Summary
Proper centromere function is critical to maintain genomic stability and to prevent aneuploidy, a hallmark of tumors and birth defects. A conserved feature of all eukaryotic centromeres is an essential histone H3 variant called CENP-A that requires a centromere targeting domain (CATD) for its localization. Although proteolysis prevents CENP-A from mislocalizing to euchromatin, regulatory factors have not been identified. Here, we identify an E3 ubiquitin ligase called Psh1 that leads to the degradation of Cse4, the budding yeast CENP-A homolog. Cse4 overexpression is toxic to psh1Δ cells and results in euchromatic localization. Strikingly, the Cse4 centromere targeting domain is a key regulator of its stability and helps Psh1 discriminate Cse4 from histone H3. Taken together, we propose that the CATD has a previously unknown role in maintaining the exclusive localization of Cse4 by preventing its mislocalization to euchromatin via Psh1-mediated degradation.
Introduction
Accurate cell division requires the proper partitioning of chromosomes, resulting in daughter cells with the correct complement of genetic material. Chromosome segregation is directed by the kinetochore, the specialized protein structure that assembles on centromeric DNA and mediates attachment to the spindle (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Although kinetochore function is conserved, the size and sequence of the underlying centromeric DNA is highly variable among organisms (Henikoff et al., 2001). In multicellular eukaryotes, centromeres are characterized by megabases of DNA repeats that lack strict sequence identity and are therefore epigenetically propagated. Budding yeast is the only organism with a defined centromeric DNA sequence that is sufficient to mediate kinetochore assembly. However, epigenetic components also contribute to yeast kinetochore assembly (Tanaka et al., 1999; Mythreye and Bloom, 2003), so to varying degrees all eukaryotic organisms propagate kinetochores using epigenetic components.
A hallmark of all centromeres is a specialized chromatin structure characterized by the presence of essential histone H3 variant CENP-A, which localizes exclusively to the centromere (Earnshaw and Rothfield, 1985; Palmer et al., 1987). CENP-A is an excellent candidate to specify centromere identity because it is present at all active centromeres (Warburton et al., 1997; Amor and Choo, 2002). In most organisms, blocks of CENP-A nucleosomes are interspersed with histone H3 nucleosomes at centromeres (Blower et al., 2002). However, a single CENP-A nucleosome exists at the budding yeast centromere, consistent with a single microtubule binding site (Winey et al., 1995; Furuyama and Biggins, 2007). Despite variation in the number of CENP-A nucleosomes at centromeres among organisms, the functions of CENP-A are conserved and the budding yeast homolog can rescue a depletion of mammalian CENP-A (Wieland et al., 2004; Allshire and Karpen, 2008). CENP-A localization to the centromere depends on a centromere targeting domain (CATD) comprised of loop 1 and α2 helix, which is necessary to target an H3 chimera to centromeres (Vermaak et al., 2002; Black et al., 2004; Black et al., 2007). Recently, this domain was also implicated in binding to a putative chaperone complex that is required for CENP-A deposition, suggesting a potential mechanism to explain the role of the CATD in CENP-A localization (Foltz et al., 2009; Dunleavy et al., Shuaib et al., 2010).
Although CENP-A is highly enriched at functional centromeres, it can also incorporate into euchromatin. Low levels of endogenous CENP-A can be detected in highly transcribed euchromatic regions of the yeast genome (Camahort et al., 2009; Lefrancois et al., 2009), and overexpression of CENP-A leads to euchromatic localization in some organisms (Van Hooser et al., 2001; Tomonaga et al., 2003; Heun et al., 2006). In addition, CENP-A can be detected at sites of DNA double strand breaks prior to removal at the time of DNA repair (Zeitlin et al., 2009). Taken together, these data suggest that CENP-A can localize to euchromatin but is not stably maintained outside of centromeres. Ectopic incorporation of CENP-A can lead to dicentric chromosomes and genomic instability (Heun et al., 2006; Au et al., 2008; Amato et al., 2009) and occurs in primary colorectal cancer tissues (Tomonaga et al., 2003), although mistargeting of CENP-A to euchromatin is not necessarily sufficient to generate neocentromeres (Van Hooser et al., 2001). Taken together, these data suggest that there are multiple controls over both CENP-A localization and kinetochore formation that are critical for genomic stability. One mechanism that regulates CENP-A localization in budding yeast and flies is the specific degradation of euchromatic but not centromere-bound CENP-A by ubiquitin-mediated proteolysis (Collins et al., 2004; Moreno-Moreno et al., 2006). CENP-A proteolysis has also been detected in human cells undergoing senescence or infection with herpes simplex virus 1 (Lomonte et al., 2000; Maehara et al., 2010), suggesting that CENP-A proteolysis is a conserved mechanism. However, the key machinery involved in CENP-A destruction has not been identified in any organism.
Ubiquitin-mediated proteolysis requires the covalent conjugation of ubiquitin monomers onto a substrate protein (for review, see (Deshaies and Joazeiro, 2009).) After E1 activation, ubiquitin is transferred to an E2 conjugating enzyme and subsequently conjugated to a substrate via an E3 ligase. The specificity of substrate recognition is largely dictated by the E3 ligases, consistent with the large number of predicted E3 relative to E1 and E2 enzymes throughout eukaryotes. Here, we identify the Psh1 protein as an E3 ubiquitin ligase that mediates the degradation of Cse4, the budding yeast CENP-A protein. The centromere targeting domain helps Psh1 distinguish Cse4 from H3, so we propose that the centromeric histone variant ensures its exclusive centromeric localization by utilizing the CATD to directly target the protein to centromeres as well as to ensure destruction of non-centromeric Cse4 protein.
