 | 39.4 |
| Reasons | 41.1 | 23.4 | 29.9 |
Table 4 details the evaluation of each test configuration compared to the baseline (the original system). Refining the extraction rules (#6) brought the highest improvement in drug name extraction, followed by lexicon filtering (#1), the addition of generic expressions (#2), and common misspellings (#5). However, adding new lists of drug names (#3) and recognizing drug names (#4) based on affixes caused the F-measure to drop substantially. Lexicon extension (#7) and rule improvement (#8) both improved the associated information extraction results.
Table 4.
F-measure (%) obtained with each test configuration
| Baseline | #1 filt | #2 gen | #3 d-lex | #4 unk | #5 missp | #6 d-rule | #7 other-lex | #8 other-rule | |
| Horizontal level | 77.3 | 78.0 | 77.4 | 75.8 | 71.5 | 77.4 | 78.3 | 77.6 | 78.1 |
| Medications | 79.8 | 81.0 | 80.3 | 77.1 | 74.3 | 80.1 | 81.7 | 80.1 | 80.3 |
| Dosages | 80.4 | 80.4 | 80.2 | 79.8 | 73.3 | 80.3 | 80.6 | 80.3 | 81.1 |
| Modes | 83.6 | 83.5 | 83.3 | 83.2 | 79.0 | 83.3 | 84.0 | 84.7 | 85.2 |
| Frequencies | 82.7 | 82.9 | 82.8 | 82.1 | 76.1 | 82.8 | 83.3 | 83.1 | 84.1 |
| Durations | 39.4 | 40.0 | 39.1 | 39.1 | 34.4 | 39.4 | 39.6 | 39.4 | 39.5 |
| Reasons | 29.9 | 31.6 | 29.8 | 28.6 | 26.7 | 29.9 | 30.1 | 29.9 | 30.0 |
1filt, drug lexicon filtering; #2 gen, generic expressions of drugs; #3 d-lex, additional drug lexicons; #4 unk, unknown drug name recognition; #5 missp, misspellings in drug names; #6 d-rule, drug name extraction rule refinement; #7 other-lex, associated information lexicon extension; #8 other-rule, associated information extraction rule improvement.
We then created an optimal configuration by selecting the modifications that improved the results (#1 + #2 + #5 + #6 + #7 + #8). This configuration achieved a global F-measure of 81% (see table 5).
Table 5.
Results obtained with the final improved system
| Precision (%) | Recall (%) | F-measure (%) | ||||
| Horizontal level | 86.3 | +3.6 | 75.5 | +3.0 | 80.5 | +3.2 |
| Medications | 84.4 | +4.2 | 82.3 | +3.0 | 83.7 | +3.9 |
| Dosages | 91.3 | +2.1 | 74.0 | +0.8 | 81.8 | +1.4 |
| Modes | 92.4 | +3.9 | 83.5 | +4.3 | 87.7 | +4.1 |
| Frequencies | 90.4 | +1.1 | 81.1 | +4.1 | 85.5 | +2.8 |
| Durations | 67.5 | +1.8 | 28.4 | +0.2 | 40.0 | +0.6 |
| Reasons | 48.2 | +7.1 | 23.7 | +0.3 | 31.7 | +1.8 |
Discussion
While we achieved a good performance level (77% F-measure) at the i2b2 2009 shared task, our system had yet to be improved compared to the top systems, especially regarding the extraction of drug names.
We studied two types of changes likely to improve medication extraction: changes affecting the lexicons and changes affecting the rules themselves. Rule improvement, resulting from a careful study of attested examples from the development corpus, systematically brought better results, while changes involving the lexicons were not always very beneficial. Filtering methods improved the results, which is consistent with the findings of Sirohi and Peissig.12 Adding new items proved to be more challenging. When those additions were limited and motivated by a corpus study they improved the system, while the addition of uncontrolled extensive lists of drug names caused the results to drop substantially. Modules designed to guess unknown entities (drug names) based on heuristics (affixes) were also unsuccessful.
Our final system achieved good results, with a global F-measure of 81%. The extraction of durations and reasons remains problematic. They show far more variability in the way they are expressed and their occurrence in association with a drug name is less systematic than other types of information, which makes it harder to detect them in a robust manner.
We had one held-out, test corpus which we used to evaluate the configurations of our system as well as its final configuration. Ideally the latter should have been done on a different test corpus and this constitutes a limitation of this study. Nevertheless, keeping the test corpus unseen ensures some degree of scalability.
Comparing our system to existing ones not participating in the challenge is difficult because the evaluation corpus and target information are different. Our system seems to yield better results than recent approaches15 16 for information associated to drugs but could be improved for drug name recognition.
