Abstract
Objective
This paper presents Lancet, a supervised machine-learning system that automatically extracts medication events consisting of medication names and information pertaining to their prescribed use (dosage, mode, frequency, duration and reason) from lists or narrative text in medical discharge summaries.
Design
Lancet incorporates three supervised machine-learning models: a conditional random fields model for tagging individual medication names and associated fields, an AdaBoost model with decision stump algorithm for determining which medication names and fields belong to a single medication event, and a support vector machines disambiguation model for identifying the context style (narrative or list).
Measurements
The authors, from the University of Wisconsin-Milwaukee, participated in the third i2b2 shared-task for challenges in natural language processing for clinical data: medication extraction challenge. With the performance metrics provided by the i2b2 challenge, the micro F1 (precision/recall) scores are reported for both the horizontal and vertical level.
Results
Among the top 10 teams, Lancet achieved the highest precision at 90.4% with an overall F1 score of 76.4% (horizontal system level with exact match), a gain of 11.2% and 12%, respectively, compared with the rule-based baseline system jMerki. By combining the two systems, the hybrid system further increased the F1 score by 3.4% from 76.4% to 79.0%.
Conclusions
Supervised machine-learning systems with minimal external knowledge resources can achieve a high precision with a competitive overall F1 score.Lancet based on this learning framework does not rely on expensive manually curated rules. The system is available online at http://code.google.com/p/lancet/.
Pharmacotherapy is an important part of a patient's medical treatment, and nearly all patient records incorporate a significant amount of medication information. The administration of medication at a specific time point during the patient's medical diagnosis, treatment, or prevention of disease is referred to as a medication event,1–3 and the written representation of these events typically comprises the name of the medication and any of its associated fields, including but not limited to dosage, mode, frequency, etc.4 Accurately capturing medication events from patient records is an important step toward large-scale data mining and knowledge discovery,5 medication surveillance and clinical decision support6 and medication reconciliation.7–10
In addition to its importance, medication event information (eg, treatment outcomes, medication reactions and allergy information) is often difficult to extract, as clinical records exhibit a range of different styles and grammatical structures for recording such information.4 Therefore, Informatics for Integrating Biology & the Bedside (i2b2) recognized automatic medication event extraction with natural language processing (NLP) approaches as one of the great challenges in medical informatics. As one of 20 groups that participated in the i2b2 medication extraction challenge, we report in this study on Lancet, which we developed for medication event extraction.
Related work
Over two decades, several approaches and systems have been developed to extract information from clinical narratives, including concept mapping,11 syntactic and semantic parsing and pattern matching12 13 and supervised machine-learning approaches.14 15
Systems for medication event extraction have been reported previously. Gold et al1 developed a rule-based system called MERKI to extract medication names and the corresponding attributes from structured and narrative clinical texts. Cimino et al16 explored the MedLEE system to extract medication information from clinical narratives. Recently, Xu et al4 built an automatic medication extraction system (MedEx) on discharge summaries by leveraging semantic rules and parsing techniques, achieving promising results for extracting medication and related fields.
There are also some commercial systems designed to extract medication information from medical records, including LifeCode, A-Life Medical, FreePharma, etc. Jagannathan et al17 evaluated the performance of four commercial NLP tools showing that these tools performed well in recognizing medication names but poorly on recognizing related information such as dosage, route and frequency.
Although the existence of such NLP systems is evidence of the progress that has been made in this area, most of these systems are not publicly available. Furthermore, different systems have been developed for different purposes and have been evaluated against different gold standards. This makes comparing these approaches with one another a challenging task. Therefore, the i2b2 project attempts to provide a common purpose and gold standard to different NLP systems.15
The i2b2 data and extraction task
Medication event extraction
The i2b2 challenge defines a medication event as an event incorporating a medication name and any of the following associated fields: dosage, frequency, mode, duration and reason. Table 1 shows the definition released by the i2b2 organizers.18 As an example, figure 1 shows a clinical narrative/list excerpt released by the i2b2 organizers in which medication events were annotated based on the i2b2 annotation guidelines.
Table 1.
Definitions of medication name and associated fields
| Fields | Definition |
| Medication | Substances for which the patient is the experiencer, excluding food, water, diet, tobacco, alcohol, illicit drugs and allergic reaction-related drugs |
| Dosage | The amount of a single medication used in each administration |
| Mode/route | Expressions describing the method for administering the medication |
| Frequency | Terms, phrases, or abbreviations that describe how often each dose of the medication should be taken |
| Duration | Expressions that indicate for how long the medication is to be administered |
| Reason | The medical reason for which the medication is stated to be given |
Figure 1.