Results
Psh1 is an E3 ligase that mediates Cse4 ubiquitination
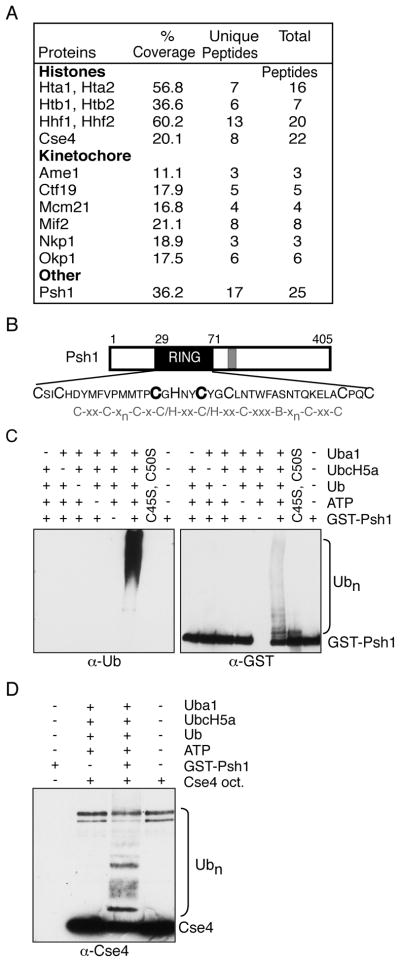
To identify proteins involved in CENP-A degradation, we purified a lysine-free mutant of the budding yeast Cse4 protein that is not ubiquitinated in vivo (Collins et al., 2004). 3xFLAG-Cse416R was overexpressed as the sole genomic copy and affinity purified by anti-FLAG immunoprecipitation. The material was eluted with FLAG peptide and subjected to anion exchange followed by mass spectrometry (LC-MS/MS) on two of the peak fractions containing Cse4. As expected (Westermann et al., 2007; Camahort et al., 2009), we detected Cse4 and histones H2A, H2B and H4, as well as six kinetochore proteins (Fig 1A). We also identified additional proteins including Psh1 (Pob3/Spt16/histone binding protein; Supplementary Table S1), a putative E3 ubiquitin ligase that was first isolated via its interaction with the FACT complex (Pob3/Spt16) (Krogan et al., 2002).
Fig 1. Psh1 is an E3 ligase that co-purifies with Cse4.
(A) List of histone and kinetochore proteins identified by MS after purification of 3xFLAG-Cse416R (SBY5442). (B) Psh1 has a consensus RING domain (residues 29–71, black) and a pair of C4-type zinc finger motifs (residues 150–171, gray). RING domain cysteine and histidine residues predicted to bind zinc are in large typeface and the mutants are indicated in bold. “B” is a bulky residue in the RING domain. (C) Psh1 autoubiquitinates in vitro. Recombinant GST-Psh1 was added to reactions in the presence (+) or absence (−) of ubiquitin-activating enzyme (Uba1), ubiquitin-conjugating enzyme (UbcH5a), ubiquitin (Ub), and ATP (lanes 1–6). A recombinant GST-Psh1 RING mutant with C45S and C50S mutations was used in a complete reaction instead of WT protein in lane 7. Lane 8 contains GST-Psh1 alone. The reactions were run on two gels and immunoblotted with either anti-ubiquitin antibody (left) or anti-GST antibody (right). (D) Recombinant Cse4 octamers were added to reactions containing ubiquitin-activating enzyme (Uba1), ubiquitin-conjugating enzyme (UbcH5a), ubiquitin (Ub), and ATP in the absence (−) or presence (+) of GST-Psh1 (lanes 2 and 3). GST-Psh1 (lane 1) or Cse4 octamers (lane 4) alone were also included in the immunoblot as controls. The blot was probed with anti-Cse4 antibodies.
Because Psh1 contains a RING domain that is a hallmark of many E3 enzymes (Fig 1B, (Deshaies and Joazeiro, 2009)), we tested whether it is a ubiquitin ligase. Recombinant GST-Psh1 exhibited robust autoubiquitination activity in vitro when incubated with ubiquitin, ATP, E1 and E2 enzymes (Fig 1C). The activity is specific to Psh1 because it requires the conserved catalytic cysteines C45 and C50 in the RING domain. We next tested whether Psh1 could facilitate the addition of ubiquitin to the Cse4 protein. When Cse4 octamers were incubated with Psh1 in a ubiquitin reaction in vitro, we detected a ladder of higher molecular weight conjugates on Cse4 (Fig 1D). Taken together, these data show that Psh1 is a ubiquitin ligase that can mediate Cse4 polyubiquitination in vitro.
Psh1 interacts with non-centromeric Cse4
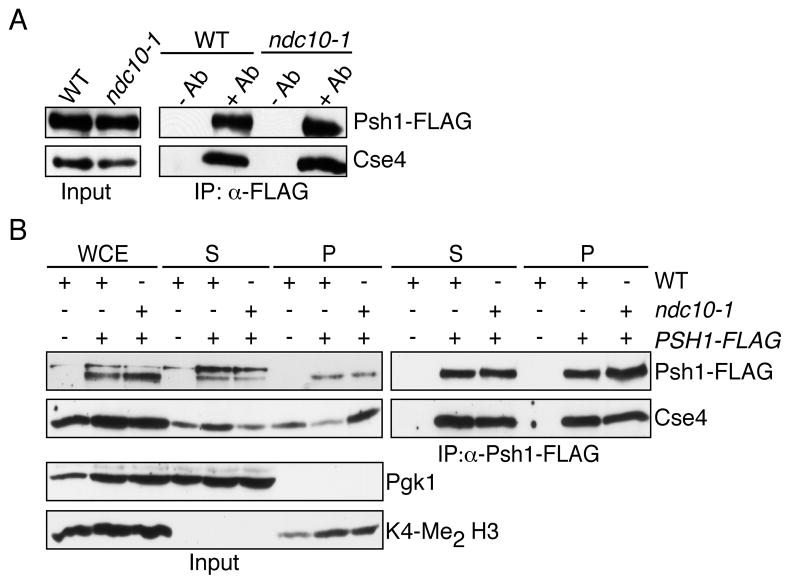
Because we identified Psh1 via its interaction with an overexpressed Cse416R mutant protein, we tested whether it also binds to the endogenous wild-type Cse4 protein. In addition, we asked whether the interaction can be detected in the absence of centromere-bound Cse4 by abolishing its centromeric localization using the ndc10-1 temperature sensitive mutant (Goh and Kilmartin, 1993). Psh1-FLAG was immunoprecipitated from wild-type and ndc10-1 mutant cells and equivalent amounts of Cse4 were detected (Fig 2A). Although this does not exclude an interaction between Psh1 and Cse4 at the centromere, it shows that Cse4 does not need to associate with centromeres for the proteins to interact. Cse4 localizes to some non-centromeric loci at low abundance (Camahort et al., 2009; Lefrancois et al., 2009), so we tested whether Psh1 and Cse4 interact in euchromatin. Yeast extracts were fractioned and Psh1-FLAG was immunoprecipitated from the soluble and chromatin fractions. Psh1 and Cse4 were associated in both chromatin and soluble fractions, and their interaction was not altered in ndc10-1 mutant cells where all chromatin-bound Cse4 is euchromatic (Fig 2B). Taken together, these data show that Psh1 and Cse4 can bind independently of the centromere.