Our system does not use any deep natural language processing, such as POS tagging, chunking or syntactic parsing. This demonstrates that a simple NLP system with surface rules can also achieve high performance to capture the regularities in the expression of medication-related information in clinical records. Nevertheless, there remains a certain degree of variation, which is difficult to capture in an exhaustive way.
Conclusion
We presented methods and a system to detect medications in narrative patient records. The challenge lies in capturing the regularities of the expression of drug prescriptions while coping with the variations found in texts. Based on the study of a development corpus and a priori knowledge, we built a semantic lexicon and hand-designed rules to encode most forms of medications. They were evaluated in the 2009 i2b2 challenge and obtained an F-measure of 77%. Shortcomings were identified and further work was performed to address some of them; in particular, drug name recognition and multiple entries. This increased the global F-measure to 81%. We explored which tactics are the most beneficial to medication extraction, and observed that controlled modifications (lexicon filtering, rule refinement) best raised the performance, while the addition of larger, less controlled drug lists, and the identification of unknown drug names based on morphological patterns, were detrimental. In further work, we plan to explore more elaborate language processing methods, namely syntactic parsing, and to introduce machine learning methods.
Acknowledgments
Unidentified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Ozlem Uzuner, i2b2 and SUNY.
Footnotes
Funding: Agence Nationale de la Recherche (ANR) 212, rue de Bercy 75012 Paris. This work was partially funded by project Akenaton under grant number ANR-07-TECSAN-001. The project described was supported in part by the i2b2 initiative, Award Number U54LM008748 from the National Library of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
https://www.i2b2.org/NLP/Medication/Main.php (accessed Jul 2010).
http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm (accessed Jul 2010).
http://www.rxlist.com/ (accessed Jul 2010).
http://whqlibdoc.who.int/hq/2002/a76618.pdf (accessed Jul 2010).
http://www.julesberman.info/abbtwo.htm (accessed Jul 2010).
qd, each day.
po, orally; qam, every day before noon; qhs, every night at bedtime.
prn, as needed.
http://wwwcf.nlm.nih.gov/umlslicense/rxtermApp/rxTerm.cfm (accessed Jul 2010).
http://dailymed.nlm.nih.gov/ (accessed Jul 2010).
http://www.drugs.com/ (accessed Jul 2010)—this website integrates a set of common misspelling drug names entries.
References
- 1.Sager N, Friedman C, Lyman M, et al. The analysis and processing of clinical narrative. In: Salamon R, Blum B, Jorgensen M, eds. Proceedings of the fifth conference on medical informatics. Washington DC, USA: ElsevierScience Publishers; B.V, 1986:1101–5 [Google Scholar]
- 2.Friedman C, Alderson P, Austin J, et al. A general natural language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meystre S, Savova G, Kipper-Schuler K, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 4.Lussier Y, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: a feasibility study. Proc AMIA Symp 2001:418–22 [PMC free article] [PubMed] [Google Scholar]
- 5.Baud R. A natural language based search engine for ICD10 diagnosis encoding. Med Arh 2004;58(1 Suppl 2):79–80 [PubMed] [Google Scholar]
- 6.Melton G, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005;12:448–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penz J, Wilcox A, Hurdle J. Automated identification of adverse events related to central venous catheters. J Biomed Inform 2007;40:174–82 [DOI] [PubMed] [Google Scholar]
- 8.Jain N, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 1997:829–33 [PMC free article] [PubMed] [Google Scholar]
- 9.Fiszman M, Chapman W, Aronsky D, et al. Automatic detection of acute bacterial pneumonia from chest x-ray reports. J Am Med Inform Assoc 2000;7:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman W, Fiszman M, Dowling J, et al. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap. Stud Health Technol Inform 2004;107(Pt 1):487–91 [PubMed] [Google Scholar]
- 11.Levin MA, Krol M, Doshi AM, et al. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Proc 2007:438–42 [PMC free article] [PubMed] [Google Scholar]
- 12.Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. Pac Symp Biocomput 2005:10:308–18 [DOI] [PubMed] [Google Scholar]
- 13.Shah A, Martinez C. An algorithm to derive a numerical daily dose from unstructured text dosage instructions. Pharmacoepidemiol Drug Saf 2006;15:161–6 [DOI] [PubMed] [Google Scholar]
- 14.Evans D, Brownlow N, Hersh W, et al. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proc AMIA Annu Fall Symp 1996:388–92 [PMC free article] [PubMed] [Google Scholar]
- 15.Gold S, Elhadad N, Zhu X, et al. Extracting structured medication event information from discharge summaries. AMIA Annu Symp Proc 2008:237–41 [PMC free article] [PubMed] [Google Scholar]
- 16.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009;78:284–91 [DOI] [PubMed] [Google Scholar]
- 17.Xu H, Stenner S, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman W, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]