Illustration of medication events in both a narrative and a list. As shown here, each event includes a medication name and any of its related medication fields. Medical-field associations are indicated by a dotted line with an arrow. Different font styles indicate different fields: bold plus underline for medication name; italic for dosage; underline for mode/route; italic plus bold for frequency; bold for duration and italic plus underline for reason. The bracket pair “[ ]” shows the narrative/list attribute.
While the challenge was to extract all medication events from both lists and narrative context, the challenge's main interest was in the extraction of medication information from the narrative medical records, as illustrated in figure 1.
Training dataset and annotation
The i2b2 organizers released 696 unannotated deidentified patient discharge summaries (available at https://www.i2b2.org/NLP/DataSets/Main.php),18 from which we randomly selected a total of 147 discharge summaries for annotation. Two authors (ZFL and LA) manually and independently annotated 75 and 72 discharge summaries. Each article was annotated by one annotator. This collection of 147 summaries incorporated the 17 ‘ground truth’ summaries,18 a community annotation effort. A post-hoc annotation of 10 summaries was used to measure interannotator agreement between ZFL and LA.
Medication event extraction systems
We first describe Lancet, a supervised machine-learning system for medication event extraction. We then describe the rule-based system jMerki and a hybrid system.
Lancet system
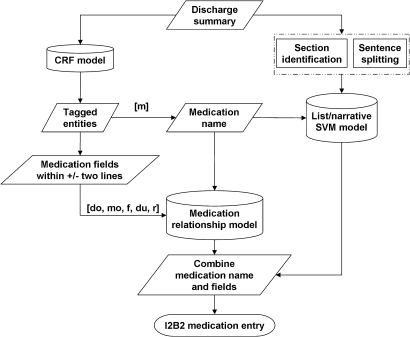
Figure 2 shows the overall Lancet system, which incorporated three supervised machine-learning models: (1) a conditional random fields (CRF) model for identifying instances of a medication name (m) and its associated fields: dosage (do), mode (mo), frequency (f), duration (du) and reason (r); (2) a medication relationship model, an AdaBoost classification model with decision stump for associating a medication name with its corresponding fields; (3) a list/narrative support vector machines (SVM) model, a SVM classifier for distinguishing lists from narratives.
Figure 2.
Flow chart of Lancet.
Preprocessing
The preprocessing includes lowering case, sentence boundary detection19 and subheading recognition. We applied the following manually curated regular expressions to detect medication-related subheadings:
‘medications\s+on\s+(admission|discharge|transfer)’,
‘(discharge|transfer|home|admi\w+|new)\s+(medication|med)s?', ‘(prn\s+)?med(ication)?s’
A CRF model for medication named entity recognition
We trained a CRF model on the 147 annotated discharge summaries to recognize the medication name and five fields (do, mo, f, du and r). We applied ABNER, an open-source biomedical named entity recognizer.20 We used the default feature set, which are the standard bag-of-words, morphology and n-gram features.
An AdaBoost model for associating a medication name with its corresponding fields
We built a supervised machine learning classifier to associate a medication with its fields. This two-way classifier attempted to determine whether a medical field was associated with a medication name or not. As the number of potential medication–name field pairs can be large, we followed a heuristic rule suggested by the i2b2 organizers in which any medication name and field within the distance of two lines (± two lines) was considered to be a candidate medication–name field pair. The features used to train the model are displayed in table 2.
Table 2.
Features for the medication relationship model
| Feature name | Meaning |
| Same sentence | Whether the medication and field are both in the same sentence, as determined by Splitta |
| Same subsection | Whether both elements in a medication field pair are located in the same subsection of the discharge summary |
| Numeral | Whether the value of the medication field contains numerals |
| Distance | The number of tokens between a medication name and medication field |
| Position | Whether the medication field appears before or after the medication name |
| Field type | The type of field, such as duration, reason, etc |
| Medication between | The number of other medication names between the pair |
For implementation, we used the AdaBoost.M1 with decision stump in the Weka toolkit, which is a well-known algorithm less susceptible to over-fitting.21
A SVM classifier for distinguishing lists from narrative text
One of the i2b2 competition requirements was to determine whether the text describing a medication is in a list or a narrative format. Using the 147 annotated discharge summaries as the training data, we built a SVM classifier (Weka toolkit)21 to determine the format of each candidate sentence. The features we explored included bag of words, bi-grams, tri-grams and subsection features.