Fig 2. Psh1 and Cse4 can associate independently of the centromere.
(A) Wild-type (SBY8281) or ndc10-1 mutant (SBY8282) cells containing Psh1-FLAG were shifted to 37 °C for 3 hrs and Psh1-FLAG was immunoprecipitated. The corresponding immunoblot was probed with anti-FLAG and anti-Cse4 antibodies. (B) WT (SBY8281) or ndc10-1 mutant (SBY8282) cells were grown as in (A). Psh1-FLAG was immunoprecipitated from the soluble (S) and chromatin (P) fractions and the resulting immunoblots were probed with anti-FLAG and anti-Cse4 antibodies. Pgk1 and K4-Me2 H3 are shown as markers for S and P fractions, respectively. Note that there is loss of material during the procedure so the levels between soluble and chromatin fractions cannot be compared.
Psh1 is required for Cse4 ubiquitination and degradation in vivo
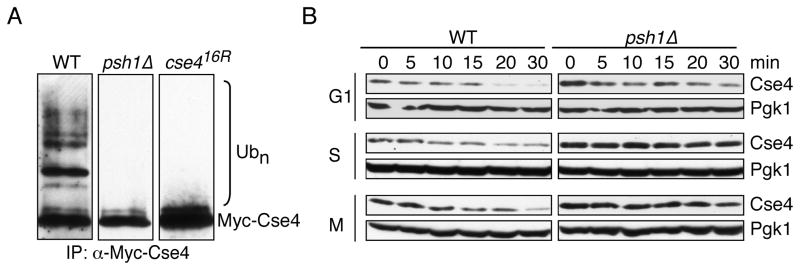
Because Psh1 is an E3 ligase that associates with Cse4 and can ubiquitinate Cse4 octamers in vitro, we asked whether it targets Cse4 for ubiquitin-mediated proteolysis in vivo. First, we analyzed ubiquitin conjugates on Cse4 in the presence and absence of Psh1. Similar to the Cse416R protein that cannot be ubiquitinated, we found that the slower-migrating forms of Cse4 that we previously showed are ubiquitin conjugates were no longer detectable on either overexpressed or endogenous Cse4 protein in psh1 mutant cells (Fig 3A, Fig S1 and (Collins et al., 2004)). We therefore analyzed Cse4 stability at various cell cycle stages in wild-type and psh1Δ cells by arresting cells in G1-, S- and M-phases and monitoring the endogenous Cse4 protein levels after repressing translation. At all cell cycle stages, Cse4 has a short half-life, which is extended when Psh1 is deleted (Fig 3B). Quantification confirmed that the Cse4 half-life increases in psh1Δ cells (data not shown), although we cannot determine the precise change because there are soluble, euchromatic and centromere-bound pools of Cse4, each with unique half-lives (Collins et al., 2004). Similar to previous observations on Cse416R (Collins et al., 2004), the total levels of Cse4 eventually decrease in psh1Δ cells. The residual degradation is not due to the Tom1-mediated histone degradation pathway ((Singh et al., 2009) and data not shown) and it is unclear whether the mechanism is ubiquitin-dependent.
Fig 3. Psh1 is required for Cse4 ubiquitination and degradation in vivo.
(A) Wild-type (SBY3570) and psh1Δ (SBY8355) cells expressing pGAL-Myc-CSE4 were grown in galactose and Cse4 was immunoprecipitated using anti-Myc antibodies. The immunoblot was probed with anti-Myc antibodies. Wild-type cells expressing pGAL-Myc-CSE416R (SBY3571) were used as a control. (B) Wild-type (SBY3) and psh1Δ (SBY8336) cells were arrested in G1-, S- or M-phase with α-factor (G1), hydroxyurea (S) or nocodazole (M), respectively. Protein synthesis was inhibited by cycloheximide addition at time zero and lysates were analyzed for endogenous Cse4 levels using anti-Cse4 antibodies at the indicated time points. Pgk1 is shown as a loading control. See also Figure S1.
Cse4 accumulates in euchromatin in psh1Δ cells
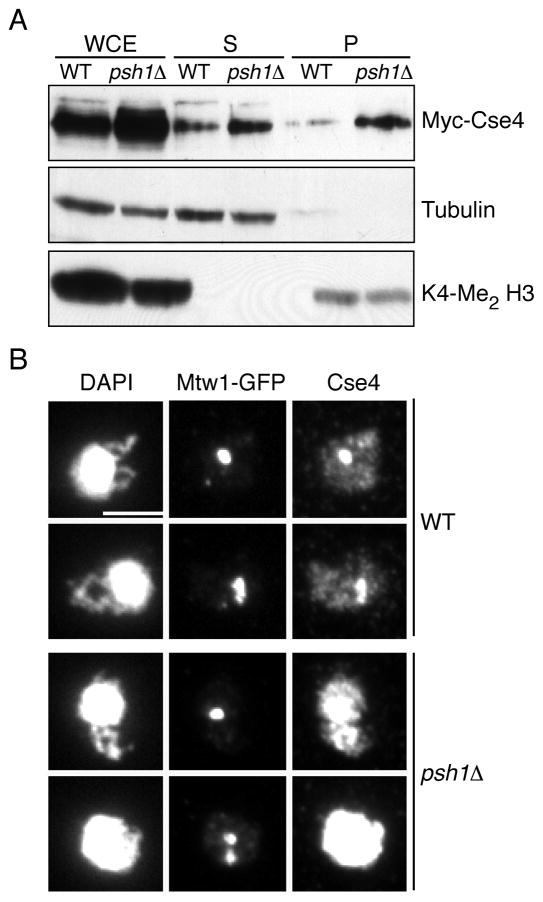
Because Psh1 associates with both soluble and chromatin-bound Cse4 (Fig 2B), we tested whether Psh1 specifically affects one of these Cse4 pools. Although there was more Cse4 in the soluble fraction prepared from psh1Δ cell extracts, there was a more substantial increase in chromatin-associated Cse4 (Fig 4A). We confirmed this by localizing Cse4 in psh1Δ cells using chromosome spreads, a technique that removes soluble material and allows the chromatin-bound Cse4 to be specifically visualized (Loidl et al., 1998). In contrast to wild-type cells where discrete kinetochore foci are observed, transient overexpression of Cse4 in psh1Δ cells results in overall euchromatic localization in 100% of the DAPI masses examined. When we analyzed the kinetochore protein Mtw1-3GFP, there was a discrete signal in 95% of the cells, suggesting that the kinetochore is largely intact in these cells (Fig 4B). Taken together, these data are consistent with Psh1-mediated degradation preventing Cse4 from accumulating in euchromatin.
Fig 4. Psh1 prevents Cse4 from accumulating in euchromatin.
Extracts (WCE) from wild-type (SBY3570) and psh1Δ (SBY8355) cells expressing pGAL-Myc-CSE4 were fractionated into soluble (S) and chromatin (P) fractions. Cse4 levels were assayed in each fraction using anti-Myc antibodies. Tubulin and K4-Me2 H3 are markers for S and P fractions, respectively. Note that there is loss of material during the procedure so the levels between soluble and chromatin fractions cannot be compared. (B) FLAG-Cse4 was transiently overexpressed for 1 hour in wild-type (SBY8918) and psh1Δ (SBY8917) cells and its chromatin localization was assayed by immunofluorescence on chromosome spreads. DAPI, Alexafluor-GFP and anti-Cse4 staining recognize the DNA, Mtw1, and Cse4, respectively. Scale bar = 5 μm.
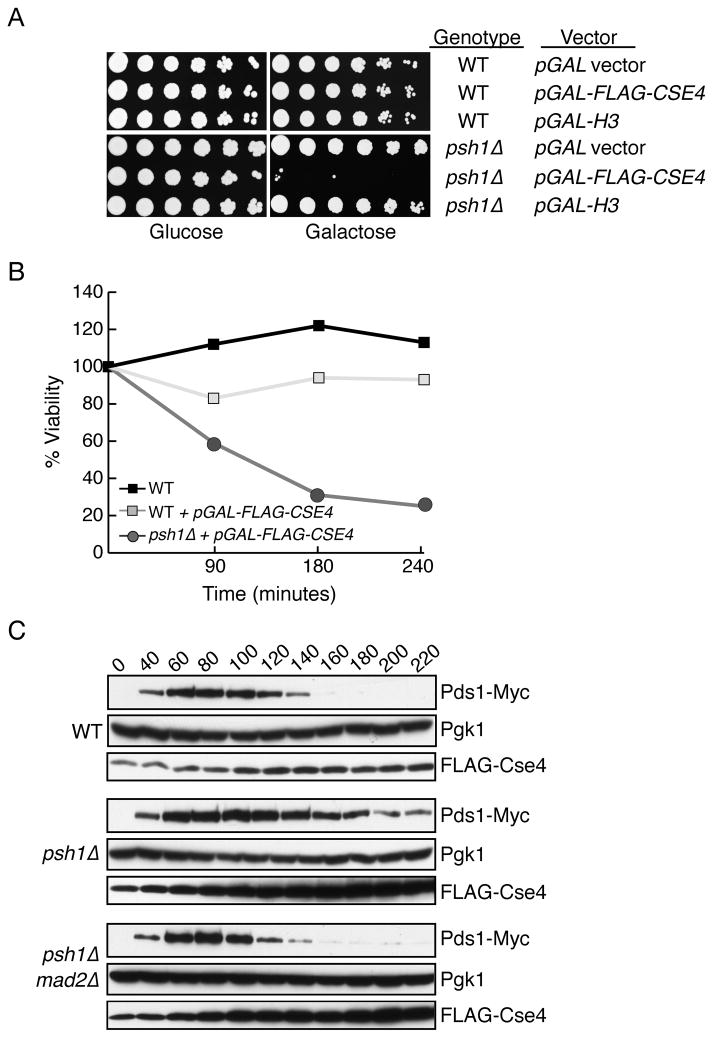
Cse4 overexpression is toxic to psh1Δ cells
Although psh1Δ cells have a genomic instability phenotype (Collins et al., 2007; Yuen et al., 2007) and display a very mild sensitivity to benomyl, the cells grow well and do not exhibit temperature sensitivity (Fig S2A and S2B). In addition, we did not detect a delay in cell cycle progression in psh1 mutants (Fig S2C). We considered the possibility that cells are viable without Psh1 because they maintain low cellular levels of Cse4 due to additional sources of regulation, such as transcriptional and post-transcriptional controls that regulate the major histones (Osley, 1991). We therefore tested the effect of Cse4 overexpression on psh1Δ mutant cells. A Myc-Cse4 construct had a moderate effect on the growth of psh1 mutant cells when overexpressed (Fig S2D), while a FLAG-Cse4 construct strongly inhibited growth (Fig 5A). The difference in growth inhibition is correlated with protein expression differences between the two contructs (Fig S2E), and it is specific to Cse4 because H3 overexpression had no effect on psh1Δ cell viability (Fig 5A). We further analyzed the phenotype by releasing psh1Δ mutant cells from G1 into conditions that overexpress FLAG-Cse4 and found that they die within a single cell cycle (Fig 5B).
Fig 5. Cse4 overexpression is toxic to psh1Δ cells.
(A) 5- fold dilutions of the indicated strains (SBY674, SBY8904, SBY4471, SBY9007, SBY8903 and SBY8857) were plated on glucose or galactose media at 23 °C. (B) WT (SBY3), WT+pGAL-FLAG-CSE4 (SBY8904) and psh1Δ+pGAL-FLAG-CSE4 (SBY8903) were released from G1 into galactose and plated at the indicated timepoints on glucose media to determine relative viability. (C) Wild-type (SBY8976), psh1Δ (SBY 8982) and psh1Δ mad2Δ (SBY8975) cells containing pGAL-FLAG-CSE4 were released from G1 into galactose media. Lysates were prepared at the indicated time points and analyzed for Pds1 levels by immunoblotting. See also Figure S2.