The integration
We integrated all three models into Lancet. Lancet first detects medication names and fields with the CRF model, and then applies the AdaBoost model to determine whether a medication field belongs to a medication name. Finally, a SVM classifier separates lists from narratives.
jMerki: a rule-based baseline system
jMerki was a rule-based system implemented in JAVA. It integrated the rules in the MERKI system,1 including rules for dosage, frequency, time and pro re nata (or as needed). We added additional rules for the i2b2 medication detection, including applying regular expressions to detect subheadings in discharge summaries. The system performed dictionary look up and regular expression matching for identifying related fields. We built a medication name dictionary with two external knowledge resources, RxNorm and DrugBank.1 22 This baseline system cannot recognize list or narrative form, so the SVM classifier of Lancet was employed for the performance evaluation.
The hybrid system
As a post-hoc experiment, we built a hybrid system to increase both recall and precision. In particular, we aligned and matched jMerki and Lancet systems' outputs. If both jMerki and Lancet detected the same medication name, but differed in other content (eg, dosage, etc), Lancet's output was chosen because it has a higher precision than jMerki. If jMerki and Lancet did not agree with a medication name, then the hybrid system kept both medication entries detected by the two systems. This step would increase recall.
Evaluation
The primary evaluation in this i2b2 medication challenge is based on the system-level horizontal metrics, as shown in the following formulas. The F1 score is calculated using precision (P) and recall (R): 2(P*R)/(P+R). For details, please refer to Uzuner et al.18
The gold standard used for the i2b2 evaluation was built as a community effort.18 We found that the gold standard medication names belonged to 295 categories, which represented 50.4% of total drug categories in DrugBank. The results suggested that the coverage of drugs in the i2b2 challenging task was reasonably broad.
Results
Evaluation of Lancet in the i2b2 challenge 2009
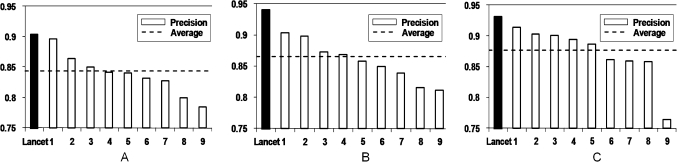
Although we report the results of three systems in this study, Lancet was the only system of the three that competed in the i2b2 challenge. Among the top 10 systems, Lancet achieved the highest precision at the system-level horizontal evaluation: 90.4% in exact matching and 94.0% in inexact matching (figure 3A,B). The corresponding F1 values were 76.4% and 76.5%. For F1 value, Lancet ranked 10th in exact matching and 9th in inexact matching. For list, Lancet achieved the highest precision of 93.1%, with an F1 of 66.0%, on exact match at the system-level horizontal evaluation (figure 3C). For narratives, Lancet achieved a precision of 36.6% with an F1 of 38.4%. Lancet ranked 10th for narratives or lists.18
Figure 3.
Precision for system-level horizontal evaluation of top 10 systems: (A) Strict evaluation with exact match; (B) Relaxed evaluation with inexact match; (C) Strict evaluation with exact match on list entries only. Dashed line indicates the average of the top 10 systems.
Comparison of the three systems
We described earlier the three systems we developed: Lancet, the rule-based jMerki and the hybrid system. Table 3 shows the results of all three systems. On horizontal level evaluation with exact matching, Lancet outperformed jMerki by 12.0% (system) and 10.4% (patient). The hybrid system further improved the performance by 3.4% (system) and 4.6% (patient), yielding the highest F1 score of 79% (system) and 77.6% (patient). For recall, both Lancet (66.1%) and jMerki (58.7%) are relatively low. On the other hand, the recall of the hybrid system increases to 74%.
Table 3.
Three systems' comparison results (F1 score, exact match)
| Two levels | Granularity | Tags | jMerki | Lancet* | Hybrid* |
| Horizontal | System | Medication event | 68.2% | 76.4% | 79.0% |
| Patient | Medication event | 67.2% | 74.2% | 77.6% | |
| Vertical | System | Medication name | 77.2% | 80.2% | 83.4% |
| Patient | Medication name | 76.6% | 79.1% | 82.9% | |
| System | Dosage | 67.9% | 80.2% | 81.8% | |
| Patient | Dosage | 66% | 78.3% | 80.6% | |
| System | Mode | 70.8% | 82.1% | 85.0% | |
| Patient | Mode | 68.2% | 74% | 81.9% | |
| System | Frequency | 66.3% | 81.3% | 82.4% | |
| Patient | Frequency | 63% | 78.8% | 80% | |
| System | Duration | 8.9% | 18% | 21.2% | |
| Patient | Duration | 5.6% | 14% | 16.5% | |
| System | Reason | 0† | 3% | 2.9% | |
| Patient | Reason | 0† | 2.4% | 2.4% |
Significant outperformance is indicated by (p<0.05, Wilcoxon rank sum test).
Caused by programming bug.
Similarly, on the vertical level evaluation, Lancet outperformed jMerki, and the hybrid system outperformed both. The hybrid system achieved good performance (F1 80–85%) in the fields of dosage, medication, mode and frequency, but achieved poor performance (F1 2.4–21.2%) in the duration and reason fields. In addition, the results show that the system-level performance was consistently better than the patient level for both horizontal and vertical level evaluation.
Error analysis and follow-up experiments
We first examined annotation inconsistency and then manually analyzed the system output. We found that errors contributed to data sparseness, multiple medication entries, grammatical errors in clinical notes and negated events. Based on the results of our error analyses, we further performed post-hoc experiments by exploring negation detection, external medication name dictionaries and others, to improve the medication event extraction based on our Lancet system. Due to space limitations, the detailed analysis and post-hoc experimental results can be found in the full-length appendix (available online only).
Discussion
The agreement between our annotation and the annotation by the i2b2 organizers was 81.5% F1 score, lower than the annotation agreement reported by the i2b2 organizers (89.7% F1 score18), but closer to the annotation agreement among the participating teams (82.4% F1 score).18 The annotation guideline was iteratively updated throughout our training data generation, we therefore speculate that inconsistency was introduced throughout the guideline refinement process. As Lancet was trained on these 147 annotated records, the annotation inconsistency introduced errors.
Table 3 shows that Lancet significantly outperformed jMerki, increasing the precision from 81.3% to 90.4% and the recall from 58.7% to 66.1% (horizontal system level with exact match). The results suggest that machine learning-based methods hold advantages over the rule-based system. Another advantage is that Lancet did not rely on expensive manually curated rules.
One post-hoc experiment (see table 4 in the full-length appendix, available online only) showed that Lancet increased both recall and precision with a high-quality external knowledge resource and that the hybrid system yielded the highest performance, both of which demonstrated that data sparseness contributed to errors. Other sources of error include multiple medication entries, misspelling and negations.
Overall, Lancet performed with the highest precision among the top 10 teams. We found that most of the top 10 systems23–31 incorporated extensively manually curated patterns and external dictionaries. In contrast, Lancet did not use any external knowledge resources, was trained on the annotated dataset and applied few curated rules, suggesting that external resources or rules may damage performance for medication event detection. On the contrary, our post-hoc experiment showed that a high-quality external knowledge resource increased Lancet's performance, suggesting that the knowledge resources used by other teams may be noisy, although more investigation is needed.
Our post-hoc experiments (see the full-length online appendix for details) showed that linguistic features (eg, affix) have mixed results. While affix increased precision, it decreased recall. Digit normalization slightly improved the overall F1 score performance.
Conclusion and further directions
We present three systems for medication event extraction from patient discharge summaries: the supervised machine learning system Lancet, the rule-based system jMerki and the hybrid system. We applied Lancet to the i2b2 medication event extraction challenge, and the evaluation results showed that it performed with the highest precision (90.4% and 94.0% F1 scores in exact and inexact match) among the top 10 teams.
Our post-hoc experiments show that Lancet and jMerki have different strengths and that the hybrid system performed the best, yielding a 79% F1 score (85% precision and 74% recall). Our error analysis showed that annotation inconsistency and data sparseness introduced errors, and we therefore speculated that a large scale of high-quality annotated data may further improve Lancet. Future work includes exploring semisupervised learning and deeper syntactic and semantic parsing.
Supplementary Material
Acknowledgments
The authors would like to thank Qing Zhang and Shashank Agarwal for valuable discussions.