To determine whether the growth defect is due to altered kinetochore function, we analyzed cell cycle progression in wild-type and psh1Δ cells overexpressing FLAG-Cse4. The anaphase inhibitor Pds1 cycled normally in the control wild-type and psh1Δ mutant cells released from G1 (Fig S2C). However, although Pds1 cycled normally in wild-type cells overexpressing Cse4, there was a transient delay in Pds1 destruction when Cse4 was overexpressed in psh1Δ mutant cells released from G1 (Fig 5C). The delay was mediated by the spindle checkpoint because it was eliminated when the Mad2 checkpoint protein was deleted, suggesting that Cse4 overexpression in psh1Δ mutant cells alters kinetochore function. Taken together, these data suggest there is an effect on kinetochore function when Cse4 is overexpressed in psh1Δ cells, although the transient checkpoint activation is unlikely to solely account for the complete lack of viability in a single cell cycle. The high level of Cse4 accumulation in euchromatin in the absence of Psh1-mediated degradation likely disrupts one or more chromatin-based processes in addition to altering kinetochore function.
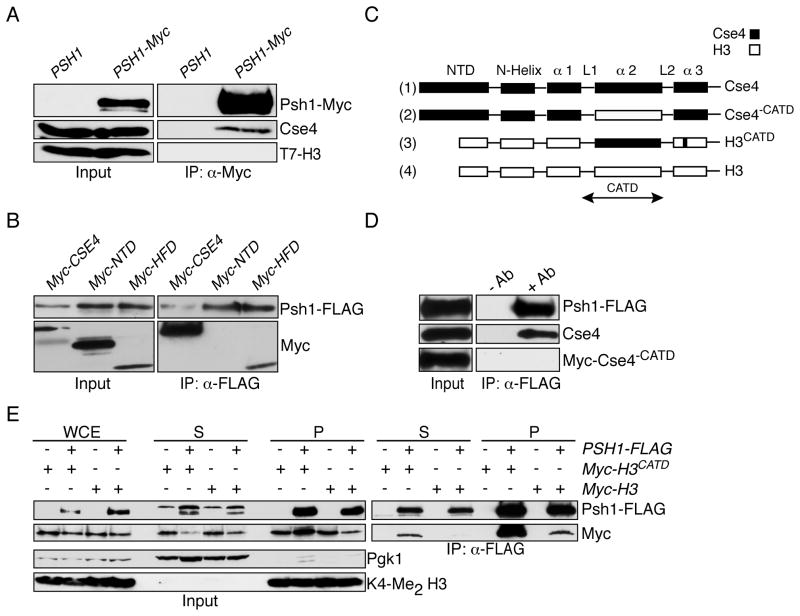
Psh1 recognizes Cse4 via the CATD
The lethality due to Cse4 but not H3 overexpression in psh1Δ mutant cells suggested that Psh1 specifically interacts with Cse4. To test this, we immunoprecipitated Psh1-Myc from cells containing T7-H3 as the sole copy of histone H3. Although the endogenous Cse4 protein co-precipitated, we did not detect H3 interacting with Psh1 despite much higher cellular protein levels (Fig 6A). We therefore asked how Psh1 distinguishes Cse4 from H3. The Histone Fold Domains (HFD) of Cse4 and H3 share 64% identity, whereas their N-terminal domains (NTD) bear no sequence similarity (Stoler et al., 1995). To identify the Psh1 binding domain in Cse4, we immunoprecipitated Psh1-FLAG from strains expressing Myc-tagged full-length Cse4, NTD-Cse4 or HFD-Cse4. While full length Cse4 and the HFD-Cse4 associate with Psh1, the NTD-Cse4 does not interact with Psh1 (Fig 6B). However, we reproducibly detected greater levels of full-length Cse4 associating with Psh1 compared to HFD-Cse4, suggesting that the N-terminus might contribute to the interaction in vivo. Because the HFD-Cse4 contains the CATD, we asked whether the CATD helps Psh1 distinguish Cse4 from H3. First, we replaced the CATD in Cse4 with loop 1 and α2 helix of H3 to generate Myc-Cse4−CATD (Fig 6C, construct 2). The Myc-Cse4−CATD no longer immunoprecipitated with Psh1-FLAG, suggesting that the CATD is required for the interaction (Fig 6D). Second, we tested the interaction of chimeric Myc-H3CATD or Myc-H3 proteins with Psh1 in both the soluble and chromatin-bound fractions (Fig 6C, constructs 3 and 4). Strikingly, the H3CATD protein co-precipitates with Psh1-FLAG in the soluble fraction whereas H3 does not (Fig 6E). We also detected a robust interaction between Psh1 and H3CATD in the chromatin fraction, although lower levels of H3 also co-precipitated with Psh1. This may reveal a decreased affinity of Psh1 for H3, or a transient interaction in the context of chromatin. Taken together, these data strongly suggest that the CATD helps Psh1 recognize Cse4.
Fig. 6. Psh1 requires the centromere targeting domain (CATD) to recognize Cse4.
(A) Anti-Myc-conjugated beads were used to immunoprecipitate Psh1-Myc (SBY5364 and SBY8027). The samples were subsequently analyzed by immunoblotting with anti-Myc, anti-Cse4 or anti-T7 antibodies. (B) Psh1-FLAG was immunoprecipitated from strains expressing Myc-tagged fusions to full length Cse4 (SBY7268), the N-terminal domain (NTD, SBY7269), or the Histone Fold Domain (HFD, SBY7270). The immunoprecipitates were subsequently probed with anti-FLAG or anti-Myc antibodies. (C) Schematic of the chimeric constructs Cse4−CATD and H3CATD. Cse4 and H3 are included as a reference. (D) Psh1-FLAG was immunoprecipitated from cells carrying pGAL-Myc-CSE4−CATD (SBY9126) and subsequently probed with anti-Myc antibodies to detect the Cse4 chimeric protein and anti-Cse4 antibodies to detect endogenous Cse4 protein as a control. (E) Extracts from cells containing Psh1-FLAG and either pGAL-Myc-H3CATD (SBY 8932) or pGAL-Myc-H3 (SBY9134) were fractionated into soluble (S) and chromatin (P) fractions. Psh1-FLAG was imunoprecipitated and subsequently probed for the Myc tagged proteins. Cells lacking Psh1-FLAG were used as controls (SBY8994 and SBY9133). The samples were immunoblotted with anti-FLAG or anti-Myc antibodies.