Footnotes
Funding: This study was supported by grants from the National Institute of Health (5R01LM009836, 5R21RR024933 and 5U54DA021519). The project described was supported in part by the i2b2 initiative, Award Number U54LM008748 from the National Library of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Gold S, Elhadad N, Zhu X, et al. Extracting structured medication event information from discharge summaries. AMIA Annu Symp Proc 2008:237–41 [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz E, Levine HB, Sullivan MC, et al. Use of the Medication Event Monitoring System to estimate medication compliance in patients with schizophrenia. J Psychiatry Neurosci 2001;26:325–9 [PMC free article] [PubMed] [Google Scholar]
- 3.de Klerk E, van der Heijde D, Landewé R, et al. The compliance-questionnaire-rheumatology compared with electronic medication event monitoring: a validation study. J Rheumatol 2003;30:2469–75 [PubMed] [Google Scholar]
- 4.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullins IM, Siadaty MS, Lyman J, et al. Data mining and clinical data repositories: insights from a 667,000 patient data set. Comput Biol Med 2006;36:1351–77 [DOI] [PubMed] [Google Scholar]
- 6.Kuperman GJ, Bobb A, Payne TH, et al. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc 2007;14:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates DW, Cohen M, Leape LL, et al. Reducing the frequency of errors in medicine using information technology. J Am Med Inform Assoc 2001;8:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JG, Jay SJ, Anderson M, et al. Evaluating the impact of information technology on medication errors: a simulation. J Am Med Inform Assoc 2003;10:292–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha AK, Kuperman GJ, Teich JM, et al. Identifying adverse drug events: development of a computer-based monitor and comparison with chart review and stimulated voluntary report. J Am Med Inform Assoc 1998;5:305–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pronovost P, Weast B, Schwarz M, et al. Medication reconciliation: a practical tool to reduce the risk of medication errors. J Crit Care 2003;18:201–5 [DOI] [PubMed] [Google Scholar]
- 11.Sager N, Lyman M, Nhan NT, et al. Medical language processing: applications to patient data representation and automatic encoding. Methods Inf Med 1995;34:140–6 [PubMed] [Google Scholar]
- 12.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DA, Brownlow ND, Hersh WR, et al. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proc AMIA Annu Fall Symp 1996:388–92 [PMC free article] [PubMed] [Google Scholar]
- 14.Bramsen P, Deshpande P, Lee YK, et al. Finding temporal order in discharge summaries. AMIA Annu Symp Proc 2006:81–5 [PMC free article] [PubMed] [Google Scholar]
- 15.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ, Bright TJ, Li J. Medication reconciliation using natural language processing and controlled terminologies. Stud Health Technol Inform 2007;129:679–83 [PubMed] [Google Scholar]
- 17.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009;78:284–91 [DOI] [PubMed] [Google Scholar]
- 18.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillick D. Sentence boundary detection and the problem with the US. Annual Conference of the North American Chapter of the Association for Computational Linguistics 2009, June 4–5 2009, Boulder, Colorado, USA. Proceedings of NAACL HLT 2009: short papers. 2009:241–4
- 20.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 2005;21:3191–2 [DOI] [PubMed] [Google Scholar]
- 21.Frank E, Hall M, Trigg L, et al. Data mining in bioinformatics using Weka. Bioinformatics 2004;20:2479–81 [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledge base for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36:D901–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doan S, Bastarache L, Klimkowski S, et al. Vanderbilt's system for medication extraction. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009. Sponsored by Informatics for Integrating Biology and the Bedside.
- 24.Grouin C, Deleger L, Zweigenbaum P. A simple rule-based medication extraction system. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 25.Hamon T, Grabar N. Concurrent linguistic annotations for identifying medication names and the related information in discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 26.Meystre SM, Thibault J, Shen S, et al. Description of the textractor system for medications and reason for their prescription extraction from clinical narrative text documents. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 27.Patrick J, Li M. A cascade approach to extract medication event (i2b2 challenge 2009). Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 28.Shooshan SE, Aronson AR, Mork JG, et al. NLM's i2b2 tool system description. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009. [Google Scholar]
- 29.Solt I, Tikk D. Yet another rule-based approach for extracting medication information from discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 30.Spasic I, Sarafraz F, Keane JA, et al. Medication information extraction with linguistic pattern matching and semantic rules. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
- 31.Yang H. A linguistic approach for medication extraction from medical discharge summaries. Proceedings of the Third i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. Third i2b2 Shared-Task and Workshop Challenges in Natural Language Processing for Clinical Data, San Francisco, California, USA, November 13 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.