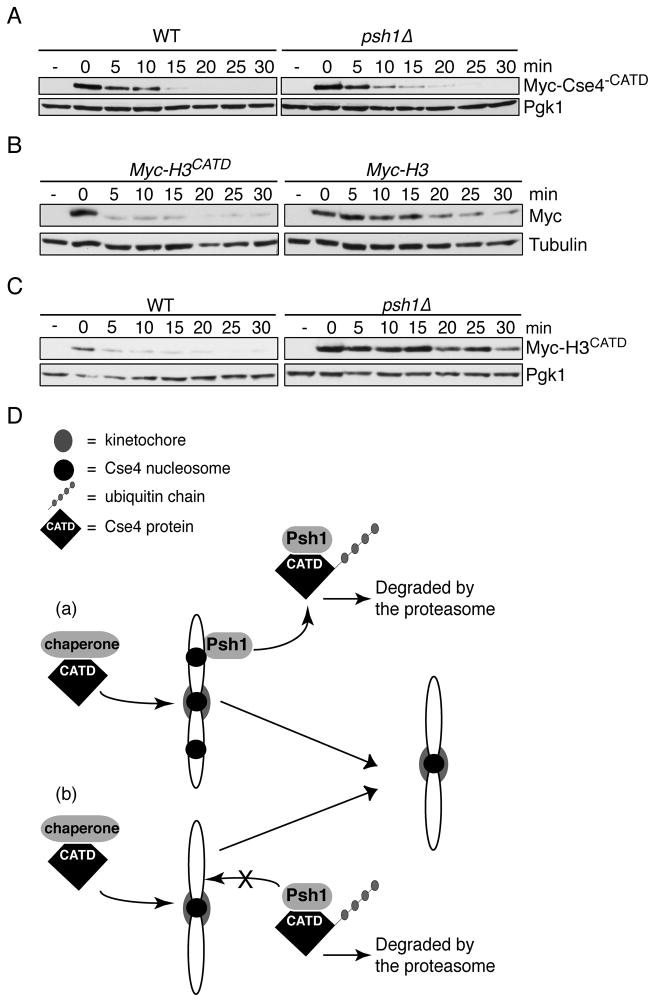
The CATD is required for Psh1-mediated degradation
Given that the CATD is important for Psh1 binding to Cse4, we analyzed the stability of the chimeric proteins. First, we analyzed Myc-Cse4−CATD levels in WT and psh1 mutant cells after repressing transcription and translation. Although Myc-Cse4−CATD is quickly degraded, the degradation does not depend on Psh1 (Fig 7A). This is consistent with Psh1-mediated degradation requiring the CATD. Next, we assayed the stability of the Myc-H3CATD construct in a similar experiment and found that the chimeric H3CATD is also rapidly degraded (Fig 7B). However, in contrast to the Cse4 chimera lacking the CATD, the degradation of H3CATD is dependent on Psh1 (Fig 7C). Taken together, these data suggest that the CATD is a key regulator of Cse4 stability via the Psh1 ubiquitin ligase.
Fig 7. The CATD regulates Cse4 stability.
(A) The stability of Myc-Cse4−CATD was assayed in WT (SBY8700) and psh1Δ (SBY8760) cells after repressing transcription and translation. The lysates were immublotted with anti-Myc antibodies. Pgk1 is a loading control. The (−) timepoint indicates samples taken before induction of the protein. (B) The stability of Myc-H3CATD (SBY8932) and Myc-H3 (SBY 9134) in WT cells as in (A). The lysates were immunoblotted with anti-Myc antibodies. Tubulin is a loading control. The (−) timepoint indicates samples taken before induction of the proteins. (C) The stability of Myc-H3CATD was assayed in wild-type (SBY8932) and psh1Δ (SBY8959) cells as in (A). (D) Model: Cse4 recognition by its chaperone occurs via the CATD, which targets it to the centromere where it is protected from degradation by kinetochore assembly. Psh1 also recognizes Cse4 via the CATD. Psh1 prevents mislocalization of Cse4 in at least two possible ways, by (a) recognizing mis-incorporated Cse4 on the chromatin and targeting it for degradation, or by (b) by regulating the level of soluble Cse4, thereby limiting its misincorporation rate into the euchromatin. The exclusive localization of Cse4 to the centromere is therefore maintained by the CATD through a combination of targeting Cse4 to the centromere and degrading mislocalized Cse4. See also Figure S3.
Discussion
We report the identification of an E3 ubiquitin ligase that has specificity for CENP-A relative to H3 and has not yet been described in any other organism. Psh1 polyubiquitinates and partially mediates Cse4 degradation in vivo. In the absence of Psh1, Cse4 overexpression leads to euchromatic mis-incorporation and lethality. Psh1 degradation of Cse4 is mediated through the CATD, suggesting that the CATD has a previously unrecognized function in degrading Cse4 to prevent its mislocalization.
Although the endogenous Cse4 protein can be detected in euchromatin at highly transcribed genes, its abundance is much lower than at centromeres (Camahort et al., 2009; Lefrancois et al., 2009). These data suggest that Cse4 transiently localizes to euchromatin but there are mechanisms that ensure it is not stably maintained. Consistent with this, the overexpression of Cse4 does not lead to detectable euchromatin localization or have growth consequences in WT cells (Collins et al., 2004). In contrast, Cse4 overexpression is toxic and leads to its accumulation in euchromatin in the absence of Psh1. These data strongly suggest that Psh1-mediated degradation of Cse4 is a key mechanism that prevents Cse4 from stably incorporating into euchromatin. Although we cannot rule out an additional role for Psh1 at the centromere, we did not detect any defects in cell cycle progression in psh1Δ mutant cells. In addition, the previously detected genomic instability in psh1Δ mutant cells is just as consistent with a function in preventing ectopic Cse4 localization as in regulating centromere function (Collins et al., 2007; Yuen et al., 2007). We hypothesize that the cellular levels of Cse4 are kept low by additional mechanisms, such as transcriptional and post-transcriptional controls that regulate the major histones (Osley, 1991), such that Cse4 mis-incorporation in the absence of Psh1 is not sufficient to manifest a gross defect under normal conditions. Consistent with this, Cse4 levels are only partially stabilized in the absence of Psh1 so additional pathways of Cse4 degradation exist that need to be identified in the future.
Our data suggest that the toxicity caused by Cse4 overexpression in psh1Δ mutant cells may be a result of multiple defects. The spindle checkpoint is eventually satisfied in these cells, yet they die within a single cell cycle. It is not possible to determine if ectopic kinetochores form de novo in budding yeast because they cannot be visualized due to the limit of resolution of the light microscope. In addition, traditional chromatin immunoprecipitation techniques cannot identify ectopic kinetochores unless the site of assembly is similar throughout the population. We favor the possibility that the toxicity is due to the mislocalization of one or more kinetochore proteins as well as the disruption of chromatin-based processes such as transcription when Cse4 accumulates in euchromatin. Consistent with this, it was previously shown that the relative stoichiometry of H3 and Cse4 is important for the proper localization of each protein (Au et al., 2008). We were not able to directly test this due to technical difficulty in achieving similar levels of Cse4 expression when H3 was simultaneously overexpressed. Regardless, our data show that Psh1 mediates Cse4 degradation and contributes to the fidelity of its cellular localization.
The CATD in Cse4 appears to be a key domain that directs Psh1-mediated degradation. A CATD deletion within Cse4 prevents its association with Psh1 and abolishes Psh1-mediated degradation, and insertion of the CATD is sufficient to mediate H3 binding to Psh1. In the future, it will be important to determine whether additional features of Cse4 are involved in mediating the interaction with Psh1, as well as to establish whether Psh1 directly recognizes Cse4 in the context of chromatin. Cse4 and Psh1 interact in both the soluble and chromatin fractions, but the precise location of the ubiquitination reaction is not known. In addition, Psh1 is not able to discriminate between Cse4 and H3 octamers in vitro using a non-specific E2 enzyme (data not shown), so additional specificity must exist in vivo. We speculate that chromatin factors that disassemble nucleosomes during chromatin-based processes expose the CATD and allow Psh1 to target Cse4 for degradation. Consistent with this idea, Cse4 and Psh1 co-purify with Spt16 and Pob3, components of the budding yeast FACT complex that disassemble and re-assemble nucleosomes to facilitate transcription and replication (Reinberg and Sims, 2006; VanDemark et al., 2006). Psh1 mediates the interaction between Cse4 and Spt16 (Fig S3), suggesting that Psh1 could link Cse4 degradation to the FACT complex. In addition, mutants in the Spt4 protein that is implicated in nucleosome assembly, nucleosome stabilization and transcription also lead to Cse4 mislocalization to euchromatin (Swanson and Winston, 1992; Hartzog et al., 2002; Crotti and Basrai, 2004).
We propose the following model (Fig 7D). Cse4 is targeted to centromeres via a chaperone-mediated interaction with the CATD where it becomes protected from degradation by kinetochore assembly (Collins et al., 2004; Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010). Cse4 also localizes to euchromatin, especially at sites of high histone turnover (Camahort et al., 2009; Lefrancois et al., 2009). Psh1 may regulate Cse4 localization by degrading Cse4 that is evicted from the chromatin when the CATD is exposed during chromatin-based processes. It is also possible that Psh1 degrades excess soluble Cse4, thereby indirectly controlling its misincorporation rate into the chromatin. Although it is not yet clear whether Psh1 homologs exist in vertebrates, CATD function is conserved throughout eukaryotes (Black et al., 2007). In addition, depletion of the HJURP chaperone that associates in a CATD-dependent manner leads to lower intracellular CENP-A levels (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010), consistent with the possibility that the proteolytic machinery targets unbound CENP-A for degradation via the CATD. In the future, it will be critical to determine whether the mechanisms that prevent the non-centromeric accumulation of CENP-A are conserved. It will also be interesting to further investigate proteolysis as a mechanism to regulate chromatin composition and ultimately maintain epigenetic states in multicellular eukaryotes (Gunjan et al., 2006).
Experimental Procedures
Strain construction and Microbial techniques
Media and microbial techniques are as described (Sherman et al., 1974; Rose et al., 1990). Yeast strains were constructed by standard genetic techniques. Specific plasmid construction and yeast strains used in this study are described in supplementary data and Table S2. To assay viability and Pds1 levels, cells were grown in YEP + lactic acid media and arrested in G1 with 1 μg/ml of α-factor for 3.5 hrs. Cse4 overexpression was induced with 2% galactose during the last 30 min of the arrest and then the cells were washed and released into 2% galactose. α-factor was added back when cells were small-budded to prevent cells from entering the second cell cycle. To assay viability, cells were plated on YPD at 23 °C after diluting 10−4 and > 100 cells were counted for each strain. To assay Pds1 levels, lysates were made as described below.
Protein purification, immunoprecipitation and immunoblotting
Plasmids containing either GST-Psh1 (pSB1535) or the GST-Psh1 RING mutant (pSB1541) were transformed into BL-21 cells. Protein was purified from 2 L of cells after inducing expression with 1 mM IPTG for 2 hrs. GST purification was carried out as described (Kellogg et al., 1995). For Cse416R purification, 500 mls of mid-log (OD600 ~ 0.6) cells carrying pCUP1-3XFLAG-CSE416R were induced with CuSO4 to a final concentration of 5μM for 30 minutes. Lysates were prepared in Buffer H/0.1M KCl (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.1 % NP-40, 15% glycerol and 100 mM KCl) supplemented with protease inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml chymostatin, and 0.2 mM PMSF) and phosphatase inhibitors (1 mM sodium pyrophosphate, 2 mM Na-beta-glycerophosphate, 0.1 mM NA3VO4, 5 mM NaF, 100 nM microcystin-LR). Cells were lysed with glass beads in a beater (Biospec Products, Inc.) for 30 seconds, three times, with 1 min on ice in between and then centrifuged for 90 minutes at 27,000 rpm at 4°C. The supernatant was incubated with 60 μl of M2 anti-FLAG agarose beads (Sigma) for 3 hrs at 4°C. The beads were washed with buffer H/0.3 M KCl 3 times, followed by 3 more washes with buffer H/0.1 M KCl. Proteins were eluted with 60μl of 0.5mg/ml FLAG peptide. The eluate from the purification was subject to anion exchange (0.2 ml of Source Q column) with a starting salt gradient of 0.1M KCl and final concentration of 1M KCl. Mass Spectrometry (MS) analysis was carried out on fractions 14 and 15 that contained a peak of Cse4 protein as described in supplementary experimental procedures.
For immunoprecipitations, 50 ml cultures were harvested at midlog phase and lysates were prepared as described (Akiyoshi et al., 2009) except that cell extracts were prepared with glass beads in a beater (Biospec Products, Inc.) for 35 sec, three times, with 1 min on ice in between and then centrifuged for 30 minutes. The supernatant was incubated with 10 μl of protein G dynabeads (Invitrogen) and either 0.5 μl of M2 anti-Flag (sigma) or 4 μl of A-14 anti-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies for 2 hrs at 4° C. The beads were washed four times with lysis buffer and the immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting as described above.
For immunoblotting, protein extracts were made and analyzed as described (Minshull et al., 1996). 9E10 anti-Myc antibodies were obtained from Covance (Richmond, CA) and used at a 1:10,000 dilution. Cse4 antibodies were used at 1:500 (Collins et al., 2004). Tubulin antibodies were obtained from Accurate Chemical and Scientific (Westbury, NY) and used at 1:1000. Pgk1 antibodies obtained from Invitrogen were used at 1:10,000. K4-Me2 H3 antibodies were obtained from Upstate Cell Signaling Solutions (NY, gift from Rich Gardner) and used at 1:3000. Quantitative immunoblotting was performed using IR Dye 800CW obtained from LI-COR (gift from Steve Hahn, FHCRC). Note that due to an increase in Cse4 levels in psh1Δ mutant cells and Cse416R, different exposures were used to detect Cse4 ubiquitin conjugates. However, ubiquitin conjugates were not detected in the psh1 mutant cells even when exposure times were increased.
Stability assays
For endogenous Cse4 stability assays, cells were grown in YEPD and subsequently arrested with 1 μg/ml factor (G1), 0.023 g/ml hydroxyurea (S) or 15 μg/ml nocodazole (M) for 3 hrs. Samples were taken for analysis after repressing protein synthesis with 50 μg/ml of cycloheximide. For all other stability assays, cells were grown in 2% raffinose media, a timepoint (−) was taken, and then cultures were induced with 2% galactose for 2.5 hrs at 23°C. Timepoints were taken after inhibiting transcription and translation by adding glucose to 2% and cycloheximide to a final concentration of 50 μg/ml respectively. Extracts were prepared and protein levels were assayed using Myc antibodies or quantified as described above.
Ubiquitination Assay
Ubiquitination assays were performed as described (Gardner et al., 2005). 15 μl reactions containing 0.4 μg ubiquitin activating enzyme (yeast Uba1), 0.4 μg ubiquitin conjugating enzyme (human UbcH5a), 36 ng purified GST-Psh1 (or C45S, C50S mutant), 2.5 μg ubiquitin, 2 mM Mg-ATP, 50 mM Tris-HCl (pH7.5), 2.5 mM MgCl2, and 0.5 mM DTT were incubated at 23°C for 30 min. Reactions were stopped by adding sample buffer and boiling for 3 minutes. Uba1, UbcH5a, ubiquitin and Mg-ATP were obtained from Boston Biochemicals. Ubiquitin conjugates were detected using anti-ubiquitin antibodies (gift from D. Gottschling, FHCRC). Cse4 and core histones were produced in E. coli (Luger et al., 1997). Histone octamers were refolded and purified by Superdex S-200 gel-filtration chromatography as described (Dyer et al., 2004). 1μg of Cse4 octamers were used in ubiquitination assays and probed for Cse4 ubiquitin conjugates with anti-Cse4 antibodies (gift from Carl Wu, NIH).
Chromatin Fractionation assays
Chromatin Fractionation experiments were performed as described (Liang and Stillman, 1997) with the following changes. 30 Units of micrococcal nuclease (Worthington Biochemical Corporation) per 50 ml of mid-log culture were used to solubilize the chromatin. The micrococcal nuclease digestion was carried out at 37°C for 15 minutes and analyzed after stopping the reaction with 1mM EDTA. For fractionation followed by immunoprecipitation, the same protocol was used to prepare soluble and chromatin fractions and immunoprecipitations were carried out as described above.
Immunofluorescence
Cells were grown in lactic acid, followed by nocodazole arrest for 3hrs. For the last hour of arrest, FLAG-Cse4 expression was induced with 2% galactose. Under this overexpression condition, cell viability remains at ~80%. Chromosome spreads were performed as described previously (Collins et al., 2004). DAPI was obtained from Molecular Probes (Eugene, OR) and used at a 1μg/ml final concentration. Lipsol was obtained from Fisher. Anti-Cse4 antibodies were used at 1:250 dilution (Collins et al., 2004), Cy3 and Cy5 secondary antibodies from Jackson Immunoresearch (West Grove, PA) were used at a 1:1000 dilution, and Alexafluor-GFP antibodies obtained from Invitrogen were used at a 1:250 dilution.
Viability and cell cycle experiments
For viability experiments, cells were grown in YEP + lactic acid and arrested in G1 for 3.5 hrs. For the last half hour of the arrest, galactose was added to induce Cse4 expression. Cells were then released from G1 into galactose and plated at indicated timepoints on glucose media to determine viability. For cell cycle experiments, cells were grown as described above except Cse4 expression was induced for the last hour during the arrest in G1. Cells were then released from G1 into galactose media and samples were taken at the indicated timepoints.
Supplementary Material
Acknowledgments
We are grateful to M. Basrai, R. Gardner, D. Gottschling, S. Hahn, L. Jansen, T. Tsukiyama and C. Wu for advice and reagents. We thank members of the Biggins’ lab, S. Henikoff and T. Tsukiyama for critical reading of the manuscript, and C. Breed for help with the Cse416R purification. This work was supported by NIH R01 GM078069 to S. B., a Chromosome Metabolism and Cancer Training Grant (NIH NCI 5 T32 CA09657) to P. R., and NIH/NCRR Funded Yeast Resource Center P41 RR011823 grant to M. J. M. S. B. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:1–14. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Choo KH. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Crotti LB, Basrai MA. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. Embo J. 2004;23:1804–1814. doi: 10.1038/sj.emboj.7600161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr Opin Genet Dev. 2006:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Speer JL, Lindstrom DL. Transcript elongation on a nucleoprotein template. Biochim Biophys Acta. 2002;1577:276–286. doi: 10.1016/s0167-4781(02)00458-x. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila Centromere-Specific Histone CID Promotes Formation of Functional Ectopic Kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray AW. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:1–18. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Klein F, Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Lomonte P, Sullivan KF, Everett RD. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem. 2000;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Straight A, Rudner A, Dernburg A, Belmont A, Murray AW. Protein Phosphatase 2A Regulates MPF Activity and Sister Chromatid Cohesion in Budding Yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K, Bloom KS. Differenctial kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional Complementation of Human Centromere Protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KW, Warren CD, Chen O, Kwok T, Hieter P, Spencer FA. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci U S A. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.